Abstract
A mutant of Saccharomyces cerevisiae deficient in the lactate-proton symport was isolated. Transformation of the mutant with a yeast genomic library allowed the isolation of the gene JEN1 that restored lactate transport. Disruption of JEN1 abolished uptake of lactate. The results indicate that, under the experimental conditions tested, no other monocarboxylate permease is able to efficiently transport lactate in S. cerevisiae.
Saccharomyces cerevisiae is able to utilize short-chain monocarboxylic acids as sole carbon and energy sources under aerobic conditions. Transport across the plasma membrane is the first step in the metabolism of these substrates. Monocarboxylate proton symporters with different specificities have been described: acetic acid or ethanol-grown cells present a permease which is shared by acetate, propionate, and formate (5), while cells grown with lactic acid have an additional permease which transports lactate, pyruvate, acetate, and propionate (4, 6). The capacity to transport monocarboxylates was not present when the cells were grown in glucose (6). In agreement with the physiological results indicating the existence of different monocarboxylate permeases in S. cerevisiae, four open reading frames (ORFs) with important similarities to mammalian monocarboxylate permeases were found in the genome of S. cerevisiae (13, 16). However, to our knowledge, no direct evidence relating a concrete gene product with a definite monocarboxylate transport activity is available. We have obtained yeast mutants unable to transport lactic acid and have cloned a gene that restores this transport capacity. The results presented in this work indicate that the product of the gene JEN1 is a permease for lactate and suggest that, under the experimental conditions analyzed, no other system for lactate transport was operating in S. cerevisiae.
S. cerevisiae W303-1A (MATa ade2 leu2 his3 trp1 ura3) (20) was used as starting material. Its isogenic strain of the opposite mating type was used in crosses. Yeasts were grown in 1% (wt/vol) yeast extract–1% (wt/vol) peptone (YP) or in 0.7% (wt/vol) Difco yeast nitrogen base (YNB) supplemented with adequate quantities of nutrients required for growth. Carbon sources were 2% (wt/vol) glucose, 0.5% (vol/vol) dl-lactic acid (pH 5.0), 0.5% (wt/vol) sodium pyruvate (pH 5.0), 0.5% (vol/vol) acetic acid (pH 5.0), 1% (wt/vol) glycerol, and 1% (wt/vol) ethanol. To prepare solid media, 2% (wt/vol) agar was added. Growth was at 28°C, and liquid cultures were grown with shaking. Cultures were always harvested during the exponential phase of growth. Escherichia coli XL-Blue and DH5α were used for plasmid amplification and preparation (19). Uptake experiments were carried out with yeast suspensions prepared from cells growing exponentially in YP-lactate washed twice with chilled water and resuspended in water. Conical centrifuge tubes containing 30 μl of 0.1 M KH2PO4 buffer at pH 5.0 and 10 μl of the yeast suspension (25 to 45 mg [dry weight]/ml) were incubated for 4 min at 25°C. Next, the reaction was started by the addition of 10 μl of an aqueous solution of labelled dl-[1-14C]lactic acid (sodium salt; Amersham) (3,000 dpm/nmol) at pH 5.0. The reaction was stopped by dilution with 5 ml of ice-cold water. Sampling times were 0, 5, 10, and 20 s, so that uptake rates remained linear. The reaction mixtures were filtered immediately through GF/C membranes (Whatman, Inc., Clifton, N.J.), and the filters were washed with 10 ml of ice-cold water and transferred to scintillation fluid (Opti-Phase HiSafe II; LKB FSA Laboratory Supplies, Loughborough, United Kingdom). Radioactivity was measured in a Packard Tri-Carb 2200 CA liquid scintillation spectrophotometer, with disintegrations per minute correction. For nonspecific 14C adsorption, labelled lactate was added at zero time after the cold water. To determine the transport kinetics best fitting the experimental initial uptake rates and to estimate the kinetic parameters, a computer-assisted nonlinear regression analysis (GraphPAD Software, San Diego, Calif.) was used. All the experiments were repeated at least three times. Proton uptake was measured in a thermostated chamber at 25°C with a standard pH meter connected to a recorder. To the chamber, 4.5 ml of 10 mM KH2PO4 and 0.5 ml of cell suspension were added, and the pH was adjusted to the desired value to obtain a baseline. The lactate solution, adjusted to the same pH value, was added and the subsequent alkalization was monitored. Rates of proton uptake were calculated from the slope of the initial part of the pH trace. Calibration was performed with HCl.
A commercial S. cerevisiae genomic library in YEp13 (ATCC 37323) was used. Transformation of yeast cells and molecular biology operations were performed according to the methods of Ausubel et al. (1). Disruption of the cloned gene was done by replacing the 1,758-bp HpaI-Eco47III sequence by a 1,472-bp PvuII-PvuII fragment from vector YDpH (2) carrying the HIS3 gene. A 2.7-kb XbaI-ScaI fragment was used to replace the genomic copy of the gene by the one-step disruption method (18) (Fig. 1). Northern blot analysis of JEN1 expression was performed according to standard procedures (19).
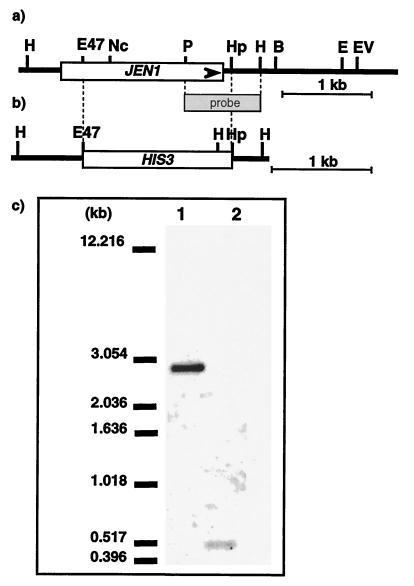
FIG. 1.
Scheme of the yeast genomic DNA region containing JEN1 and Southern blot analysis of its disruption. (a) Schematic restriction map of the yeast genomic DNA fragment contained in the plasmid pT12. This plasmid, which contains the complete sequence of JEN1 (YKL 217w), was isolated from the original YEp13 genomic library. (b) Disruption of JEN1 was carried out as described in the text. The 2.7-kb SacI-XbaI fragment was used to replace the genomic wild-type copy. (c) Southern blot analysis of the JEN1 disruption. Genomic DNA was digested with HindIII, subjected to electrophoresis in a 0.8% agarose gel, transferred to Hybond-N membrane, and hybridized with the 855-bp probe indicated in panels a and b. Lanes: 1, DNA from wild type; 2, DNA from BLC 203 strain carrying the jen1::HIS3 disruption. Sizes of the fragments are indicated at left.
Isolation and characterization of a mutant unable to grow on dl-lactate.
A suspension of about 107 yeast cells/ml was irradiated with a UV germicidal lamp, plated on YP-glycerol, and replica plated onto YNB-lactate. Four colonies unable to grow in this medium were found, but only one appeared affected in its capacity to transport lactate. A diploid isolated from a cross of this mutant with a wild-type strain grew on lactate, indicating that the mutation was recessive. The segregation of the negative growth on lactate was 2+:2− in 10 complete tetrads analyzed, indicating that the phenotype was caused by a nuclear, monogenic mutation. The original mutant was named BLC 55, and a spore from a cross presenting the lactate-negative phenotype was termed BLC 142. In both cases, faint, residual growth on lactate was always observed. A strain mutated in the PCK1 gene, encoding phosphoenolpyruvate carboxykinase, or a double mutant with alterations in the CYB2 and DLD genes (12), encoding the d- and l-lactate ferricytochrome c oxidoreductases, respectively, gave a complete absence of growth in the same medium. Strains BLC 55 and BLC 142 grew in glycerol, ethanol, or acetate as did the wild type, but they did not grow in pyruvate.
Cells from the BLC 142 mutant grown in glucose and incubated in YP-lactate for 4 h showed at 2.0 mM lactate a transport activity that was only one-fifth that of the wild type. Moreover, the transient extracellular alkalization indicative of proton uptake was absent after addition of dl-lactate or pyruvate (not shown). The observed lactate uptake could be due to diffusion of the undissociated acid. This behavior was similar to that found in glucose-repressed cells of the wild type, where no measurable activity of the lactate permease is found (6).
Isolation of the gene responsible for the lactate-negative phenotype.
The gene whose mutation caused inability to grow in lactate was isolated by functional complementation of strain BLC 142. The yeast was transformed to leucine prototrophy, and transformants were screened for growth on lactate. Among 20,000 transformants, 20 were able to grow efficiently on dl-lactate. When these clones were grown under nonselective conditions, the ability to grow in lactate and the Leu+ phenotypes were lost together in one clone. Plasmid DNA was prepared from this transformant and used to transform again the original BLC 142 strain. The plasmid (pT12) restored the ability to grow in lactate, the permease activity, and the capacity to alkalinize the medium after addition of the acid (not shown). By restriction analysis, a yeast genomic DNA insert of 4.4 kb was identified in pT12 (Fig. 1a). It seems, therefore, that the DNA on the plasmid carries the gene encoding the lactate permease. A 0.9-kb EcoRI-BamHI yeast genomic DNA fragment contained in the pT12 plasmid was sequenced. It was found to have a sequence of nucleotides in the vicinity of ORF YKL217w (21). This ORF corresponds to a gene termed JEN1 that in the YPD database is said to be implicated in the transport of lactate. The restriction analysis of pT12 indicated that it contained JEN1. This gene has been described as a protein member of the major facilitator superfamily belonging to the sialate:H+ family (13, 15). It is also apparently similar to the E. coli genes encoding the osmoregulatory proline-betaine transporter proP and the α-ketoglutarate transporter (10). It is likely that different search algorithms recognize diverse features and produce different assignments within a superfamily, in this case the major facilitator superfamily (15).
Effects of the disruption of the JEN1 gene.
To ascertain the physiological role of the JEN1 gene, we disrupted it and placed the disrupted copy into the genome (Fig. 1b). Southern blot analysis demonstrated that the integration was correct (Fig. 1c). The disruptant, BLC 203, did not grow on lactate and did not show measurable permease activity (Fig. 2a). In contrast, transport of acetate was operational in the mutant BLC 203 under all derepressed conditions tested (data not shown), confirming previous results which indicated that the transport system for acetate was always present in derepression conditions but could not transport lactate.
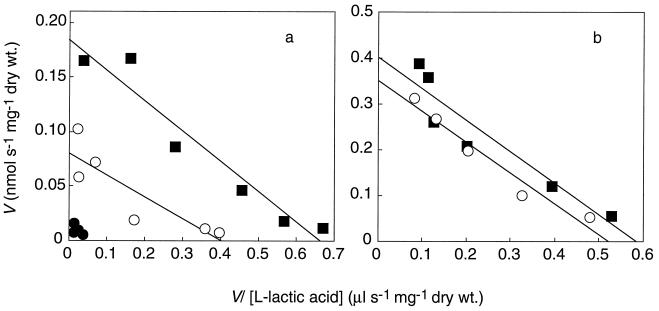
FIG. 2.
Eadie-Hoffstee plots of the initial uptake rates of lactate by the wild type and a jen1::HIS3 disrupted strain. Uptake studies were done at pH 5.0 as described in the text. (a) Cells growing exponentially in YNB-glucose were harvested by centrifugation, washed twice with deionized water, and incubated in YP-lactate for 4 h. (b) Cells growing exponentially in YP-lactate. Symbols: ■, wild type; ●, mutant BLC 203; ○, mutant BLC 203 transformed with plasmid pT12.
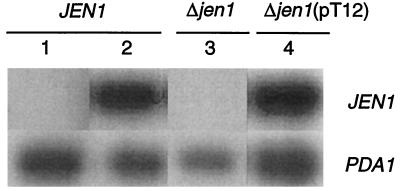
The mutant BLC 203 transformed with plasmid pT12 was able to grow in YP-lactate and recovered the activity of the lactate carrier (Fig. 2b). From the data in Fig. 2b, the following kinetics parameters were estimated: for the wild-type, strain Vmax was estimated to be 0.40 nmol s−1 mg (dry weight)−1 and Km was estimated to be 0.69 mM; for the BLC 203 transformed strain, Vmax was estimated to be 0.36 nmol s−1 mg (dry weight)−1 and Km was estimated to be 0.68 mM. The fact that the transformed strain did not transport lactate faster than the wild-type strain could be due either to a lack of appropriate expression or to problems with insertion of the protein in the membrane. To distinguish between these possibilities, a Northern analysis of the expression of JEN1 was performed (Fig. 3). After induction in YP-lactate medium, the wild-type strain exhibited a strong signal against a JEN1 probe while no signal was detected in noninduced cells. Also, no signal was visible with strain BLC 203, which carries an interrupted JEN1 gene. BLC 203 cells transformed with pT12 plasmid recovered JEN1 expression. However, in these cells, the level of JEN1 mRNA after induction was of the same order of magnitude as that found in the wild-type strain. This finding points to the existence of some regulatory element in the promoter that is missing in the plasmid; in fact, it has only about 500 bp in front of the putative ATG. The results of the Northern analysis indicate therefore that the lack of increased transport activity in the transformants is due to an inefficient expression.
FIG. 3.
Northern blot analysis of JEN1 transcripts from strains differing in locus state. JEN1, wild type; Δjen1, disruptant mutant BLC 203; Δjen1(pT12), BLC 203 strain transformed with pT12 plasmid. Total cellular mRNA was prepared from cells growing exponentially in YNB-glucose (lane 1) and subsequently incubated in YP-lactate for 4 h (lanes 2, 3, and 4) (14). Total RNA (20 μg) was separated on 1.5% agarose morpholinepropanesulfonic acid (MOPS)-formaldehyde gels and blotted to nylon membranes by vacuum transference. Hybridization was carried out using the 844-bp NcoI-PstI fragment as a probe for JEN1 (Fig. 1a), and PDA1 (22) served as internal standard.
The transport of lactate is an important step in the metabolism of several organisms. In mammals, it is a key step in the reutilization by liver and kidney of lactate produced in muscle glycolysis (the Cori cycle). A family of monocarboxylate transporters that also transport pyruvate and other ketoacids exists with different tissue specificities (17). In erythrocytes, three different systems with different mechanisms appear to operate in lactate transport, although proton symport accounts for about 90% of it (7). In E. coli, the lct locus, involved in lactate metabolism, encompasses three overlapping genes, one of which (lctP) appears to encode a lactate transporter (8). However, to our knowledge no further information on this system is available. Among yeasts, the mechanisms of lactate transport are different; while Candida utilis (11), S. cerevisiae (4, 6), and Torulaspora delbrueckii (3) transport lactate by proton symports, Kluyveromyces marxianus does it via a uniport (9). It will be interesting to compare the structures of the respective proteins involved in the process.
Our initial aim was to isolate the genes encoding the H+ symport that takes up lactate in S. cerevisiae. The results presented here clearly show that the gene JEN1 encodes a lactate permease and that in its absence diffusion alone does not allow efficient growth of the yeast in lactate.
Acknowledgments
We acknowledge the help of Maria J. Lafuente in the DNA sequencing and analysis and the critical reading of the manuscript by Juana M. Gancedo.
This study was supported by Portuguese grant PRAXIS/2/2.1/BIO/1068/95. Work in the laboratory of C.G. was supported by grant PB97-1213-CO2-01 from the Spanish CICYT. M.C. had a FEBS short-term fellowship during the early stages of this work. S.P. received a fellowship from PRAXIS XXI-BM4229. R.P.A. received a fellowship from PRAXIS XXI-BD15737/98.
REFERENCES
- 1.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1998. [Google Scholar]
- 2.Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 3.Casal M, Leão C. Utilization of short-chain monocarboxylic acids by the yeast Torulaspora delbrueckii: specificity of the transport systems and their regulation. Biochim Biophys Acta. 1995;1267:122–130. doi: 10.1016/0167-4889(95)00067-3. [DOI] [PubMed] [Google Scholar]
- 4.Casal M, Blázquez M, Gamo F-J, Gancedo C, Leão C. Lack of lactate-proton symport activity in pck1 mutants of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1995;128:279–282. doi: 10.1111/j.1574-6968.1995.tb07536.x. [DOI] [PubMed] [Google Scholar]
- 5.Casal M, Cardoso H, Leão C. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology. 1996;142:1385–1390. doi: 10.1099/13500872-142-6-1385. [DOI] [PubMed] [Google Scholar]
- 6.Cássio F, Leão C, van Uden N. Transport of lactate and other short-chain monocarboxylic acids in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 1987;53:509–513. doi: 10.1128/aem.53.3.509-513.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deuticke B, Beyer E, Forst B. Discrimination of three parallel pathways of lactate transport in the human erythrocyte membrane by inhibitors and kinetic properties. Biochim Biophys Acta. 1982;684:96–110. doi: 10.1016/0005-2736(82)90053-0. [DOI] [PubMed] [Google Scholar]
- 8.Dong J M, Taylor J S, Latour D J, Iuchi S, Lin E C. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fonseca A, Spencer-Martins I, van Uden N. Transport of lactic acid in Kluyveromyces marxianus: evidence for a monocarboxylate uniport. Yeast. 1991;7:775–780. doi: 10.1002/yea.320070803. [DOI] [PubMed] [Google Scholar]
- 10.Garrel J I. The yeast proteome handbook. 2nd ed. Beverly, Mass: Proteome Inc.; 1997. [Google Scholar]
- 11.Leão C, van Uden N. Transport of lactate and other monocarboxylates in the yeast Candida utilis. Appl Microbiol Biotechnol. 1986;23:389–393. doi: 10.1128/aem.53.3.509-513.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lodi T, Ferrero I. Isolation of the gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme d-lactate ferricytochrome c oxidoreductase. Mol Gen Genet. 1993;238:315–324. doi: 10.1007/BF00291989. [DOI] [PubMed] [Google Scholar]
- 13.Nelissen B, De Wachter R, Goffeau A. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol Rev. 1997;21:113–134. doi: 10.1111/j.1574-6976.1997.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 14.Newman A. Analysis of pre-mRNA splicing in yeast. In: Higgins S J, Hames B D, editors. RNA processing: a practical approach. I. Oxford, England: IRL Press; 1994. pp. 182–183. [Google Scholar]
- 15.Pao S S, Paulsen I T, Saier M H. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulsen I T, Sliwinski M K, Nelissen B, Goffeau A, Saier M H. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998;430:116–125. doi: 10.1016/s0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 17.Price N T, Jackson V N, Halestrap A P. Cloning and sequencing of four new mammalian monocarboxylate transporter (MCT) homologues confirms the existence of a transporter family with an ancient past. Biochem J. 1998;329:321–328. doi: 10.1042/bj3290321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothstein R J. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;58:819–830. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 21.Tzermia M, Horaitis O, Alexandraki D. The complete sequencing of a 24.6 kb segment of yeast chromosome XI identified the known loci URA1, SAC1 and TRP3, and revealed 6 new open reading frames including homologues to the threonine dehydratases, membrane transporters, hydantoinases and the phospholipase A2-activating protein. Yeast. 1994;10:663–679. doi: 10.1002/yea.320100511. [DOI] [PubMed] [Google Scholar]
- 22.Wenzel T J, Teunissen A W R H, Steensma H Y. PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res. 1995;23:883–884. doi: 10.1093/nar/23.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]