Abstract
Two catalases, KatA and KatB, have been detected in Sinorhizobium meliloti growing on rich medium. Here we characterize a new catalase gene encoding a third catalase (KatC). KatC activity was detectable only at the end of the stationary phase in S. meliloti growing in minimum medium, whereas KatA activity was found during the exponential phase. Analysis with a katC-lacZ fusion demonstrated that katC expression is mainly regulated at the transcription level. An increase of catalase activity correlating with KatA induction was detected in bacteroids. A dramatic decrease of nitrogen fixation capacity in a katA katC double mutant was observed, suggesting that these catalases are very important for the protection of the nitrogen fixation process.
Cellular metabolism of molecular oxygen produces reactive and potentially toxic oxygen species such as superoxide radicals, hydrogen peroxide, and hydroxyl radicals (16). For defense against these reactive oxygen species, organisms contain antioxidants and enzymes that repair oxidative damage. Catalases (H2O2:H2O2 oxidoreductase; EC 1.11.1.6) are heme-containing enzymes involved in the dismutation of H2O2 in O2 and H2O. These enzymes play an important role in reducing the formation of the highly reactive hydroxyl radical which arises from H2O2 degradation via the Fenton reaction (16). The response of bacteria to oxidative stress has been most extensively studied in the enteric bacterium Escherichia coli (reviewed in reference 7) which synthesizes two types of catalase enzyme, a bifunctional catalase/peroxidase (HPI) encoded by katG (32) and a monofunctional catalase (HPII) encoded by katE (34). These two kat genes are regulated differently in terms of growth phase and response to oxidative stress (reviewed in reference 24).
Sinorhizobium meliloti is a soil bacterium able to establish a symbiosis with alfalfa (Medicago sativa). This symbiosis leads to the formation of nodules on the alfalfa roots following a flow of signals transmitted between plant and bacteria (13). In several aspects, this symbiosis can be considered a controlled incompatible reaction (29). Most infections can be aborted by a hypersensitivity-like response mediated by an H2O2 oxidative burst which results in the plant exhibiting control over the number of nodules formed (33). Inside the nodules, the bacteria differentiate into their endosymbiotic, bacteroid forms. The microsymbiont is able to reduce nitrogen to ammonia, which can subsequently be metabolized by the plant. Nitrogenase, the key enzyme in this fixation of atmospheric nitrogen, is quickly and irreversibly inactivated by oxygen, but the energy for N2 fixation requires a high bacteroid respiration rate. To solve this seeming paradox, a variable diffusion barrier controls the entry of O2 into the infected region (36), and a large amount of the O2-carrying protein leghemoglobin facilitates the supply of O2 to the microsymbiont (1). Additionally, an important antioxidant defense occurs in the peripheral cell layers of legume root nodules (5). Bacteroids produce a high concentration of reactive oxygen species because of the stringent conditions required to reduce N2, the potential of nitrogenase to directly reduce O2 (4), and the high rates of bacteroid respiration; it has been suggested that these oxygen-derived species have a role in the inactivation of nitrogenase (28).
To elucidate the role of catalases in the establishment and/or maintenance of Medicago-Sinorhizobium functional nodules, we have previously cloned the S. meliloti katA gene, which encodes the H2O2-inducible catalase KatA (18). We showed that free-living S. meliloti bacteria in stationary phase on rich medium produce two catalases, namely a monofunctional catalase (KatA) and a bifunctional catalase/peroxidase (KatB). A katA::Tn5 mutant showed a drastic sensitivity to H2O2, and KatA appeared to be the major component of an H2O2-adaptative response. Neither nodulating capacity nor nitrogen fixing activity were impaired in the katA mutant, suggesting that KatA is not essential for the nodulation and nitrogen fixation processes.
Cloning and analysis of the katA-homologous gene in S. meliloti.
We previously took advantage of the high homology between regions of E. coli HPII and several catalases from various phyla to clone the katA gene of S. meliloti by nested PCR (18). Southern analysis of genomic DNA, digested with different restriction enzymes, showed a pattern of two bands (7.1- and 14-kb ApaI-ApaI fragments), suggesting the presence of a second, katA-homologous, catalase gene. The 1.4-kb ApaI-EcoRI fragment corresponding to the katA coding region was radiolabeled by using the Prime-a-Gene labeling system (Promega, Charbonnières, France) and used as a probe to hybridize with an S. meliloti genomic cosmid library (12) under low stringency (55°C). Restriction analysis of five positive clones carrying a DNA insert of 22 kb indicated that three cosmids represented the same genomic region but were different from katA. Restriction enzyme mapping, Southern blotting, and deletion analysis of one of these three cosmids, pLRK2, were performed to localize the katA-homologous gene. A 3.3-kb EcoRI-PstI katA-hybridizing fragment from pLRK2 was subcloned into a pBluescript vector (pBSKC1) and fully sequenced by the dideoxy chain termination method in accordance with the U.S. Biochemicals protocol for the Sequenase 2.0 enzyme with α-35S-dATP (ICN, Orsay, France). Analysis of the sequence revealed one major open reading frame encoding 687 amino acid residues, corresponding to a protein with an Mr of 76,000 and a pI of 6.5. The ATG was preceded by a potential ribosome binding site (AAGGAG) located 7 bp upstream. Inverted repeat sequences are present downstream of the stop codon and might serve as a transcription terminator. Southern analysis of digested genomic DNA with a radiolabeled 2.2-kb EcoRI-ApaI fragment of this katA-homologous gene, kat2, confirmed that this probe hybridized with the 14-kb ApaI-ApaI fragment of the S. meliloti chromosome previously detected (data not shown). As expected, a search of the current nonredundant DNA and protein databases with the BLAST algorithm (Beckman Center for Molecular and Genetic Medicine, Stanford, Calif.) revealed that the deduced amino acid sequence of kat2 had regions of high homology with monofunctional catalases from mammals, plants, and bacteria but not with bifunctional catalases. A multiple alignment of KatA (U59271) and Kat2 amino acid sequences from S. meliloti was performed with Xanthomonas oryzae KatX (X97673), E. coli hyperoxidase II (M55161), and Rhizobium sp. strain SNU003 (U56239), using the Genetics Computer Group programs PILEUP and PRETTY (data not shown). Considering both identical and conservative replacement of amino acids, Kat2 showed a high degree of identity with X. oryzae KatX (identity of 56.9%) and E. coli HPII (identity of 48.9%). Surprisingly, the Kat2 sequence showed very low amino acid identity with the S. meliloti KatA (28.2%) and with the Rhizobium sp. strain SNU003 catalase (28%), since a large divergence in the C-terminal region was observed between Kat2 and these two catalases. However, the amino acid residues thought to be involved in the active-site and the proximal- and distal-heme-site ligands (23) were highly conserved in the five sequences.
Induction of a new catalase during stationary phase in minimum medium.
Strains and plasmids used in this study are listed in Table 1. Previously, we detected only KatA and KatB in protein extracts of stationary-phase cells grown at 30°C in rich medium (Luria broth [LB]-MC: yeast extract, 5 g/liter; tryptone, 10 g/liter; NaCl, 10 g/liter; 2.5 mM MgSO4; and 2.5 mM CaCl2), (18). Sequence analysis indicates that the kat2 product could not correspond to a bifunctional, HPI-like catalase/peroxidase. To test this, a recombinant Rm5000 strain carrying an interposon (Ω) on the kat2 gene (MK5002) was constructed. The 1.5-kb SacI-ApaI subclone (pBSKC2) containing the 5′ end of kat2 was cleaved at the neighboring SmaI sites within the coding region, creating a deletion. The kat2 was disrupted by ligation within an Ω-interposon cassette containing a Spr/Smr element flanked by transcriptional terminators translational stops in all three reading frames (8). The SacI-KpnI fragment containing this null allele was subcloned into pRK415. Recombinant plasmids were transferred to the rifampin-resistant SU47 derivative Rm5000 by triparental mating with E. coli MT616 as a helper in accordance with the protocol previously described (14). The recipient strains containing the recombinant plasmid were selected by growth on medium complemented with rifampin (20 μg/ml), spectinomycin (100 μg/ml), and tetracycline (10 μg/ml). The plasmid incompatibility technique (30) was then used to detect strains in which the insertion had recombined from the plasmid to the S. meliloti genome. Recombinants carrying Ω on the kat2 gene (MK5002) were selected for the gentamicin-resistant plasmid pGM2, on LB-MC medium containing gentamicin (70 μg/ml), spectinomycin, and rifampin. Southern analysis of the MK5002 genomic DNA confirmed the insertion of the Ω-spectinomycin cassette in the coding region of kat2. Analysis of the catalase pattern of the kat2 mutant revealed no difference between it and the wild-type profile, confirming that this gene does not encode KatB. These results suggested the presence of at least three catalase genes in S. meliloti. Three catalases have been detected in Rhizobium leguminosarum bv. phaseoli free-living bacteria grown in YEM medium (3). Analysis of the transcription of katE in E. coli by using lacZ fusion clearly showed that the katE promoter gave rise to low expression during growth in rich medium but elevated expression during growth in poor medium (27).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm5000 | S. meliloti SU47, rif-5 | 9 |
| MK5001 | Same as Rm5000 but with katA::Tn5 | 18 |
| MK5002 | Same as Rm5000 but with katC::ΩSmr/Spr | This work |
| MK5003 | Same as MK5002 but with katA::Tn5 | This work |
| RKCZ01 | Same as Rm5000 but with pkatC-lacZ fusion | This work |
| E. coli | ||
| DH5α | F−supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 (rK− mK+) recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| MT607 | pro-82 thi-1 hsdR17 supE44 recA56 | 10 |
| MT616 | MT607(pRK600) | 10 |
| Plasmids | ||
| pLAFR1 | IncP cosmid cloning vector, Tcr | 12 |
| pRK415 | IncP cosmid cloning vector, Tcr | 21 |
| pRK600 | ColE1 replicon with RK2 transfer region, Cmr | 10 |
| pHP45Ω | Ampr pBR322 derivative with ΩSmr/Spr | 8 |
| R751-pGM2 | IncP Gmr Smr | 19 |
| pBluescriptKS(+) | Derivative of pUC19 with f1(+), oriR Ampr | Stratagene |
| pSUP202 | ColE1, Mob+ Tcr Ampr Cmr | 31 |
| pKOK5 | Ampr Kmr, pSUP202 derivative; source of lacZ-Kmr cartridge | 22 |
| pLRK2 | pLAFR1, S. meliloti cosmid clone with katC | This work |
| pBSKC1 | pBluescript, 3.3-kb EcoRI-PstI fragment with partial katC | This work |
| pBSKC2 | pBluescript, 1.5-kb SacI-ApaI fragment with partial katC | This work |
| pBSKC3 | pBluescript, 2,190-bp EcoRI-ApaI fragment with partial katC | This work |
| pSUPKC | pSUP202, PstI-PstI fragment of pBSKC3 | This work |
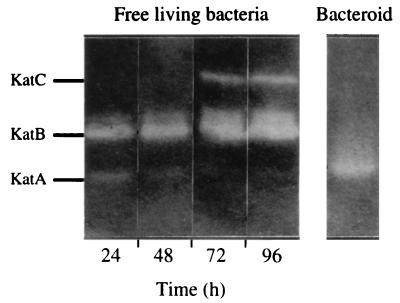
To analyze the effect of growth medium on the catalase profile, protein extracts from bacteria grown in M9 medium (42.5 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, 18.7 mM NH4Cl, 2.5 mM MgSO4, 2.5 mM CaCl2, and 4 g of glucose per liter) were analyzed for catalase activity on 7% native polyacrylamide gels (Fig. 1). Electrophoresis was performed at 100 V for 3 h in a Miniprotean II cell (Bio-Rad), and catalase activity was visualized via the inhibition of diaminobenzidine oxidation by H2O2 as described before (18). A new upper band, designated KatC, was detected in bacterial extracts at the end of the stationary phase. No corresponding peroxidase activity was detected, indicating that KatC is an additional monofunctional catalase enzyme (data not shown). Moreover, a perfectly inverted activity profile for KatA and KatC was observed during bacterial growth. The highest levels of KatA activity were found during the exponential phase, with a gradual decrease during stationary phase, and the KatC activity increased as KatA activity decreased. To confirm that KatC was the kat2 product, the catalase activity of the MK5002 strain was analyzed on native gel after 96 h of growth on M9 medium. The KatC activity band was not detected in this strain (data not shown); therefore, kat2 was renamed katC.
FIG. 1.
Expression of KatA, KatB, and KatC during growth of free-living S. meliloti on M9 minimal medium and catalase profile in 5-week-old bacteroids. Catalase activity was detected in samples (25 μg of proteins) after electrophoresis on native polyacrylamide gels.
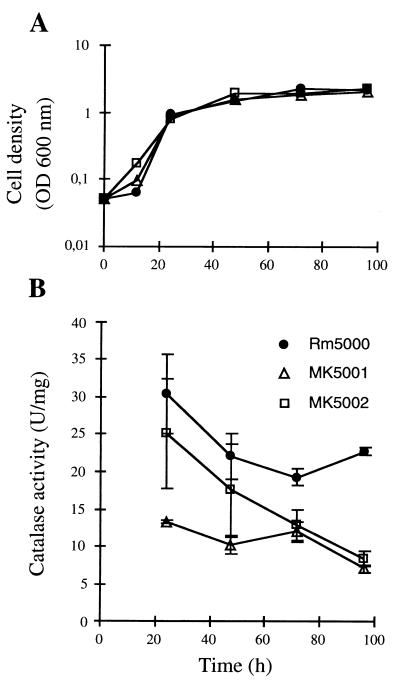
The total catalase activities in wild-type strain Rm5000 and in strains lacking either KatA (MK5001) or KatC (MK5002) were measured during growth in M9 medium (Fig. 2B) by using the protocol described previously (18). The katA and katC mutations had no effect on growth rate compared to the wild type (Fig. 2A). Levels of catalase activity in Rm5000 bacteria were high during exponential phase, decreased during late exponential phase, and slowly increased again during stationary phase. MK5001 showed lower total catalase activity at all stages of growth than the wild-type Rm5000 strain. The most significant decrease was observed at the end of the exponential phase (24 h of growth), confirming the contribution of KatA to total catalase activity during this stage. KatA is the unique catalase which is inducible by H2O2 in S. meliloti. In E. coli, the increase in catalase activity during the exponential growth, which is mainly due to the H2O2-inducible HPI, was correlated with an increase in H2O2 production (15). Assuming that the regulation of KatA is performed in the same way, this would imply that S. meliloti has to deal with the same burst of H2O2 production during late exponential growth. However, the induction of KatA seems to be weak in comparison with HPI induction. This difference could be explained by the constant presence of KatB, which could maintain H2O2 concentration at low levels without the need for a strong induction of KatA. Measurement of MK5002 catalase activity showed that the katC mutation had an effect at all stages of growth, indicating that katC might be expressed even during exponential growth, despite the nondetection of KatC on native gels at that stage. However, the impact of the mutation remained limited at 24 and 48 h of growth. In contrast, from 72 h of growth, catalase activity decreased strongly, showing a dramatic effect of the katC mutation. These results are consistent with the hypothesis of a weak KatC contribution to total catalase activity during exponential and early stationary phases and are consistent with the observation that the increase in total catalase activity during late stationary phase was mainly due to a rise in KatC activity. Very similar kinetics of catalase induction have been observed in cultures of other organisms such as E. coli (17), Salmonella typhimurium (11), Haemophilus parainfluenzae (35), and Rhodobacter sphaeroides (2), indicating a conserved strategy for surviving in starvation conditions. High similarity was observed between KatC and HPII regulations in that both catalases are upregulated by stationary phase but not by H2O2 (25). However, the HPII enzyme represents the dominant form during late-stationary-phase growth in E. coli (15), whereas the presence of KatB in S. meliloti considerably reduces the impact of KatC induction on total catalase activity. Unexpectedly, a very different pattern is found in the S. meliloti-related bacterium R. leguminosarum bv. phaseoli (3). In this organism, the catalase activity rises to a maximum during early-exponential-phase growth and falls to a minimum during late-exponential-phase growth. This trend was confirmed on catalase activity gels: the bands corresponding to the three catalase isoenzymes are observed in all stages during growth, and changes in total catalase activity are seen in all three bands (3). This apparent coregulation of the three catalases of R. leguminosarum bv. phaseoli differs from the differential regulation that we observed in S. meliloti free-living bacteria.
FIG. 2.
Effect of a katA or katC mutation on total catalase activity during growth on M9 medium. Stationary cultures of RM5000, MK5001, and MK5002 were inoculated into fresh M9 medium to an initial optical density at 600 nm (OD600) of 0.05. Samples were taken from cultures at the indicated times. Bacterial growth was monitored as OD600 (A), and total catalase activity was measured (B). Values represent averages of at least two experiments; error bars show standard deviations.
Regulation of the katC expression.
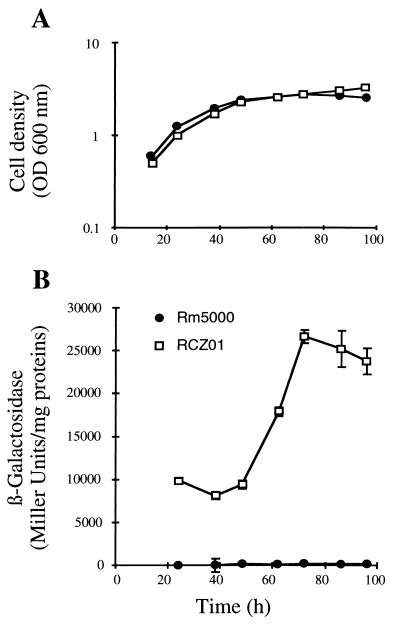
To determine if KatC induction during stationary phase is controlled at the transcriptional level, a katC::lacZ transcriptional fusion was constructed. Plasmid pBSKC3 carries a 2,190-bp EcoRI-ApaI fragment with a segment of katC purified from pBSKC1 and subcloned into pBluescript KS(+). pSupKC carries a 2.2-kb PstI-PstI fragment purified from pBSKC3 and subcloned at the PstI site of the vector pSUP202. To construct a transcriptional lacZ fusion to the katC promoter, a BamHI-BamHI lacZ-Kmr cartridge purified from plasmid pKOK5 was inserted in BglII sites of pSupKC. After transfer by conjugation to S. meliloti Rm5000, a simple recombinant clone, designated RKCZ01, was isolated on LB-MC medium containing rifampin and neomycin (200 μg/ml). Recombination at the correct location was checked by hybridization with the specific DNA fragment from pBSKC1. The bacterial growth in M9 medium and the β-galactosidase activity of the katC::lacZ recombinant strain RKCZ01 and wild-type strain RM5000 as a control are shown in Fig. 3. β-Galactosidase activity was determined by measuring the hydrolysis of o-nitrophenyl-β-d-galactoside as described by Miller (26), and protein concentration was determined as described previously (18). The growth curves of the two strains were very similar (Fig. 3A). However, a threefold increase of the β-galactosidase activity was observed during the stationary phase for strain RKCZ01 (Fig. 3B), which could be correlated with the induction of KatC (Fig. 1). These data suggest that growth-dependent KatC regulation is mainly or solely transcriptional.
FIG. 3.
Growth stage induction of katC::lacZ fusion. Stationary cultures of Rm5000 and RCZ01 (katC::lacZ transcriptional fusion) were inoculated in fresh M9 medium to an initial optical density at 600 nm (OD600) of 0.05. Bacterial growth was monitored as OD600 (A). At the indicated times, samples were taken to assay β-galactosidase activity (B). Values represent averages of at least two experiments; error bars show standard deviations.
To characterize the regulation of katC expression, we determined the β-galactosidase activity in RKCZ01 bacteria after the organisms had been subjected to different types of stress (Table 2). The β-galactosidase assays revealed that an induction (threefold increase) of katC was observed after bacteria had been exposed to heat stress (37°C), salt stress (NaCl), or ethanol for 1 h. In addition, no induction was observed when bacteria were exposed to exogenous H2O2, whereas a threefold increase of β-galactosidase activity was detected after treatment with the superoxide generator paraquat. Thus, the katC expression profile in S. meliloti is very similar to many catalase genes encoding HPII-homologous catalases, such as the Bacillus subtilis katE (6) and the Aspergillus nidulans katB (20) genes.
TABLE 2.
Effect of different stresses on the regulation of katC expression determined by measuring β-galactosidase activity
| Stressora | β-Galactosidase activityb |
|---|---|
| None | 10,309 ± 1,087 |
| Heat (37°C) | 41,253 ± 1,233 |
| Salt (4% NaCl) | 33,485 ± 941 |
| Ethanol (4%) | 27,189 ± 1,562 |
| H2O2 | |
| 1 mM | 10,112 ± 620 |
| 100 μM | 9,116 ± 943 |
| 10 μM | 8,960 ± 1,021 |
| Paraquat (10−3 M) | 29,777 ± 779 |
S. meliloti RKCZ01 bacteria were grown for 36 h in M9 medium and subjected to indicated stressor for 1 h.
Values were obtained by using duplicate samples from two independent experiments and are given as Miller units per milligram of protein ± the standard errors of the means.
Catalase expression pattern in bacteroids.
To further investigate the role of catalases in the development of functional nodules, M. sativa plants were inoculated with the wild-type Rm5000 strain, and bacteroids were isolated from 5-week-old nodules. Around 1.5 g of freshly harvested nodules was crushed in a mortar at 4°C in 3 ml of homogenization medium containing 50 mM phosphate buffer (pH 7.4) and insoluble polyvinylpyrrolidone. The homogenate was filtered through a 200-μm nylon mesh. A first centrifugation (1,500 × g, 5 min, 4°C) eliminated cell debris and polyvinylpyrrolidone residues. The bacteroids were sedimented during a second centrifugation (8,000 × g, 8 min, 4°C). The pellet was washed twice in 50 mM phosphate buffer (pH 7.4) containing 2 mM magnesium sulfate and 0.3 M sucrose. The pellet was resuspended in 1 ml of extraction buffer containing 50 mM phosphate buffer (pH 7) and 1 mM EDTA. The bacteroids were then sonicated, and membrane debris was eliminated by centrifugation (8,000 × g, 10 min, 4°C). Analysis of bacteroid protein extracts from five independent experiments showed that a higher catalase activity (sevenfold) was detected in bacteroids (214.9 ± 38.6 U/mg of protein) than in free-living bacteria (30.3 ± 5.3 U/mg of protein), suggesting that a large amount of H2O2 was directly or indirectly generated in the microsymbiont.
We showed previously that the katA mutation had no effect on nodulation efficiency and nitrogen fixation, suggesting that KatA has a minor protective role in the nitrogen fixation process (18). Surprisingly, analysis on a native gel of the catalase profile of a wild-type bacteroid isolated from 5-week-old nodules showed that KatA was induced, in contrast to results for free-living bacteria, whereas neither KatC nor KatB was detectable (Fig. 1). Moreover, when plants were inoculated with MK5001, analysis of bacteroids revealed no increase in total catalase activity and no induction of KatB and KatC (data not shown). So the null katA bacteroid can still cope with a potential H2O2 problem without detectable induction of KatB and KatC.
Reduction of nitrogen fixation capacity in a katA katC double mutant.
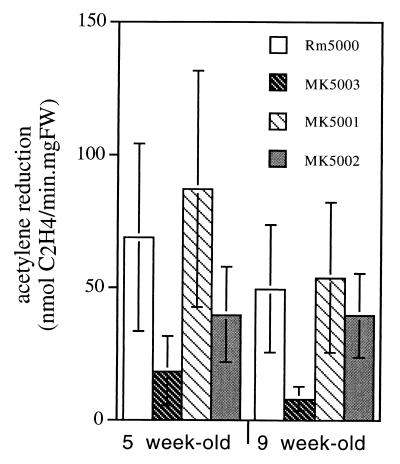
To test the effect of a katA and/or katC mutation on nodulation and nitrogen fixation, a katA katC double mutant (MK5003) was constructed by transduction. The katA::Tn5 mutation from strain MK5001 was transferred to MK5002 by general transduction by using the φM12 phage, in accordance with a standard protocol (14). Transductants (MK5003) were selected on LB-MC medium containing kanamycin (50 μg/ml) and spectinomycin (100 μg/ml). M. sativa host plants were inoculated with MK5001, MK5002, and MK5003 mutant strains and with the wild-type Rm5000 strain as a control. Seventy-two plants were grown in sterile tubes (three plantlets per tube) containing 20 ml of a nitrogen-free nutrient medium with 0.8% agarose prepared as a slant. Plants were inoculated with the appropriate S. meliloti strains 1 week after germination. Nitrogen fixation activity was determined by C2H2 reduction by using a gas chromatograph (ATI-Unicam, model 610) equipped with a column of Porapak T (80/100 mesh) as described previously (18). Incubations of these nodulated plantlets were made at 25°C, in rubber-cap tubes containing O2 (20 kPa) and C2H2 (10 kPa) in argon. The fresh weight of nodules per tube was measured, and the phenotypes of the bacteria recovered from the nodules were checked on the appropriate media. Plants were visually screened for nodule formation by observing the root system 5 weeks after Sinorhizobium inoculation. A high efficiency of nodulation (87 to 97%) was observed for the plants inoculated with the single mutants MK5001 and MK5002 and with Rm5000. In contrast, only 75% of the plants inoculated with the double-mutant katA katC MK5003 strain showed a nodulating phenotype. Acetylene reduction activity was assayed for M. sativa nodulated with the different strains at 5 and 9 weeks after bacterial infection (Fig. 4). The level of C2H2 reduction observed in MK5001-nodulated plants was not significantly different from that of Rm5000-nodulated plants, confirming our previous results (18). There was also no significant reduction of nitrogen fixation in MK5002-nodulated plants compared to Rm5000-nodulated plants, which is consistent with the nondetection of katC in bacteroids. However, a drastic decrease in C2H2 reduction, especially in 9-week-old nodules, was observed for MK5003-nodulated plants.
FIG. 4.
Effect of catalase mutations on nitrogen fixation. C2H2 reduction activity was measured in a tube containing three plants at 5 weeks and 9 weeks after inoculation with the wild-type Rm5000 strain or the MK5001, MK5002, or MK5003 mutant strain. Values are means ± standard errors (n = 24). Experimental data were assessed for statistical significance by means of Student’s t test. FW, fresh weight.
Thus, suppression of both HPII-like monofunctional catalases has a severe effect on the maintenance of functional nodules despite the presence of KatB. These results indicate that the presence of at least one of these catalases is absolutely necessary for the protection of the nitrogen fixation process. We are in the process of cloning the katB gene to allow us to define the role of KatB in free-living bacteria and bacteroids. The construction of lacZ fusions with the promoter from each catalase gene will help us to determine the expression patterns of these genes in planta during the different steps of the symbiosis.
Nucleotide sequence accession number.
The 3.3-kb EcoRI-PstI katA-hybridizing fragment from pLRK2 has been assigned GenBank accession no. AF121348.
Acknowledgments
We thank Magne Osteras and Karine Mandon for helpful discussions. We also thank the colleagues whose works are cited in Table 1 for generously providing the strains used in this study.
This work was supported by the Centre National de la Recherche Scientifique and by the Human Capital and Mobility program (contract CT94-0605). S. Sigaud acknowledges the generous support of the Fondation Dufrenoy (Académie d’Agriculture de France).
REFERENCES
- 1.Appleby C A. Leghemoglobin and Rhizobium respiration. Annu Rev Plant Physiol. 1984;35:443–478. [Google Scholar]
- 2.Clayton R K. The induced synthesis of catalase in Rhodopseudomonas sphaeroides. Biochim Biophys Acta. 1960;37:503–512. doi: 10.1016/0006-3002(60)90507-2. [DOI] [PubMed] [Google Scholar]
- 3.Crockford A J, Davis G A, Williams H D. Evidence for cell-density-dependent regulation of catalase activity in Rhizobium leguminosarum bv. phaseoli. Microbiology. 1995;141:843–851. [Google Scholar]
- 4.Dalton D A. Antioxidant defenses of plant and fungi. In: Ahmad S, editor. Oxidative stress and antioxidant defenses in biology. New York, N.Y: Chapman and Hall; 1995. pp. 298–342. [Google Scholar]
- 5.Dalton D A, Joyner S L, Becana M, Iturbe-Ormaetxe I, Chatfield J M. Antioxidant defenses in the peripheral cell layers of legume root nodules. Plant Physiol (Rockville) 1998;116:37–43. doi: 10.1104/pp.116.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a ςB-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Finan T M, Hartwieg E, LeMieux K, Bergman K, Walker G C, Signer E R. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn G J, Condon S. Regulation of catalase synthesis in Salmonella typhimurium. J Bacteriol. 1975;123:570–579. doi: 10.1128/jb.123.2.570-579.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 13.Geurts R, Franssen H. Signal transduction in Rhizobium-induced nodule formation. Plant Physiol (Rockville) 1996;112:447–453. doi: 10.1104/pp.112.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glazebrook J. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- 15.González-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halliwell B, Gutteridge J M C. Oxygen free-radicals and iron in relation to biology and medicine. Some problems and concepts. Arch Biochem Biophys. 1986;246:501–508. doi: 10.1016/0003-9861(86)90305-x. [DOI] [PubMed] [Google Scholar]
- 17.Hassan H M, Fridovich I. Regulation of the synthesis of catalase and peroxidase in Escherichia coli. J Biol Chem. 1978;253:6445–6450. [PubMed] [Google Scholar]
- 18.Hérouart D, Sigaud S, Moreau S, Frendo P, Touati D, Puppo A. Cloning and characterization of the katA gene of Rhizobium meliloti encoding a hydrogen peroxide-inducible catalase. J Bacteriol. 1996;178:6802–6809. doi: 10.1128/jb.178.23.6802-6809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacoby G A, Jacob A E, Hedges R W. Recombination between plasmids of incompatibility groups P-1 and P-2. J Bacteriol. 1976;127:1278–1285. doi: 10.1128/jb.127.3.1278-1285.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki L, Wysong D, Diamond R, Aguirre J. Two divergent catalase genes are differentially regulated during Aspergillus nidulans development and oxidative stress. J Bacteriol. 1997;179:3284–3292. doi: 10.1128/jb.179.10.3284-3292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 22.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 23.Loewen P C. Probing the structure of catalase HPII of Escherichia coli—a review. Gene. 1996;179:39–44. doi: 10.1016/s0378-1119(96)00321-6. [DOI] [PubMed] [Google Scholar]
- 24.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 25.Loewen P C, Switala J, Triggs-Raine B L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 27.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puppo A, Rigaud J. Superoxide dismutase: an essential role in the protection of the nitrogen fixation process? FEBS Lett. 1986;201:187–189. [Google Scholar]
- 29.Rolfe B G, Gresshoff P M. Genetic analysis of legume nodule initiation. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:297–319. [Google Scholar]
- 30.Ruvkun G B, Ausubel F M. A general method for site-directed mutagenesis in prokaryotes. Nature (London) 1981;189:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- 31.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 32.Triggs-Raine B L, Doble B W, Mulvey M R, Sorby P A, Loewen P C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J Bacteriol. 1988;170:4415–4419. doi: 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasse J, de Billy F, Truchet G. Abortion of infection during a Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 1993;4:555–566. [Google Scholar]
- 34.von Ossowski I, Mulvey M R, Leco P A, Borys A, Loewen P C. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991;173:514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White D C. Cytochrome and catalase patterns during growth of Haemophilus parainfluenzae. J Bacteriol. 1962;83:851–859. doi: 10.1128/jb.83.4.851-859.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witty J F, Skot L, Revsbech N P. Direct evidence for changes in the resistance of legume root nodules to O2 diffusion. J Exp Bot. 1987;38:1129–1140. [Google Scholar]