Abstract
This systematic review explores the use of artificial intelligence (AI) in the analysis of biofluid markers in age-related macular degeneration (AMD). We detail the accuracy and validity of AI in diagnostic and prognostic models and biofluid markers that provide insight into AMD pathogenesis and progression. This review was conducted in accordance with the Preferred Reporting Items for a Systematic Review and Meta-analysis guidelines. A comprehensive search was conducted across 5 electronic databases including Cochrane Central Register of Controlled Trials, Cochrane Database of Systematic Reviews, EMBASE, Medline, and Web of Science from inception to July 14, 2021. Studies pertaining to biofluid marker analysis using AI or bioinformatics in AMD were included. Identified studies were assessed for risk of bias and critically appraised using the Joanna Briggs Institute Critical Appraisal tools. A total of 10,264 articles were retrieved from all databases and 37 studies met the inclusion criteria, including 15 cross-sectional studies, 15 prospective cohort studies, five retrospective cohort studies, one randomized controlled trial, and one case–control study. The majority of studies had a general focus on AMD (58%), while neovascular AMD (nAMD) was the focus in 11 studies (30%), and geographic atrophy (GA) was highlighted by three studies. Fifteen studies examined disease characteristics, 15 studied risk factors, and seven guided treatment decisions. Altered lipid metabolism (HDL-cholesterol, total serum triglycerides), inflammation (c-reactive protein), oxidative stress, and protein digestion were implicated in AMD development and progression. AI tools were able to both accurately differentiate controls and AMD patients with accuracies as high as 87% and predict responsiveness to anti-VEGF therapy in nAMD patients. Use of AI models such as discriminant analysis could inform prognostic and diagnostic decision-making in a clinical setting. The identified pathways provide opportunity for future studies of AMD development and could be valuable in the advancement of novel treatments.
Keywords: artificial intelligence, biofluid, age-related macular degeneration, diagnosis, pathogenesis
Plain Language Summary
Age-related macular degeneration (AMD) is the leading cause of blindness in developed countries and has a projected global prevalence of 288 million in 2040. Despite its prevalence, there are no well-established causes of AMD and no easy way of predicting its progression. Artificial intelligence (AI) allows for the study of the thousands of molecules within ocular fluids, which could enable a better understanding of the causes of AMD. This, in turn, could support the development of clinical tools and spur therapeutic advances. In this systematic review, we present studies that used AI to analyze ocular biofluid markers in AMD. Our results indicate that biological processes such as altered lipid metabolism, inflammation, oxidative stress, glycerophospholipid pathway, and protein and mineral absorption were involved in AMD development and progression. However, variability in the studies and the patient populations prevented identification of a singular characteristic marker for AMD. AI tools were able to differentiate between AMD patients and controls, an application that could be used in both screening and diagnosis of AMD. Further, AI was able to predict how well a patient may respond to AMD therapy, another application that could augment existing clinical tools and inform decision-making by healthcare professionals.
Introduction
Age-related macular degeneration (AMD) is the leading cause of blindness in developed countries, with a projected prevalence of 288 million in 2040.1,2 With a growing elderly population in many parts of the world, solutions to improve screening, prevention, and management of AMD are crucial.1,2 Novel technologies have been applied in AMD clinical tools and research efforts to address this growing need and have demonstrated strong initial results.3–7 While research has identified molecular etiologies in AMD development, including lipofuscin accumulation in retinal pigmented epithelial cells, choroidal ischemia with vascular endothelial growth factor (VEGF) involvement, oxidative stress, and genetic factors, no clear pathogenic mechanism to direct treatment or prevention has emerged.8–10 Pathogenic biomarkers are contained in biofluids such as serum, tears, aqueous humour and vitreous humour, which can be obtained in both clinical and surgical settings. As the relationships between fluid biomarkers and clinical characteristics are complex and often present within highly dimension data sets, artificial intelligence (AI) provides an opportunity to uncover meaningful associations not possible by traditional analytical methods.
AI has already been applied in AMD research and clinical tool development, with compelling research efforts focused on screening, treatment, prognosis, and structure-function mapping of the retina.3–7,11,12 Supervised AI techniques, including discriminant analysis or artificial neural networks, are trained using defined cases and learn to classify groups or predict outcomes.10,13–15 In contrast, unsupervised AI techniques such as hierarchical cluster analysis and principal component analysis (PCA) are adept at determining trends in highly dimensional data as they can group unlabeled data based on similarities or differences and find associations between variables in large data sets.16,17 Bioinformatics tools such as pathway analysis translate complex findings into interpretable information. To date, AI use in the context of AMD has been primarily for review of fundus photography, optical coherence tomography, and other ocular imaging modalities.3,5,18–21 However, AI application is expanding to biofluid biomarker analysis, enabling improved exploration of molecular AMD etiology, which could support an array of AMD clinical tools and spur therapeutic advances.22–24
Clinical tools built with AI could allow for mass screening of AMD, earlier intervention and monitoring, and subsequently, improved personalized treatments for better patient outcomes.1,2 Imaging-based AI tools could be augmented by the inclusion of biofluid biomarkers, allowing for more robust tool creation, further exploration of disease pathogenesis, and the development of targeted therapeutics.22–24 Herein we aim to systematically review the available literature and describe the application of AI and bioinformatics in the analysis of biofluid biomarkers in AMD. This review will provide a detailed analysis of the types of AI and bioinformatics tools applied to biofluid markers in AMD, synthesize evidence regarding biomarkers implicated in AMD development and progression, and identify areas for future studies.
Methods
This systematic review was conducted in accordance with the Preferred Reporting Items for a Systematic Review and Meta-analysis (PRISMA) guidelines.25 The protocol was registered in PROSPERO (reg. CRD42020196749). Ethics approval from our Institutional Review Board was not required as this is a systematic review of published studies and does not involve human subjects. This systematic review is part of a series of systematic reviews on AI/bioinformatic analysis of biofluid biomarkers in ophthalmology. The primary outcome was to report the applications of AI in the analysis of biofluid markers in AMD.
Eligibility Criteria for Considering Studies for This Review
Study selection inclusion criteria were: (1) original peer-reviewed studies analyzing biomarker concentrations to predict or modify patient therapy or outcome/diagnosis in ophthalmic conditions; (2) biomarker analysis utilized AI and/or bioinformatics approaches; (3) biomarker samples were gathered from vitreous fluid, aqueous fluid, tear fluid, plasma, serum, or ophthalmic biopsies; (4) biomarker samples were of a protein, lipid, or metabolite; (5) studies using regression analysis (the simplest form of AI) were either longitudinal or applied their findings to change treatment or prognosis in the study populations. Note, studies that combined biofluid biomarkers with other types of biomarkers (eg imaging) in their analysis were included, as were studies using complex AI (supervised, unsupervised, bioinformatics) that were not longitudinal or did not directly apply findings to the study population. Study selection exclusion criteria were: (1) study only examined ophthalmic diseases that affect pediatric patients (eg retinopathy of prematurity); (2) studies on non-human subjects (animal or cell studies); (3) studies only analyzing post-mortem samples from eyes; (4) non-English studies; (5) abstracts, reviews, systematic reviews, and meta-analyses. Lastly, all articles in which AMD was the main disease of interest were selected for analysis in this review.
Search Methods for Identifying Studies
The search strategy was developed in consultation with an experienced librarian. A comprehensive search was conducted across five electronic databases (EMBASE, Medline, Cochrane Central Register of Controlled Trials, Cochrane Database of Systematic Reviews, and Web of Science) for all articles meeting the inclusion and exclusion criteria from inception to August 11, 2020, and was updated on July 14, 2021. The search used both controlled vocabulary terms and synonymous free-text words to capture the concepts of “ophthalmology”, “AI/bioinformatics”, and “proteomics, metabolomics, lipidomics.” No language or study design restrictions were placed on the search, although non-English studies were excluded manually during the article selection process. Gray literature indexes were searched through EMBASE. Supplemental Materials 1 details the full search strategy for all databases. Additionally, hand searching of references of the included studies for relevant articles which may have not been captured in the search was performed. Covidence software (Melbourne, Australia) was used to manage studies and eligibility status.
Study Selection
Abstracts and titles and subsequent full-text review were screened by two independent reviewers. Disagreements between the reviewers were resolved and adjudicated by a third independent reviewer. Data extraction was performed by one reviewer, with 10% of the extractions verified by a second independent reviewer to ensure agreement and consistency between data extractors.
Data Collection and Risk of Bias Assessment
Data extraction for each article included study population demographics, biofluid markers analysed, description of the AI or bioinformatics tool(s) used in the study, the reason for AI or bioinformatics tool selection, and the overall findings/conclusions of each study. The Joanna Briggs Institute Critical Appraisal Tools (Faculty of Health and Medical Sciences at the University of Adelaide, South Australia) were utilized for risk of bias and quality assessment.26 The Joanna Briggs Institute Critical Appraisal Tools consists of questions that can be scored as “yes”, “no”, “unclear”, or “not applicable”, with “yes” indicating that the study adequately addressed a specific domain of bias. Study risk of bias was assessed by one reviewer, with 10% of the risk of bias assessments verified by a second, independent reviewer to ensure consistency between assessors. Articles were scored as high risk of bias if they had <49% of questions scored “yes”, moderate risk of bias if 50–79% of questions scored “yes”, and low risk of bias if >80% of questions scored “yes.”27
Data Synthesis and Analysis
Descriptive synthesis of evidence was undertaken for all included studies. Meta-analytic methods were not employed given the heterogeneity of study designs, the AI tools used, and the biomarkers implicated. The results detailed the proportions of study type, country of publication, and type of AI analysis used. We also synthesized the accuracy of any predictive AI models and the common applications of each AI class. Further, the biomarkers and pathways that are implicated in AMD development, progression, and treatment were described.
Included studies were categorized according to study objectives and type of AI methodologies utilized. Studies were classified based on study purpose into the following categories: 1) Disease Characteristics; 2) Risk Factors; and 3) Treatment Decisions. Studies characterized as Disease Characteristics detailed untargeted exploration of AMD biomarkers with the intention of exploring the pathogenic mechanism of AMD or the factors that influence AMD progression. Amongst the studies classified under Risk Factors, the impact of a specific biomarker or set of biomarkers on AMD development, progression, or diagnosis were examined. Finally, the Treatment Decisions studies sought to predict outcomes following treatment selection or guide selection of therapeutic or surgical options using biomarkers.
Results
Study Characteristics
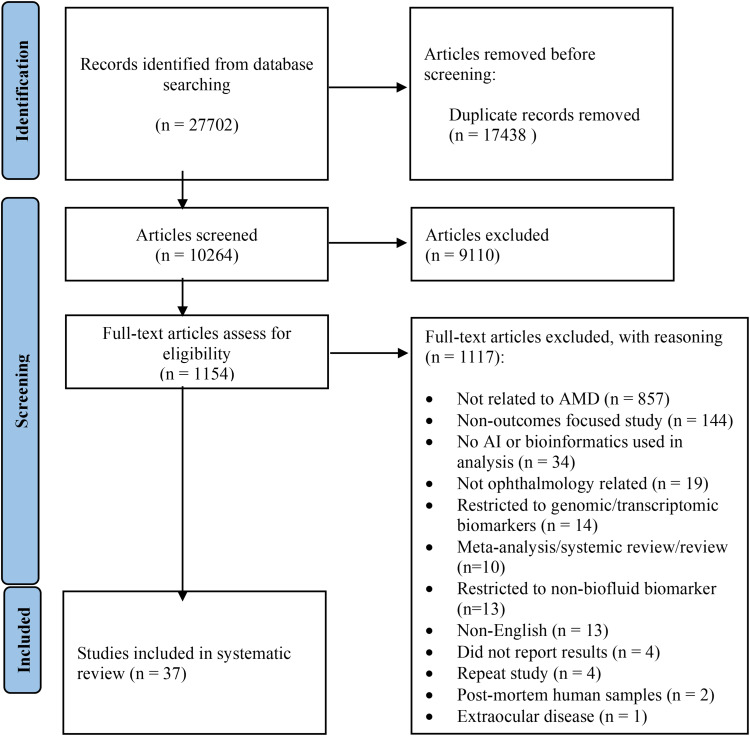
A total of 10,264 articles were retrieved by the search from all databases combined. After removal of duplicates, 37 papers met the inclusion criteria (Figure 1). Study designs included 15 cross-sectional studies (40.5%), 15 prospective cohort studies (40.5%), five retrospective cohort studies (13.5%), one randomized controlled trial (2.7%), and one case–control study (2.7%; Table 1). There was a global distribution of the included studies, with the largest shares conducted in the USA (30%), the Netherlands (14%), Japan (11%), and China (11%). With regard to study design, 15 studies examined Disease Characteristics, 15 studied Risk Factors, and seven guided Treatment Decisions. The majority of studies focused on AMD more generally (58%), while neovascular AMD (nAMD) was the focus in 11 studies (30%), and geographic atrophy (GA) was highlighted by three studies (7%). Additional study characteristics are contained within Supplemental Table 1.
Figure 1.
PRISMA flowchart diagram for study identification and selection.
Notes: PRISMA figure adapted from Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. Creative Commons.25
Abbreviations: AMD, age-related macular degeneration; AI, artificial intelligence.
Table 1.
Summary Characteristics of Included Studies
| First Author, Publication Year | Study Design | Disease Type (Other Diseases Studied) | Country of Publication | Study Purpose | Sample Size | Classes of AI | Statistical, AI, Bioinformatics Methods | Biofluids | Biomarker(s) Analyzed | Significant Biomarker(s) and Key Pathways |
|---|---|---|---|---|---|---|---|---|---|---|
| Acar,28 2020 | Cross-sectional | AMD | Netherlands | Disease characteristics | 6608 | 2 | Unsupervised: PCA Statistical method: Univariate logistic regression, linear regression | Plasma | Metabolic profile | 146 metabolites, including those involved in large and extra-large HDL subclasses, VLDL, amino acid 73, citrate, complement activation |
| Arai,60 2020 | Prospective cohort | AMD, nAMD, PCV | Japan | Treatment decisions | 48 | 1 | Statistical method: Multiple regression analysis | Aqueous humor | Cytokines | MCP-1, IL-10 (baseline BCVA); MCP-1, CXCL13 (better BCVA in 12 months); MMP-9, CXCL12, IL-10 (increased number of injections required) |
| Boekhoorn,29 2007 | Prospective cohort | AMD | Netherlands | Risk factors | 4606 | 1 | Statistical method: Cox proportional hazards regression | Serum | CRP | CRP |
| Buch,30 2005 | Prospective cohort | AMD | Denmark | Risk factors | 359 | 1 | Statistical method: Univariate logistic regression, multivariate logistic regression | Serum | Limited lipid profile | Total cholesterol, apolipoprotein A1, apolipoprotein B |
| Chaker,31 2015 | Prospective cohort | AMD | Netherlands | Risk factors | 5573 | 1 | Statistical method: Cox proportional hazards regression | Serum | Thyroid markers | Free thyroxine |
| Cougnard-Gregoire,32 2014 | Prospective cohort | AMD | France | Risk factors | 963 | 1 | Statistical method: Generalized estimating equation logistic regression | Serum | Lipids | HDL |
| Gao,16 2020 | Prospective cohort | nAMD | Singapore | Treatment decisions | 100 | 4 | Supervised: PCA Unsupervised: OPLS-DA Bioinformatics: Pathway analysis Statistical method: Logistic regression | Serum | Metabolic profile | LysoPC (18:2), PS (18:0/20:4), glycerophosphocholine |
| Han,33 2020 | Cross-sectional | wAMD | China | Disease characteristics | 46 (20 cataract controls, 26 wAMD) | 3 | Supervised: PCA Unsupervised: OPLS-DA Bioinformatics: KEGG | Aqueous humor | Metabolic profile | Deoxycarnitine, N6, N6, N6-trimethyl-L-lysine, glycine betaine, itaconic acid, cis-aconitate, 5-aminopentanoic acid, norleucine, L-phenylalanine, carnitine, γ-glutamylglutamine, hetisine, 3-phenyllactic acid, LPC 18:2, coumaroyl agmatine, N-acetylhistidine, creatine, N-fructosyl isoleucine, L-proline (Carnitine-associated mitochondrial oxidation pathway, carbohydrate metabolism pathway, activated osmoprotection pathway) |
| Joachim,34 2015 | Prospective cohort | AMD | Australia | Risk factors | 3654 | 1 | Statistical method: Discrete logistic regression models | Serum | Limited lipid profile | None |
| Jonasson,35 2014 | Prospective cohort | AMD, GA | Iceland | Risk factors | 2868 | 1 | Statistical model: Multivariate logistic regression | Serum | Cardiovascular health profile | HDL-cholesterol |
| Kersten,36 2019 | Cross-sectional | AMD | Netherlands | Disease characteristics | 144 (72 cases, 72 control) | 2 | Supervised: sPLS-DA Statistical method: Logistic regression | Serum | Metabolic profile | Glutamine, glutamate, glutaminolysis, phosphatidylcholine diacyl C28:1 (PC aa C28.1) |
| Klein,37 2019 | Prospective cohort | AMD | USA | Risk factors | 4972 | 1 | Statistical method: Logistic regression, linear regression, multi-state Markov | Serum | Limited lipid profile | None |
| Kuiper,38 2017 | Cross-sectional | AMD (idiopathic non-infectious uveitis, primary vitreoretinal lymphoma, rhegmatogenous retinal detachment) |
Netherlands | Disease characteristics | 175 | 3 | Supervised: Decision tree Unsupervised: Hierarchical cluster analysis Statistical method: SMOTE, k-nearest neighbors | Aqueous humor | Proteomic profile | IL-10, IL-21, ACE |
| Lai,22 2009 | Randomized control trial | nAMD | Hong Kong | Treatment decisions | 50 | 1 | Statistical method: Multivariate logistic regression | Aqueous humor | VEGF, PEDF | Baseline VEGF |
| Laíns,39 2018 | Cross-sectional | AMD | USA | Disease characteristics | 120 | 2 | Unsupervised: PCA Statistical method: Multivariate logistic regression Bioinformatics: Pathway analysis | Plasma | Metabolic profile | 87 differentially expressed metabolites (48 across all AMD stages), including linoleoyl-arachidonoyl-glycerol, stearoyl-arachidonoyl-glycerol, oleoyl-arachidonoyl-glycerol, 1-Palmitoyl-2-arachidonoyl-GPC, 1-stearoyl-2-arachidonoyl-GPC, adenosine, glycerophospholipid pathway |
| Laíns,24 2019 | Cross-sectional | AMD | USA | Disease characteristics | 491 (196 with 47 controls in Boston, 295 with 53 controls in Portugal) | 3 | Unsupervised: PCA Bioinformatics: Pathway analysis, KEGG Statistical method: Multivariate logistic regression | Plasma | Proteomic profile | 28 metabolites, including those from the lycerophospholipid, purine, taurine, hypotaurine, and nitrogen metabolism pathways |
| Luo,40 2017 | Cross-sectional | wAMD | China | Disease characteristics | 40 (20 AMD, 20 controls) | 3 | Supervised: PLS-DA Unsupervised: PCA, hierarchical cluster analysis Bioinformatics: KEGG | Plasma | Metabolomic profile | N-Acetyl-L-alanine, N1-Methyl-2-pyridone-5-carboxamide, L-tyrosine, L-phenylalanine, L-palmitoylcarnitine, L-methionine, L-Arginine, isomaltose, hydrocortisone, biliverdin |
| Lynch,41 2019 | Cross-sectional | AMD | USA | Disease characteristics | 30 (10 AMD, 10 GA, 10 cataract controls) | 2 | Bioinformatics: Pathway analysis Statistical method: Linear regression | Plasma | Proteomic profile | AMD: Vinculin, CD177 AMD pathways: Cargo trafficking to the periciliary membrane, FGFR3b ligand binding and activation, VEGF binds to VEGFR leading to receptor dimerization/VEGF ligand-receptor interactions, common pathway of fibrin clot formation GA: Neuroregulin 4, soluble intercellular adhesion molecule-1 GA pathways: SHC1 events in ERBB4 signaling, PI3K events in ERBB4 signaling, SHC1 events in ERBB2 signaling, GRB2 events in ERBB2 signaling, nuclear signaling by ERBB4, NADE modulates death signaling, PI3K events in ERBB2 signaling, signaling by BMP, interleukin receptor SHC signaling, regulation of beta-cell development, regulation of commissural axon pathfinding by SLIT and ROBO, reversible hydration of carbon dioxide, tetrasaccharide linker, cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding, ERBB4 |
| Lynch,42 2020 | Cross-sectional | AMD | USA | Disease characteristics | 109 | 2 | Bioinformatics: Pathway analysis Statistical method: Linear regression, Cox proportional hazards regression, univariate logistic regression | Plasma | Proteomic profile | TCL1A, CNDP1, lysozyme C, TFF3, RNAS6, SAP3 Pathways: Tumor necrosis factor binding, digestion and absorption, activin signaling, TGF-β family signaling |
| Mendez,43 2021 | Cross-sectional | AMD | USA | Disease characteristics | 71 | 1 | Statistical method: Unspecified multilevel mixed-effects linear model | Plasma | Metabolic profile | Metabolites: linolenate, mannitol, sorbitol, glycosyl ceramide (d18:2/24:1, d18:1/24:2), beta-alanine, 3-methyl-2-oxovalerate, 3-methylglutaconate, isoleucine Pathways: polyunsaturated fatty acid (n3 and n6), fructose, mannose and galactose Metabolism, fatty acid metabolism (acyl choline), hexosylceramides (HCER), pyrimidine metabolism, uracil, leucine, isoleucine and valine metabolism |
| Millen,44 2015 | Retrospective cohort | AMD | USA | Risk factors | 913 | 1 | Statistical method: Logistic regression | Serum | Vitamin D, CRP | Vitamin D |
| Millen,45 2017 | Retrospective cohort | AMD | USA | Risk factors | 9734 | 1 | Statistical method: Logistic regression | Serum | Vitamin D, lipid profile | None |
| Mitchell,52 2018 | Cross-sectional | nAMD | USA | Disease characteristics | 292 | 4 | Supervised: PLS-DA, SV_RFE, random forest Unsupervised: Hierarchical cluster analysis Bioinformatics: Pathway analysis Statistical method: Linear regression, linear models for microarray data, variable importance for projection | Plasma | Metabolic profile | 159 metabolites from the carnitine shuttle pathway (fatty acid metabolism) and bile acid biosynthesis pathway |
| Ngai,46 2011 | Prospective cohort | AMD | UK | Risk factors | 934 | 1 | Statistical method: Logistic regression, multivariable regression | Serum | Limited metabolomic and lipidomic profile | Total triglycerides, CRP |
| Nielsen,53 2019 | Prospective cohort | GA, nAMD | Denmark | Risk factors | 110 | 1 | Statistical method: Linear regression | Plasma | Cytokines, inflammatory markers | IL-6, IL-8, IL-10, TNF-R2, CRP |
| Osborn,54 2013 | Cross-sectional | nAMD | USA | Disease characteristics | 45 (26 AMD, 19 control) | 3 | Supervised: OPLS-DA, SVM Unsupervised: PCA Bioinformatics: KEGG | Serum | Metabolomic profile | 52 metabolites including those from the tyrosine, sulfur amino acid, and urea metabolism pathways |
| Robman,47 2007 | Case–control | AMD | Australia | Risk factors | 630 (197 AMD, 433 control) | 1 | Statistical method: Logistic regression | Serum | Chlamydia pneumoniae related markers | None |
| Robman,48 2010 | Case–control, retrospective cohorta | AMD | Australia | Risk factors | Case–control: 5.44 (312 AMD, 232 control) Cohort: 254 | 1 | Statistical method: Univariate and multivariate logistic regression | Serum | CRP | CRP |
| Sato,56 2018 | Prospective cohort | nAMD | Japan | Treatment decisions | 43 (21 nAMD patients, 22 cataract controls) | 1 | Statistical method: Logistic regression | Aqueous humor | Inflammatory cytokines and growth factors | IL-6, IP-10, VEGF |
| Sato,55 2019 | Cross-sectional | nAMD | Japan | Disease characteristics | 82 (62 AMD, 20 cataract control) | 2 | Unsupervised: PCA, hierarchical cluster analysis Statistical method: Binomial logistic regression, EFA | Aqueous humor | Cytokines | IL-6, IL-7, IP-10 MCP-1, MIP-1β, VEGF |
| Schori,17 2018 | Cross-sectional | AMD, nAMD (PDR, ERM) | Switzerland | Disease characteristics | 34 (6 dry AMD, 10 nAMD, 9 PDR, 9 ERM) | 2 | Unsupervised: Hierarchical Pearson clustering Bioinformatics: GO | Vitreous humor | Proteomic profile | 677 proteins including cholinesterase and oxidative stress pathway in dry AMD, focal adhesion in nAMD, and serine carboxypeptidase in all AMD |
| Subhi,57 2019 | Prospective cohort | nAMD, PCV | Denmark | Treatment decisions | 81 | 1 | Statistical method: Multiple linear regression, Pearson correlation coefficient | Plasma | CD11b+ circulating monocytes | CD11b+ circulating monocytes |
| Ueda-Consolvo,58 2017 | Retrospective cohort | nAMD | Japan | Treatment decisions | 64 | 1 | Statistical method: Multiple regression | Unknown | HbA1c | Unknown |
| Vanderbeek,49 2013 | Retrospective cohort | AMD | USA | Risk factors | 486,124 (107,007 for nonexudative AMD analysis, 113,111 for exudative AMD analysis, 10,753 for progression to exudative AMD analysis) | 1 | Statistical method: Cox proportional hazards regression | Serum | Lipid profile | HDL, LDL |
| Yao,50 2013 | Cross-sectional | wAMD | China | Disease characteristics | 12 | 1 | Bioinformatics: GO | Aqueous humor | Proteomic profile | 68 proteins, including those from the inflammation, apoptosis, angiogenesis, and oxidative stress pathways |
| Yi,59 2020 | Prospective cohort | nAMD (CRVO, DME, BRVO, pmCNV) | China | Treatment decisions | 144 | 1 | Statistical method: Multivariate linear regression | Aqueous humor | Cytokines | ICAM-1, IL-6, VEGF |
| Yip,51 2015 | Prospective cohort | AMD, GA | UK | Risk factors | 5344 | 1 | Statistical model: Multivariable logistic regression | Serum | Cardiovascular health profile | HDL-cholesterol, CRP |
Notes: aRelevant study phase.
Abbreviations: AMD, age-related macular degeneration; AI, artificial intelligence; PCA, principal component analysis; nAMD, neovascular age-related macular degeneration; PCV, polypoidal Choroidal Vasculopathy; CRP, c reactive protein; OPLS-DA, Orthogonal Projections to Latent Structures Discriminant Analysis; wAMD, wet age-related macular degeneration; KEGG, Kyoto Encyclopedia of Genes and Genomes; GA, geographic atrophy; sPLS-DA, Sparse Partial Least Squares Discriminant Analysis; SMOTE, Synthetic Minority Oversampling Technique; EFA, exploratory factor analysis; PDR, proliferative diabetic retinopathy; ERM, epiretinal membrane; GO, gene ontology; CRVO, central retinal vein occlusion; DME, diabetic macular edema; pmCNV, pathologic myopia associated choroidal neovascularization.
Biofluid Markers
Blood-derived biofluids made up most of the biofluids analyzed, with 17 studies (46%) using serum samples and 10 analyzing plasma (27%). Ocular biofluid analysis was predominantly of aqueous humour (8 studies, 22%), with only one study analyzing vitreous humour (3%). One study did not specify the biofluid that markers were derived from. A large proportion of the included studies examined biofluid markers with the goal of determining markers associated with AMD development or progression, however there was heterogeneity in the findings. Across 26 studies over 250 markers found to be significantly associated with AMD and over 70 pathways associated with AMD development were identified.17,24,28–51 Studies each identified between one and 677 differentially expressed biomarkers. The significance of biomarkers conflicted across multiple studies.28,30,32,34,35,37,45,46,49,51 This heterogeneity was noted by Mitchell et al, who stated that 49% of participants displayed great variability in their metabolic profiles.52 Studies using unsupervised AI and/or bioinformatics for untargeted biomarker exploration typically focused on significant results only and did not report non-significant biomarkers.
The most commonly implicated biomarker in AMD development or progression was HDL-cholesterol, found to be a significant predictor in six studies.28,32,35,45,51 However, four studies had conflicting findings, indicating that HDL-cholesterol was not significantly associated with AMD.30,34,37,46 Similarly, c-reactive protein (CRP) was found to be associated with AMD in four studies with conflicting findings reported in two studies, and total serum triglycerides were found associated with AMD in three studies with conflicting findings reported in four.28–30,32,34,35,37,46,48,49,51 The most commonly reported insignificant biomarker was total cholesterol, reported to be insignificant to AMD development in five studies, although two studies found it to be significant.30,32,34,35,37,44,51 Pathways implicated in AMD development by more than one study were oxidative stress, the glycerophospholipid pathway, 2-oxocarboxylic acid metabolism, ABC transportation, protein digestion and absorption, and mineral absorption.17,24,33,39,40,50
Differentiating factors associated with nAMD were examined in four studies, and progression of nAMD was studied in one.17,52–55 With the goal of identifying biomarker changes in nAMD, two studies examined at cytokine profiles and two examined comprehensive metabolic profiles.17,52,54,55 While over 20 biomarkers and 10 pathways were found to be significantly altered in nAMD, none were confirmed by more than one study. All of the studies characterized as “Treatment Decisions” examined patients undergoing anti-vascular endothelial growth factor therapy (anti-VEGF) for nAMD treatment.16,22,56–60 Of these studies, one examined comprehensive metabolic profiles and four examined cytokine levels or inflammatory markers.16,22,56–60 While no significant biomarkers were confirmed by multiple studies, three studies concluded that biofluid markers could be used to predict responsiveness to anti-VEGF therapy and prognosis following anti-VEGF therapy.16,59,60
AI and Bioinformatics Algorithms
A total of 23 studies used a single class of AI in their analyses, with 22 of these studies using a statistical method (regression analysis, a simple form of AI) and one study using a bioinformatics approach in gene ontology. Fourteen studies used two or more types of AI analysis. Six studies developed predictive models using AI.16,24,36,38,39,52 Four predictive models used metabolic or proteomic profiles to distinguish AMD cases from non-AMD controls.24,36,38,39 Kersten et al developed two models using sparse partial least squares discriminant analysis (sPLS-DA), one using the complete metabolic profile and the other using the complete metabolic profile plus derived variables such as ratios of metabolites. The accuracy of these models was reported as area under receiver operating curve (AUROC), a graphical description of sensitivity and specificity; the former had an AUROC of 0.71, while the latter demonstrated an AUROC of 0.66. Kuiper et al developed and validated a decision tree based predictive model for AMD, idiopathic non-infectious uveitis, primary vitreoretinal lymphoma, and rhegmatogenous retinal detachment using IL-10, IL-21, and angiotensin converting enzyme, with the AMD model demonstrating a sensitivity of 85.70%, a specificity of 87.50%, a positive predictive value of 54.30%, and an accuracy of 87.20% (balanced accuracy of 86.60%).38 Lains et al developed predictive models over two studies using multivariate logistic regression, demonstrating AUROCs from 0.645 to 0.850 that varied given the clinical variables used, markers available, and the patient cohort.24,39 These models exhibited the value of including biofluid markers in predictive models, with the predictive model that incorporated metabolites outperforming a baseline model including only age, gender, body mass index, and smoking status with an AUROC of 0.8 compared to 0.71 for the baseline model.39 Another study sought to differentiate between nAMD patients and controls using support machine vector (SVM) and was able to achieve a balanced accuracy of 75.6% and an AUROC of 0.83 in the test set.52 Gao et al developed a model to differentiate between anti-VEGF treatment responders and non-responders using metabolic profiles, achieving an AUROC of 0.762 in their test set.16
Findings from unsupervised AI analyses were used to either select differentiating biomarkers for use in subsequent predictive algorithms or to explain AMD pathogenesis.16,17,24,28,33,38–40,52,54,55 Eleven studies used unsupervised AI in combination with supervised AI, bioinformatics, or statistical methods.16,17,24,28,33,38–40,52,54,55 Bioinformatics is commonly often used alongside unsupervised AI to provide insight into higher level physiological processes by translating individual metabolites, proteins, or lipids into information about physiological pathways.16,17,24,33,39–42,50,52,54 Potential therapeutic targets were also examined in bioinformatic analysis. Only one study used supervised AI without unsupervised AI.36
AI statistical methods, primarily regression analysis, were the most commonly employed class of AI. Twenty-two papers exclusively used AI statistical methods to identify independent factors related to an outcome of interest or control for known risk factors.22,29–32,34,35,37,43–49,51,53,56–60 Regression was typically used to determine longitudinal association between a biomarker and a clinical outcome or condition, with the goal of risk factor identification or identifying if an intervention was effective in preventing an outcome or disease progression.22,29–32,34,35,37,43–49,51,53,56–60 As regression is less useful in the study of highly dimensional data, studies using only regression analysis tended to focus on specific risk factor analysis rather than entire biomarker profiles; for AMD, these studies often examined the influence of lipids, CRP, VEGF, inflammatory markers such as cytokines, cardiovascular health profile, and clinical characteristics on AMD development or progression.22,29–32,34,35,37,43–49,51,53,56–60 No studies using only AI statistical methods used validation procedures such as test and validation sets or bootstrapping in their analysis.
Quality Appraisal
The included studies were generally of high quality, with 20 having low risk of bias, 15 having moderate risk of bias, and two having high risk of bias (Supplemental Figure 1). Given the exploratory nature of many included studies, non-significant findings were often omitted, introducing reporting bias. Cross-sectional studies often failed to provide robust descriptions of the study setting (38%). The cross-sectional studies also did not reference guidelines for AMD diagnosis, instead describing findings on exam that were used to make the diagnosis of AMD (50%). In contrast, cohort studies provided excellent description of participant inclusion protocols and exposure criteria, but many did not describe loss to follow-up (50%) and had unclear strategies to address incomplete follow-up (50%). The vast majority of studies described their biomarker measurement protocols (assays, laboratory parameters) in detail but used small volumes of biofluids, potentially introducing measurement error. Importantly, the studies using supervised AI, unsupervised AI and bioinformatics did not comprehensively explain algorithm activities or the rationale for AI selection; these black-box models reduce study reproducibility and compromise external validity.
Discussion
This systematic review describes the current research and applications of AI and bioinformatics in the analysis of biofluid markers in AMD. The majority of included studies sought to identify markers associated with AMD development or progression, with HDL-cholesterol, total serum triglycerides, and CRP emerging as significant markers over multiple studies. AI models that sought to discriminate AMD or nAMD patients from controls using biofluid markers demonstrated excellent predictive accuracy, with models including discriminant analysis, decision tree, logistic regression, and SVM. Notably, some of these models outperformed predictive models that only used clinical and demographic characteristics.
A total of six studies identified elevated HDL-cholesterol as a risk factor for AMD development or progression, while elevated CRP was implicated by four studies and elevated total serum triglycerides were implicated by three. These findings suggest that altered lipid metabolism may play a pathogenic role in AMD, a relationship that was previously identified.61–63 This association is biologically plausible, as the hallmark of AMD, drusen, are composed of lipids, and there have been previously established genetic links to lipid dysregulation and systemic dyslipidemia as risk factors for AMD.61 The association between AMD and inflammation that was suggested by elevated CRP has also been previously studied.64 Other pathways detailed by multiple studies that have been significantly altered in AMD included oxidative stress, the glycerophospholipid pathway, 2-oxocarboxylic acid metabolism, ABC transportation, protein digestion and absorption, and mineral absorption.17,24,33,39,40,50 With some studies focusing on the predictive value of a single biomarker, and others using complex data sets of as many as 677 proteins, some variability in findings can be expected. Additionally, differences in study populations, demographics, and fluid extraction protocols can contribute to heterogeneous findings. All of the biomarkers implicated in AMD development were also found to be insignificant in other studies. Further, as non-significant biomarkers were often not reported studies with highly dimensional data sets, discrepancies in biomarker significance are likely greater than reported. Despite the lack of a singular characteristic biomarker limiting the immediate utility of these findings in a clinical setting, lipid metabolism and inflammation present compelling topics for future studies of AMD development.
The biomarker variations noted amongst studies could be attributed to the variabilities in study design. As many of the cross-sectional studies did not describe their study populations and demographics in detail, patient characteristics such as medication use, comorbidities, and lifestyle could all alter marker concentrations.38,41,42,50,54,55 These factors would have been particularly confounding in studies with small sample sizes. Beyond patient selection and confounding, metabolic variability within AMD subjects was noted, with stating that 49% of participants had variability in their metabolic profiles.52 Additionally, many cross-sectional studies did not reference AMD diagnostic guidelines and could have selected patients with differing severities of disease or different visible pathologies.17,33,36,38,40,50,54,55 While the included studies generally provided thorough explanation of their biofluid extraction and analysis techniques, the small quantities of biofluid analyzed could introduce bias. Volumes of ocular biofluids extracted ranged from 50uL to 500uL, with the average being approximately 135uL.17,22,33,38,50,55,56,59,60 While commercial assays are able to analyze these volumes accurately, small aliquots are susceptible microenvironment changes, exacerbated by dilution of samples for analysis and variation in storage techniques and sample handling. Finally, the algorithm activities and rationale for AI selection, particularly in the studies using supervised, unsupervised, and bioinformatics analysis, were not explained rigorously, also known as a “black-box” approach.65 As studies with similar objectives and data sets could be using entirely different biomarker selection parameters, variation in algorithms could account for some disagreement in biomarker significance. Future efforts should also describe analytical methods in detail and ensure a more comprehensive description of study population.
While AI may not have provided definitive insight into AMD pathogenesis, AI tools did display utility in prognostic and diagnostic tools. Anti-VEGF injections are a safe and effective therapy to reduce nAMD progression, but also represent an invasive, time-intensive intervention.66–68 AI algorithms were able to accurately predict responsiveness to therapy and prognosis and could be used to inform decision-making regarding appropriate nAMD management and more judicious selection of patients for injections.16,59,60 As nAMD patients have their ocular biofluids accessed frequently through anti-VEGF injections, there is an opportunity to use this information in treatment planning. Algorithms that used biomarkers to differentiate AMD cases from controls demonstrated excellent accuracy, with AUROC and diagnostic accuracy as high as 0.850 and 87.50%, respectively. While no biomarker set or AI algorithm was a clear forerunner in terms of accuracy and none of these tools were tested in a clinical setting, this level of accuracy could be satisfactory for use in screening or primary care settings. Notably, biofluid markers strengthened predictive models when compared to predictive models using clinical characteristics alone, as Lains et al demonstrated with an AUROC that was 0.09 higher after inclusion of biofluid markers.39
Despite the promise of application of AI in tools that have diagnostic or prognostic power in AMD, none of these tools have been integrated or tested in clinical workflow. Difficulties in appropriate implementation or poor technical understanding could prevent appropriate use.69 While many of these technologies remain investigational, it is important to establish their role and accuracy in a clinical context before more widespread use by clinicians. A specific AI tool could be deployed in different settings, each with different utility, for example, in a screening setting it may be less viable to access ocular biofluids, yet a lower algorithm accuracy could be acceptable, while in a specialist ophthalmology setting one might have better access to biofluids but require higher algorithm accuracy or merely use AI tools to augment the existing diagnostic process.
Conclusion
In this systematic review, we present studies that use AI or bioinformatics to analyze biofluid markers in AMD. AI analysis implicated altered lipid metabolism, inflammation, oxidative stress, glycerophospholipid pathway, and protein and mineral absorption in AMD development. However, experimental design and biological variability prevented identification of a singular characteristic marker in AMD. AI tools were able to accurately differentiate between AMD patients and controls and predict responsiveness to anti-VEGF therapy in nAMD patients, applications that augment existing clinical tools and inform clinical decision-making. Future studies should seek to test AI models in clinical settings with the goal of identifying appropriate opportunities for implementation.
Acknowledgments
We would like to acknowledge Arshpreet Bassi, Shaily Brahmbhatt, Priyanka Singh, Ishita Aggarwal, Amy Basilous, Jasmine Bhatti, and Karthik Manickavachagam, who participated in article screening.
Funding Statement
This research was in-part funded by Fighting Blindness Canada. This project did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
No conflicting relationship exists for any author. The authors do not have any proprietary interests in the materials described in the article.
References
- 1.Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Heal. 2014;2(2):e106–e116. doi: 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- 2.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379(9827):1728–1738. doi: 10.1016/S0140-6736(12)60282-7 [DOI] [PubMed] [Google Scholar]
- 3.Guymer R, Wu Z. Age-related macular degeneration (AMD): more than meets the eye. The role of multimodal imaging in today’s management of AMD—A review. Clin Exp Ophthalmol. 2020;48(7):983–995. doi: 10.1111/ceo.13837 [DOI] [PubMed] [Google Scholar]
- 4.Gong D, Kras A, Miller JB. Application of deep learning for diagnosing, classifying, and treating age-related macular degeneration. Semin Ophthalmol. 2021;36(4):198–204. doi: 10.1080/08820538.2021.1889617 [DOI] [PubMed] [Google Scholar]
- 5.Dong L, Yang Q, Zhang RH, Bin WW. Artificial intelligence for the detection of age-related macular degeneration in color fundus photographs: a systematic review and meta-analysis. EClinicalMedicine. 2021;35:100875. doi: 10.1016/j.eclinm.2021.100875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai L, Hinkle JW, Arias D, et al. Applications of artificial intelligence for the diagnosis, prognosis, and treatment of age-related macular degeneration. Int Ophthalmol Clin. 2020;60(4):147–168. doi: 10.1097/IIO.0000000000000334 [DOI] [PubMed] [Google Scholar]
- 7.Cheung R, Chun J, Sheidow T, Motolko M, Malvankar-Mehta MS. Diagnostic accuracy of current machine learning classifiers for age-related macular degeneration: a systematic review and meta-analysis. Eye. 2021;36:994–1004. doi: 10.1038/s41433-021-01540-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grisanti S, Tatar O. The role of vascular endothelial growth factor and other endogenous interplayers in age-related macular degeneration. Prog Retin Eye Res. 2008;27(4):372–390. doi: 10.1016/j.preteyeres.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 9.Ding X, Patel M, Chan CC. Molecular pathology of age-related macular degeneration. Prog Retin Eye Res. 2009;28(1):1–18. doi: 10.1016/j.preteyeres.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhuiyan A, Wong TY, Ting DSW, Govindaiah A, Souied EH, Smith RT. Artificial intelligence to stratify severity of age-related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol. 2020;9(2):1–12. doi: 10.1167/TVST.9.2.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von der Emde L, Pfau M, Holz FG, et al. AI-based structure-function correlation in age-related macular degeneration. Eye. 2021;35(8):2110–2118. doi: 10.1038/s41433-021-01503-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perepelkina T, Fulton AB. Artificial Intelligence (AI) Applications for Age-Related Macular Degeneration (AMD) and other retinal dystrophies. Semin Ophthalmol. 2021;36(4):304–309. doi: 10.1080/08820538.2021.1896756 [DOI] [PubMed] [Google Scholar]
- 13.Reiter GS, Grechenig C, Vogl WD, et al. Analysis of fluid volume and its impact on visual acuity in the fluid study as quantified with deep learning. Retina. 2021;41(6):1318–1328. doi: 10.1097/IAE.0000000000003023 [DOI] [PubMed] [Google Scholar]
- 14.Schmidt-Erfurth U, Vogl WD, Jampol LM, Bogunović H. Application of automated quantification of fluid volumes to anti–VEGF therapy of neovascular age-related macular degeneration. Ophthalmology. 2020;127(9):1211–1219. doi: 10.1016/j.ophtha.2020.03.010 [DOI] [PubMed] [Google Scholar]
- 15.Hwang DK, Hsu CC, Chang KJ, et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics. 2019;9(1):232–245. doi: 10.7150/thno.28447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Teo YCK, Beuerman RW, Wong TY, Zhou L, Cheung CMG. A serum metabolomics study of patients with nAMD in response to anti-VEGF therapy. Sci Rep. 2020;10(1):1–10. doi: 10.1038/s41598-020-58346-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schori C, Trachsel C, Grossmann J, Zygoula I, Barthelmes D, Grimm C. The proteomic landscape in the vitreous of patients with age-related and diabetic retinal disease. Investig Ophthalmol Vis Sci. 2018;59(4):AMD31–AMD40. doi: 10.1167/iovs.18-24122 [DOI] [PubMed] [Google Scholar]
- 18.Romond K, Alam M, Kravets S, et al. Imaging and artificial intelligence for progression of age-related macular degeneration. Exp Biol Med. 2021;246(20):2159–2169. doi: 10.1177/15353702211031547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor R, Whigham BT, Al-Aswad LA. The role of artificial intelligence in the diagnosis and management of glaucoma. Curr Ophthalmol Rep. 2019;7(2):136–142. doi: 10.1007/s40135-019-00209-w [DOI] [Google Scholar]
- 20.Zapata MA, Royo-Fibla D, Font O, et al. Artificial Intelligence to identify retinal fundus images, quality validation, laterality evaluation, macular degeneration, and suspected glaucoma. Clin Ophthalmol. 2020;14:419–429. doi: 10.2147/OPTH.S235751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aslam TM, Zaki HR, Mahmood S, et al. Use of a neural net to model the impact of optical coherence tomography abnormalities on vision in age-related macular degeneration. Am J Ophthalmol. 2018;185:94–100. doi: 10.1016/j.ajo.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 22.Lai TYY, Liu DTL, Chan KP, Luk FOJ, Pang CP, Lam DSC. Visual outcomes and growth factor changes of two dosages of intravitreal bevacizumab for neovascular age-related macular degeneration: a randomized, controlled trial. Retina. 2009;29(9):1218–1226. doi: 10.1097/IAE.0b013e3181b32c45 [DOI] [PubMed] [Google Scholar]
- 23.Sivagurunathan S, Selvan LN, Khan A, et al. Proteomics-based approach for differentiation of age-related macular degeneration sub-types. Indian J Ophthalmol. 2021;69(3):647. doi: 10.4103/ijo.IJO_470_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laíns I, Chung W, Kelly RS, et al. Human plasma metabolomics in age-related macular degeneration: meta-analysis of two cohorts. Metabolites. 2019;9(7):127. doi: 10.3390/metabo9070127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moola S, Munn Z, Tufanaru C, et al. Systematic reviews of etiology and risk. JBI. 2020. doi: 10.46658/JBIMES-20-08 [DOI] [Google Scholar]
- 27.Valesan LF, Da-Cas CD, Réus JC, et al. Prevalence of temporomandibular joint disorders: a systematic review and meta-analysis. Clin Oral Investig. 2021;25(2):441–453. doi: 10.1007/s00784-020-03710-w [DOI] [PubMed] [Google Scholar]
- 28.Acar İE, Lores-Motta L, Colijn JM, et al. Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration: the EYE-RISK consortium. Ophthalmology. 2020;127:1693–1709. doi: 10.1016/j.ophtha.2020.06.020 [DOI] [PubMed] [Google Scholar]
- 29.Boekhoorn SS. C-reactive protein level and risk of aging macula disorder. Arch Ophthalmol. 2007;125(10):1396. doi: 10.1001/archopht.125.10.1396 [DOI] [PubMed] [Google Scholar]
- 30.Buch H, Vinding T, la Cour M, Jensen GB, Prause JU, Nielsen NV. Risk factors for age-related maculopathy in a 14-year follow-up study: the Copenhagen city eye study. Acta Ophthalmol Scand. 2005;83(4):409–418. doi: 10.1111/j.1600-0420.2005.00492.x [DOI] [PubMed] [Google Scholar]
- 31.Chaker L, Buitendijk GHS, Dehghan A, et al. Thyroid function and age-related macular degeneration: a prospective population-based cohort study - the Rotterdam study. BMC Med. 2015;13(1):1–8. doi: 10.1186/s12916-015-0329-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cougnard-Grégoire A, Delyfer MN, Korobelnik JF, et al. Elevated high-density lipoprotein cholesterol and age-related macular degeneration: the Alienor study. PLoS One. 2014;9:3. doi: 10.1371/journal.pone.0090973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han G, Wei P, He M, Teng H, Chu Y. Metabolomic profiling of the aqueous humor in patients with wet age-related macular degeneration using UHPLC-MS/MS. J Proteome Res. 2020;19(6):2358–2366. doi: 10.1021/acs.jproteome.0c00036 [DOI] [PubMed] [Google Scholar]
- 34.Joachim NDL, Mitchell P, Kifley A, Jinwang J. Incidence, progression, and associated risk factors of medium drusen in age-related macular degeneration findings from the 15-year follow-up of an Australian cohort. JAMA Ophthalmol. 2015;133(6):698–705. doi: 10.1001/jamaophthalmol.2015.0498 [DOI] [PubMed] [Google Scholar]
- 35.Jonasson F, Fisher DE, Eiriksdottir G, et al. Five-year incidence, progression, and risk factors for age-related macular degeneration: the age, gene/environment susceptibility study. Ophthalmology. 2014;121(9):1766–1772. doi: 10.1016/j.ophtha.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kersten E, Dammeier S, Ajana S, et al. Metabolomics in serum of patients with non-advanced age-related macular degeneration reveals aberrations in the glutamine pathway. PLoS One. 2019;14(6):1–12. doi: 10.1371/journal.pone.0218457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein R, Lee KE, Tsai MY, Cruickshanks KJ, Gangnon RE, Klein BEK. Oxidized Low-density Lipoprotein and the Incidence of Age-related Macular Degeneration. Ophthalmology. 2019;126(5):752–758. doi: 10.1016/j.ophtha.2018.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuiper JJW, Beretta L, Nierkens S, et al. An ocular protein triad can classify four complex retinal diseases. Sci Rep. 2017;7:1–9. doi: 10.1038/srep41595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laíns I, Kelly RS, Miller JB, et al. Human plasma metabolomics study across all stages of age-related macular degeneration identifies potential lipid biomarkers. Ophthalmology. 2018;125(2):245–254. doi: 10.1016/j.ophtha.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo D, Deng T, Yuan W, Deng H, Jin M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017;17(1):1–9. doi: 10.1186/s12886-017-0555-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lynch AM, Wagner BD, Weiss SJ, et al. Proteomic profiles in advanced age-related macular degeneration using an aptamer-based proteomic technology. Transl Vis Sci Technol. 2019;8:1. doi: 10.1167/tvst.8.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch AM, Wagner BD, Palestine AG, et al. Plasma biomarkers of reticular pseudodrusen and the risk of progression to advanced age-related macular degeneration. Transl Vis Sci Technol. 2020;9(10):1–13. doi: 10.1167/tvst.9.10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez KM, Kim J, Laíns I, et al. Association of human plasma metabolomics with delayed dark adaptation in age-related macular degeneration. Metabolites. 2021;11(3):1–13. doi: 10.3390/metabo11030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Millen AE, Meyers KJ, Liu Z, et al. Association between vitamin D status and age-related macular degeneration by genetic risk. JAMA Ophthalmol. 2015;133(10):1171–1179. doi: 10.1001/jamaophthalmol.2015.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millen AE, Nie J, Sahli MW, et al. Vitamin D status and prevalent early age-related macular degeneration in African Americans and Caucasians: the Atherosclerosis Risk in Communities (ARIC) study. J Nutr Health Aging. 2017;21(7):772–780. doi: 10.1007/s12603-016-0827-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngai LY, Stocks N, Sparrow JM, et al. The prevalence and analysis of risk factors for age-related macular degeneration: 18-year follow-up data from the Speedwell eye study, United Kingdom. Eye. 2011;25(6):784–793. doi: 10.1038/eye.2011.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robman L, Mahdi OS, Wang JJ, et al. Exposure to Chlamydia pneumoniae infection and age-related macular degeneration: the blue mountains eye study. Investig Ophthalmol Vis Sci. 2007;48(9):4007–4011. doi: 10.1167/iovs.06-1434 [DOI] [PubMed] [Google Scholar]
- 48.Robman L, Baird PN, Dimitrov PN, Richardson AJ, Guymer RH. C-reactive protein levels and complement factor H polymorphism interaction in age-related macular degeneration and its progression. Ophthalmology. 2010;117(10):1982–1988. doi: 10.1016/j.ophtha.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 49.Vanderbeek BL, Zacks DN, Talwar N, Nan B, Stein JD. Role of statins in the development and progression of age-related macular degeneration. Retina. 2013;33(2):414–422. doi: 10.1097/IAE.0b013e318276e0cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao J, Liu X, Yang Q, et al. Proteomic analysis of the aqueous humor in patients with wet age-related macular degeneration. Proteomics Clin Appl. 2013;7(7–8):550–560. doi: 10.1002/prca.201200012 [DOI] [PubMed] [Google Scholar]
- 51.Yip JLY, Khawaja AP, Chan MPY, et al. Cross sectional and longitudinal associations between cardiovascular risk factors and age related macular degeneration in the EPICNorfolk eye study. PLoS One. 2015;10(7):1–11. doi: 10.1371/journal.pone.0132565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell SL, Uppal K, Williamson SM, et al. The carnitine shuttle pathway is altered in patients with neovascular age-related macular degeneration. Investig Ophthalmol Vis Sci. 2018;59(12):4978–4985. doi: 10.1167/iovs.18-25137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen MK, Subhi Y, Molbech CR, Falk MK, Nissen MH, Sørensen TL. Systemic levels of interleukin-6 correlate with progression rate of geographic atrophy secondary to age-related macular degeneration. Investig Ophthalmol Vis Sci. 2019;60(1):202–208. doi: 10.1167/iovs.18-25878 [DOI] [PubMed] [Google Scholar]
- 54.Osborn MP, Park Y, Parks MB, et al. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS One. 2013;8:8. doi: 10.1371/journal.pone.0072737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato T, Takeuchi M, Karasawa Y, Takayama K, Enoki T. Comprehensive expression patterns of inflammatory cytokines in aqueous humor of patients with neovascular age-related macular degeneration. Sci Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-55191-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato T, Takeuchi M, Karasawa Y, Enoki T, Ito M. Intraocular inflammatory cytokines in patients with neovascular age-related macular degeneration before and after initiation of intravitreal injection of anti-VEGF inhibitor. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-19594-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subhi Y, Krogh Nielsen M, Molbech CR, et al. Association of CD11b+ Monocytes and Anti-Vascular Endothelial Growth Factor Injections in Treatment of Neovascular Age-Related Macular Degeneration and Polypoidal Choroidal Vasculopathy. JAMA Ophthalmol. 2019;137(5):515–522. doi: 10.1001/jamaophthalmol.2019.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueda-Consolvo T, Hayashi A, Ozaki M, Nakamura T, Yagou T, Abe S. The relationship between vascular endothelial dysfunction and treatment frequency in neovascular age-related macular degeneration. Jpn J Ophthalmol. 2017;61(4):347–353. doi: 10.1007/s10384-017-0515-z [DOI] [PubMed] [Google Scholar]
- 59.Yi QY, Wang YY, Chen LS, et al. Implication of inflammatory cytokines in the aqueous humour for management of macular diseases. Acta Ophthalmol. 2020;98(3):e309–e315. doi: 10.1111/aos.14248 [DOI] [PubMed] [Google Scholar]
- 60.Arai Y, Takahashi H, Inoda S, et al. Aqueous humour proteins and treatment outcomes of anti-VEGF therapy in neovascular age-related macular degeneration. PLoS One. 2020;15(3):1–14. doi: 10.1371/journal.pone.0229342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin JB, Halawa OA, Husain D, Miller JW, Vavvas DG. Dyslipidemia in age-related macular degeneration. Eye. 2022;36(2):312–318. doi: 10.1038/s41433-021-01780-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Betzler BK, Rim TH, Sabanayagam C, Cheung CMG, Cheng C-Y. High-density lipoprotein cholesterol in age-related ocular diseases. Biomolecules. 2020;10(4):645. doi: 10.3390/biom10040645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan Q, Maranville JC, Fritsche L, et al. HDL-cholesterol levels and risk of age-related macular degeneration: a multiethnic genetic study using Mendelian randomization. Int J Epidemiol. 2017;46(6):1891–1902. doi: 10.1093/ije/dyx189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan W, Zou J, Yoshida S, Jiang B, Zhou Y. The role of inflammation in age-related macular degeneration. Int J Biol Sci. 2020;16(15):2989–3001. doi: 10.7150/ijbs.49890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poon AIF, Sung JJY. Opening the black box of AI-Medicine. J Gastroenterol Hepatol. 2021;36(3):581–584. doi: 10.1111/jgh.15384 [DOI] [PubMed] [Google Scholar]
- 66.Bakri SJ, Thorne JE, Ho AC, et al. Safety and efficacy of anti-vascular endothelial growth factor therapies for neovascular age-related macular degeneration: a report by the American Academy of ophthalmology. Ophthalmology. 2019;126(1):55–63. doi: 10.1016/j.ophtha.2018.07.028 [DOI] [PubMed] [Google Scholar]
- 67.Corazza P, D’Alterio FM, Kabbani J, et al. Long-term outcomes of intravitreal anti-VEGF therapies in patients affected by neovascular age-related macular degeneration: a real-life study. BMC Ophthalmol. 2021;21(1):300. doi: 10.1186/s12886-021-02055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khanna S, Komati R, Eichenbaum DA, Hariprasad I, Ciulla TA, Hariprasad SM. Current and upcoming anti-VEGF therapies and dosing strategies for the treatment of neovascular AMD: a comparative review. BMJ Open Ophthalmol. 2019;4(1):1–8. doi: 10.1136/bmjophth-2019-000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pucchio A, Del PJ, de Moraes FY. Artificial intelligence in the medical profession: ready or not, here AI comes. Clinics. 2022;77:100010. doi: 10.1016/j.clinsp.2022.100010 [DOI] [PMC free article] [PubMed] [Google Scholar]