Abstract
The photosynthetic organelles (cyanelles) of the protist Cyanophora paradoxa are surrounded by a peptidoglycan wall, modified through amidation with N-acetylputrescine. Cyanelle envelope membrane preparations were shown to catalyze the lipid-linked steps of peptidoglycan biosynthesis as well as the putrescinylation and subsequent acetylation, occurring at the stage of lipid I and/or lipid II.
Among eukaryotes, peptidoglycan has been found in cyanelle-containing glaucocystophyte algae only (1, 11, 12, 22). This prokaryotic wall constitutes part of the envelope of cyanelles (which therefore are also called muroplasts) and is one of the cyanobacterial features that render cyanelles a living example for an origin of photosynthetic organelles from endosymbiotic cyanobacteria. The best-investigated member of this group of obligatorily photoautotrophic protists is Cyanophora paradoxa (for a review, see reference 15). Cyanelle peptidoglycan, which is of the A1γ type (1), is partially amidated with N-acetylputrescine at the 1-carboxyl group of d-glutamic acid (18, 20). The same modification has been found in the two other glaucocystophytes examined in this respect but not in the cyanobacterium Synechocystis sp. strain PCC 6714 (19).
Modifications of peptidoglycan at the 1-carboxylic group of d-glutamic acid with cadaverine and putrescine have been found in some anaerobic gram-negative bacteria (8, 9, 24) and might serve there as a connection to the negatively charged outer membrane by introducing a positive charge (7), while in some gram-positive species, alanine, glycine, or glycinamide occupies this position (23). Amidation with ammonia is frequent among gram-negative and gram-positive bacteria (23). The reason for these modifications is unknown. Amidation with glycine in Micrococcus luteus, with ammonia in Staphylococcus aureus, and with cadaverine in Selenomonas ruminantium has been shown to occur at the membrane stage of peptidoglycan biosynthesis, i.e., the amidating group is incorporated into the undecaprenol-linked mono- or disaccharide pentapeptide (6, 10, 26).
The biosynthesis of the UDP-N-acetylmuramyl pentapeptide precursor of C. paradoxa has already been localized to the cyanelle stroma (21). Because of the apparent uniqueness of peptidoglycan modification with N-acetylputrescine to glaucocystophyte algae, we decided to investigate the subsequent steps of cyanelle wall biosynthesis of C. paradoxa in detail. An incorporation of N-acetylputrescine at the lipid precursor level before cross-linking of peptide side chains was anticipated from the muropeptide pattern of the C. paradoxa cyanelles (18), which shows a concentration of the substituent on tetrapeptide side chains (pentapeptide side chains are below the detection limit). In this case, the necessary enzymes would be associated with the cyanelle inner envelope membrane.
Cyanelles were obtained by shock freezing a pellet of whole cells from 3 liters of exponentially growing culture (3), thereby disrupting the cell wall of C. paradoxa, followed by a washing step in ice-cold 0.3 M sucrose in 50 mM Tris-HCl, pH 7.5 (1,500 rpm for 5 min in a Falcon centrifuge). The washed cyanelles were resuspended in 7 ml of the same sucrose solution and sonicated for 20 min in a 20-ml beaker (cycle, 30%; amplitude, 20%; Bandelin Sonopuls HD 70/UW 70). A total of 1.8 volumes of 60% (wt/wt) sucrose in 50 mM Tris-HCl, pH 7.5, was added, resulting in a concentration of about 45% (wt/wt). The lysate was transferred into four SW40 Ti (Beckman) rotor tubes; overlaid with 3 ml of 35%, 2 ml of 30%, and 2 ml of 10% (wt/wt) sucrose solution; and centrifuged for 16 h at 26,000 rpm. The yellow membrane fraction at the boundary between 35 and 30% sucrose was collected, diluted threefold with 50 mM Tris-HCl (pH 7.5), and centrifuged for 1 h at 35,000 rpm in the same rotor. The pellet was resuspended in 0.3 M sucrose in the same buffer to a protein concentration of about 15 mg/ml. Ether extracts of cyanelle envelope preparations had a ratio of carotenoid absorption (485 nm) to chlorophyll a absorption (680 nm) of approximately 5, pointing to a slight contamination of the inner envelope membranes with thylakoid membranes (16, 17).
These envelope preparations were used as a source of enzymes catalyzing the transfer of phospho-N-acetylmuramyl pentapeptide from UDP-N-acetylmuramyl pentapeptide to undecaprenol phosphate (thereby generating lipid I), the addition of N-acetylglucosamine to lipid I from UDP-N-acetylglucosamine (generating lipid II), and the modification of lipid I or II with N-acetylputrescine. In a first step, the specificity of the modifying enzyme was determined by adding either [1,4-14C]putrescine or N-acetyl[1,4-14C]putrescine to the reaction mixture. N-Acetyl[1,4-14C]putrescine was prepared by acetylation (29) of [1,4-14C]putrescine dihydrochloride (117 mCi/mmol; DuPont New England Nuclear, Boston, Mass.) and subsequent preparative separation by descending paper chromatography on Whatman 3MM paper in ethanol-water (4:1, by volume). In contrast to reaction mixtures containing [1,4-14C]putrescine, mixtures containing only N-acetyl[1,4-14C]putrescine did not incorporate measurable amounts of radioactivity into the lipid fraction. Further experiments were therefore conducted with [1,4-14C]putrescine as a substrate for the modification reaction.
UDP-N-acetylmuramyl pentapeptide was prepared (14) and quantified through determination of its diaminopimelic acid content as described elsewhere (30). The reaction mixture for the in vitro synthesis of modified peptidoglycan precursors contained the following in a total volume of 15 μl: 50 μg of protein (cyanelle envelope membrane preparation [4]) in 8 μl of 0.3 M sucrose in Tris-HCl (pH 7.5), 15 nmol of UDP-N-acetylmuramyl pentapeptide, 1.5 nmol of UDP-N-acetylglucosamine (Sigma), 0.65 nmol of undecaprenol phosphate diammonium salt (Larodan Fine Chemicals, Malmö, Sweden) in 1.5 μl of 0.1% Triton X-100, 6 mM ATP neutralized with sodium bicarbonate, 40 mM MgCl2, 50 mM KCl, 0.02 mM cysteine hydrochloride, 100 μg of ampicillin per ml for inhibition of potential dd-carboxypeptidase action, 100 mM Tris-HCl (pH 8), and 25 nCi of [1,4-14C]putrescine dihydrochloride. The reaction mixture was incubated for 2 h at room temperature, and the reaction was terminated by addition of 0.2 ml of water.
Lipid-linked peptidoglycan precursors were extracted with 0.4 ml of 1-butanol–6 M pyridinium acetate, pH 4 (13), and freed of unreacted putrescine by backwashing of the butanol phase with water. Radioactivity incorporated into the butanol-extractable lipid fraction was determined by scintillation counting with Ecolume (ICN, Irvine, Calif.) as cocktail. An amount of butanol phase corresponding to approximately 600 cpm was lyophilized. Fifty microliters of 0.1 N HCl was added to the remaining material, and the suspension was incubated for 15 min at 100°C to split off the lipid moiety. Insoluble material was eliminated by centrifugation. After lyophilization, the sample was reduced with sodium borohydride as described by Glauner (5), reduced N-acetylmuramyl pentapeptide (prepared in an analogous way by acid cleavage of UDP-N-acetylmuramyl pentapeptide) and reduced C. paradoxa muropeptides (prepared as described in reference 18) were added as standards, and the sample was submitted to high-pressure liquid chromatography (HPLC) in 50 mM potassium phosphate buffer (pH 5.5) with a gradient of 0 to 20% methanol (18). Fractions of 1 ml were collected, lyophilized, resuspended in a small amount of water, and spotted onto Whatman 3MM chromatography paper. After drying, the paper was covered with Saran wrap and exposed with a PhosphorImager screen (Molecular Dynamics) for a couple of days.
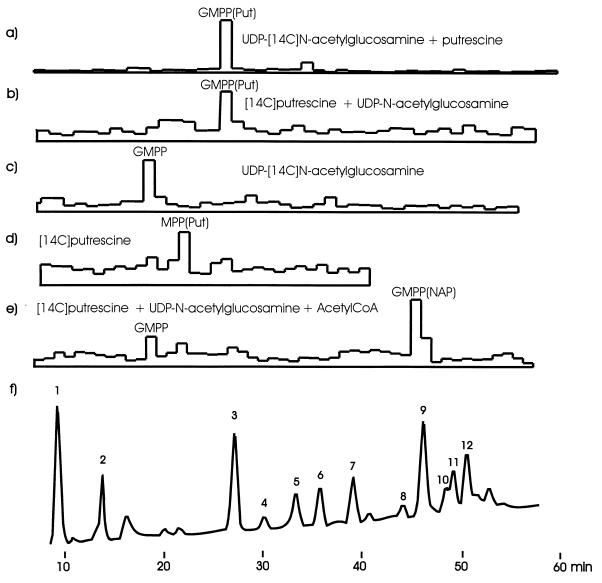
Alternatively, the labelling was done with 25 nCi of UDP-N-acetyl-[U-14C]glucosamine (265 mCi/mmol; DuPont New England Nuclear) in the presence of 1 mM unlabelled putrescine in the reaction mixture. Both labelled substrates yielded a compound with a retention time of 26 min after hydrolysis and reduction (Fig. 1a and b). Omission of putrescine from the reaction mixture resulted in the synthesis of a compound with a retention time of 18 min after hydrolysis and reduction (Fig. 1c), which fits well with the retention time expected for reduced disaccharide pentapeptide considering the retention time of the reduced N-acetylmuramyl pentapeptide standard and published data (13).
FIG. 1.
HPLC of reduced muropeptides prepared from butanol extracts of reaction mixtures for precursor synthesis. The dimension of the y axis is the relative signal intensity produced on the PhosphorImager screen for panels a to e and absorption at 210 nm for panel f. (a) Labelling with UDP-N-acetyl-[14C]glucosamine, with putrescine added to the reaction mixture; (b) labelling with [14C]putrescine, with N-acetylglucosamine added to the reaction mixture; (c) labelling with UDP-N-acetyl-[14C]glucosamine, with reaction mixture without putrescine; (d) labelling with [14C]putrescine, with reaction mixture without N-acetylglucosamine; (e) labelling with [14C]putrescine, with N-acetylglucosamine and acetyl coenzyme A (CoA) added to the reaction mixture; (f) HPLC of reduced cyanelle muropeptides: 1, disaccharide tripeptide; 2, reduced N-acetylmuramyl pentapeptide (added); 3, bis-disaccharide tetra-tripeptide; 4, bis-disaccharide tetra-tetrapeptide; 5, disaccharide tripeptide modified with N-acetylputrescine (NAP); 6, tris-disaccharide tetra-tetra-tripeptide; 7, disaccharide tetrapeptide modified with N-acetylputrescine; 8 and 9, bis-disaccharide tetra-tripeptide modified with N-acetylputrescine; 10, bis-disaccharide tetra-tetrapeptide modified with N-acetylputrescine; 11, bis-disaccharide tetra-tetrapeptide modified with N-acetylputrescine and tris-disaccharide tetra-tetra-tripeptide modified with N-acetylputrescine; 12, tris-disaccharide tetra-tetra-tripeptide modified with N-acetylputrescine. MPP(Put), putative putrescinylated reduced N-acetylmuramyl pentapeptide; GMPP(Put), putative putrescinylated reduced disaccharide pentapeptide; GMPP(NAP), putative reduced disaccharide pentapeptide modified with N-acetylputrescine; GMPP, putative reduced disaccharide pentapeptide.
Putrescine could be added to lipid I as well as to lipid II. Addition to lipid I was demonstrated by incubation with labelled putrescine in the absence of UDP-N-acetylglucosamine (Fig. 1d). The main product of the reaction, after hydrolysis and reduction, had a retention time between 22 and 23 min, which is in accordance with what would be expected for reduced N-acetylmuramyl pentapeptide modified with putrescine. Addition of putrescine to lipid II was shown by incubating the reaction mixture for 30 min without putrescine for the synthesis of unmodified lipid II. Membranes were then recollected by centrifugation and resuspended in 0.3 M sucrose in 50 mM Tris-HCl, pH 7.5, containing 10 mM UMP in order to eliminate unreacted lipid I (2). After 20 min, the membranes were again spun down and resuspended in reaction mixture containing labelled putrescine but neither UDP-N-acetylmuramyl pentapeptide nor UDP-N-acetylglucosamine. The usual amount of radioactivity was incorporated into the lipid fraction.
Since N-acetylputrescine was not a substrate, an acetylation of putrescinylated peptide side chains in a subsequent step was anticipated. Accordingly, lipid II modified with N-acetylputrescine was formed upon addition of 3 mM acetyl coenzyme A to the reaction mixture. The product obtained after hydrolysis and reduction showed the retention time in HPLC expected for reduced disaccharide pentapeptide modified with N-acetylputrescine (Fig. 1e).
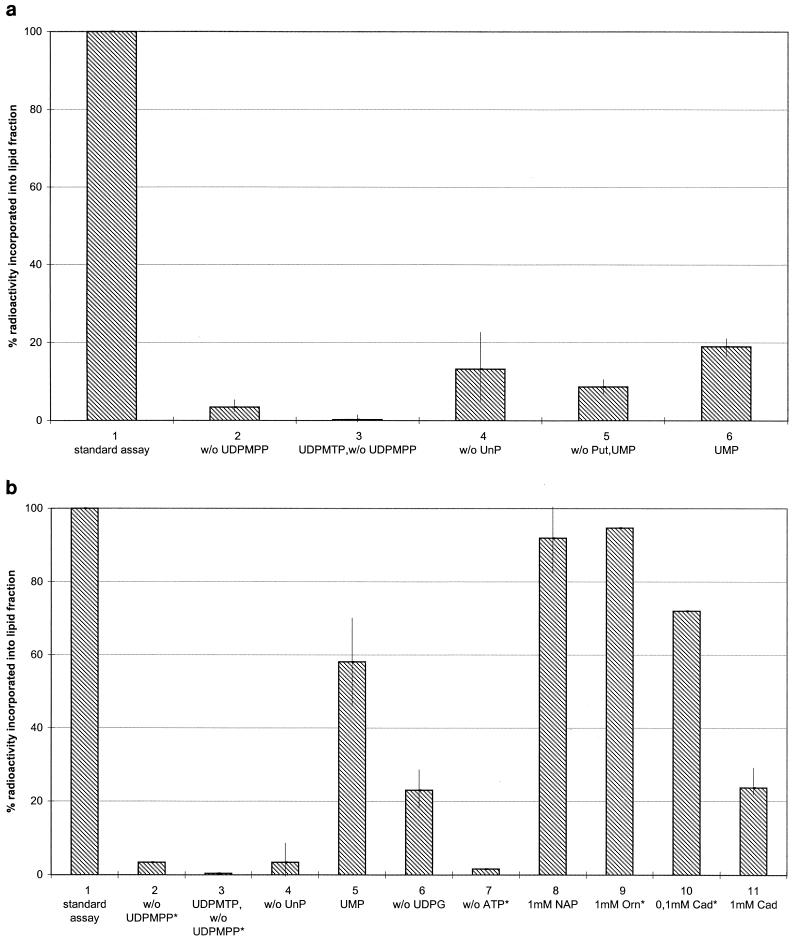
Figure 2 shows the dependence of the reaction sequence on substrates and cofactors. The synthesis of lipid-linked precursors was absolutely dependent on the presence of UDP-N-acetylmuramyl pentapeptide (Fig. 2, lane 2). In contrast to observations made with Escherichia coli (28), UDP-N-acetylmuramyl tripeptide (concentration and preparation as described for UDP-N-acetylmuramyl pentapeptide) was not accepted as substrate for the synthesis of lipid I (Fig. 2, lane 3). Omission of external undecaprenol phosphate considerably reduced the amount of product formed (Fig. 2, lane 4). The modification of lipid-linked precursors with putrescine was strictly dependent on the presence of ATP (Fig. 2b, lane 7).
FIG. 2.
Incorporation of radiolabelled substrates into cyanelle lipid-linked peptidoglycan precursors by cyanelle envelope membranes. (a) Incorporation of UDP-N-acetyl-[U-14C]glucosamine; (b) incorporation of [1,4-14C]putrescine. Standard deviations are derived from two or more experiments with the exception of experimental results marked with asterisks, which were not repeated. The amount of radioactivity incorporated into lipid precursors in the standard assay (100%) was about 600 and 1,500 cpm for [1,4-14C]putrescine and UDP-N-acetyl-[U-14C]glucosamine, respectively. UDPMPP, UDP-N-acetylmuramyl pentapeptide; UDPMTP, UDP-N-acetylmuramyl tripeptide; UnP, undecaprenol phosphate; UDPG, UDP-N-acetylglucosamine; Cad, cadaverine; Orn, ornithine; NAP, N-acetylputrescine; w/o, without.
Once modified with putrescine, lipid I shows no significant back reaction induced by UMP, in contrast to what has been observed for unmodified lipid I or lipid I amidated with ammonia in in vitro assays with S. aureus and M. luteus membranes (2, 27). This was demonstrated by incubation for 30 min at room temperature in the usual reaction mixture with labelled putrescine but without UDP-N-acetylglucosamine. After a washing step in 0.3 M sucrose in 50 mM Tris-HCl (pH 7.5), the membranes were divided into three aliquots. As a control for the continued activity of the membrane-bound enzymes, one of the aliquots was resuspended in fresh reaction mixture also containing UDP-N-acetylglucosamine and [1,4-14C]putrescine. The second aliquot was incubated with 10 mM UMP in fresh reaction mixture devoid of UDP-N-acetylmuramyl pentapeptide, UDP-N-acetylglucosamine, ATP, and [1,4-14C]putrescine. The third aliquot was treated like the second one but without addition of UMP. Incubation was continued for 2 h. While the aliquot with fresh reaction mixture incorporated 420 cpm (background corrected) into the lipid fraction, the second aliquot contained 144 cpm and the third aliquot contained 162 cpm extractable with butanol, showing that no significant UMP-specific back reaction had taken place. This explains the only weak inhibition (∼40% [Fig. 2b, lane 5]) of putrescine incorporation into the lipid fraction by 10 mM UMP, a concentration which in the absence of putrescine inhibits the synthesis of unmodified lipid II by 90% (Fig. 2a, lane 5).
The specificity of the putrescine adding enzyme was tested by adding an excess of unlabelled putative competitors of putrescine to the reaction mixture. N-Acetylputrescine (1 mM) or ornithine (1 mM), which upon decarboxylation would yield putrescine, did not noticeably inhibit the putrescinylation of lipid precursors (Fig. 2b, lanes 8 and 9). Cadaverine (diaminopentane) had a slight inhibitory effect at a 0.1 mM concentration and a pronounced effect at a 1 mM concentration, which was a 70-fold excess over the amount of putrescine in the reaction mixture (Fig. 2b, lanes 10 and 11). Whether cadaverine is actually used for the amidation of d-glutamic acid has not been investigated. In S. ruminantium, cadaverine is preferentially used for the amidation, while in Veillonella spp. both putrescine and cadaverine are equally incorporated into peptidoglycan (8, 9).
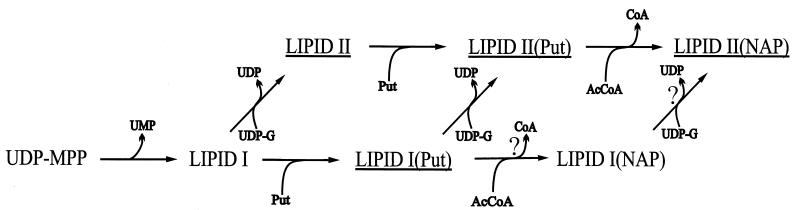
In conclusion, our cyanelle envelope membrane preparation was able to catalyze the whole reaction sequence from UDP-N-acetylmuramyl pentapeptide to lipid II modified with N-acetylputrescine, occurring in four steps (Fig. 3). As in S. aureus, S. ruminantium, and M. luteus (6, 10, 26), the amidation of d-glutamic acid is strictly dependent on ATP as an energy source. Lipid I as well as lipid II can serve as a substrate for putrescinylation. The reaction seems to be more efficient with lipid II as judged from the lower incorporation of putrescine into the lipid fraction in the absence of UDP-N-acetylglucosamine (Fig. 2b, lane 6). The likely pathway to lipid II(NAP) is that via lipid II(Put). However, the end product might also be formed via lipid I(Put) and lipid I(NAP) (Fig. 3).
FIG. 3.
Proposed pathway of cyanelle peptidoglycan biosynthesis. Steps not directly proven by experimental data are indicated by question marks. For abbreviations, see the legend to Fig. 1.
The overall similarity of the mechanism of incorporation of N-acetylputrescine into cyanelle peptidoglycan with that of ammonia, glycine, or diamine incorporation into bacterial peptidoglycan (6, 10, 26) suggests that the potential for putrescinylation has already been present in the free-living cyanobacterial ancestor of cyanelles. This could even have been the prerequisite for the evolution to a semiautonomous endosymbiont. The capability to reduce peptidoglycan polarity by putrescinylation and subsequent acetylation might have facilitated the necessary protein import across the cyanelle envelope by decreasing the impeding interaction of the positively charged transit sequences with the negatively charged peptidoglycan (25).
Acknowledgments
This work was supported by a grant from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (P10860-MOB, to W.L.).
We thank J.-V. Höltje (Tübingen) for providing a UDP-N-acetylmuramyl pentapeptide standard and M. Melkonian (Cologne) for a culture of axenic C. paradoxa.
REFERENCES
- 1.Aitken A, Stanier R Y. Characterization of peptidoglycan from cyanelles of Cyanophora paradoxa. J Gen Microbiol. 1979;112:219–223. [Google Scholar]
- 2.Anderson J S, Matsuhashi M, Haskin M A, Strominger J. Biosynthesis of the peptidoglycan of bacterial cell walls. II. Phospholipid carriers in the reaction sequence. J Biol Chem. 1967;242:3180–3190. [PubMed] [Google Scholar]
- 3.Bohnert H J, Crouse E J, Pouyet J, Mucke H, Löffelhardt W. The subcellular location of DNA components from Cyanophora paradoxa, a flagellate containing endosymbiotic cyanelles. Eur J Biochem. 1982;126:381–388. doi: 10.1111/j.1432-1033.1982.tb06791.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Glauner B. Separation and quantification of muropeptides with HPLC. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 6.Kamio Y, Terawaki Y, Izaki K. Biosynthesis of cadaverine-containing peptidoglycan in Selenomonas ruminantium. J Biol Chem. 1982;257:3326–3333. [PubMed] [Google Scholar]
- 7.Kamio Y, Pösö H, Terawaki Y, Paulin L. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J Biol Chem. 1986;261:6585–6589. [PubMed] [Google Scholar]
- 8.Kamio Y, Nakamura K. Putrescine and cadaverine are constituents of peptidoglycan in Veillonella alcalescens and Veillonella parvula. J Bacteriol. 1987;169:2881–2884. doi: 10.1128/jb.169.6.2881-2884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamio Y. Structural specificity of diamines covalently linked to peptidoglycan for cell growth of Veillonella alcalescens and Selenomonas ruminantium. J Bacteriol. 1987;169:4837–4840. doi: 10.1128/jb.169.10.4837-4840.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz W, Matsuhashi M, Dietrich C P, Strominger J L. Biosynthesis of peptidoglycan of bacterial cell walls. IV. Incorporation of glycine in Micrococcus lysodeikticus. J Biol Chem. 1967;242:3207–3217. [PubMed] [Google Scholar]
- 11.Kies L. The effect of penicillin on the morphology and ultrastructure of Cyanophora, Gloeochaete and Glaucocystis (Glaucocystophyceae) and their cyanelles. Endocyt Cell Res. 1988;5:361–372. [Google Scholar]
- 12.Kies L. Glaucocystophyceae and other protists harbouring procaryotic endocytobionts. In: Reisser W, editor. Algae and symbioses. Bristol, United Kingdom: Biopress Limited; 1992. pp. 353–377. [Google Scholar]
- 13.Kohlrausch U, Wientjes F B, Höltje J-V. Determination of murein precursors during the cell cycle of Escherichia coli. J Gen Microbiol. 1989;135:1499–1506. doi: 10.1099/00221287-135-6-1499. [DOI] [PubMed] [Google Scholar]
- 14.Kohlrausch U, Höltje J-V. One-step purification procedure for UDP-N-acetylmuramyl-peptide murein precursors from Bacillus cereus. FEMS Microbiol Lett. 1991;78:253–258. doi: 10.1016/0378-1097(91)90166-8. [DOI] [PubMed] [Google Scholar]
- 15.Löffelhardt W, Bryant D A, Bohnert H J. The cyanelles of Cyanophora paradoxa. Crit Rev Plant Sci. 1997;16:393–413. [Google Scholar]
- 16.Norling B, Mirzakhanian V, Nilsson F, Morré D J, Andersson B. Subfractional analysis of cyanobacterial membranes and isolation of plasma membranes by aqueous polymer two-phase partitioning. Anal Biochem. 1994;218:103–111. doi: 10.1006/abio.1994.1147. [DOI] [PubMed] [Google Scholar]
- 17.Norling B, Zak E, Andersson B, Pakrasi H. 2D-isolation of pure plasma membrane and thylakoid membranes from the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1998;436:189–192. doi: 10.1016/s0014-5793(98)01123-5. [DOI] [PubMed] [Google Scholar]
- 18.Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martinez-Torrecuadrada J, Schmid E R, de Pedro M A, Löffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfanzagl B, Allmaier G, Schmid E R, de Pedro M A, Löffelhardt W. N-Acetylputrescine as a characteristic constituent of cyanelle peptidoglycan in glaucocystophyte algae. J Bacteriol. 1996;178:6994–6997. doi: 10.1128/jb.178.23.6994-6997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pittenauer E, Schmid E R, Allmaier G, Pfanzagl B, Löffelhardt W, Quintela C, de Pedro M A, Stanek W. Structural characterization of the cyanelle peptidoglycan of Cyanophora paradoxa by 252Cf-plasma desorption mass spectrometry and fast atom bombardment/tandem mass spectrometry. Biol Mass Spectrom. 1993;22:524–536. doi: 10.1002/bms.1200220906. [DOI] [PubMed] [Google Scholar]
- 21.Plaimauer B, Pfanzagl B, Berenguer J, de Pedro M A, Löffelhardt W. Subcellular distribution of enzymes involved in the biosynthesis of cyanelle murein in the protist Cyanophora paradoxa. FEBS Lett. 1991;284:169–172. doi: 10.1016/0014-5793(91)80677-u. [DOI] [PubMed] [Google Scholar]
- 22.Schenk H E A. Nachweis einer lysozymempfindlichen Stützmembran der Endocyanellen von Cyanophora paradoxa (Korschikoff) Z Naturforsch. 1970;25b:656. [Google Scholar]
- 23.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schleifer K H, Leuteritz M, Weiss N, Ludwig W, Kirchhof G, Seidel-Rüfer H. Taxonomic study of anaerobic, gram-negative, rod-shaped bacteria from breweries: emended description of Pectinatus cerevisiphilus and description of P. frisingensis sp. nov., Selenomonas lacticifex sp. nov., Zymophilus raffinosovorans gen. nov., sp. nov. and Zymophilus paucivorans sp. nov. Int J Syst Bacteriol. 1990;40:19–27. doi: 10.1099/00207713-40-1-19. [DOI] [PubMed] [Google Scholar]
- 25.Schwartzbach S D, Osafune T, Löffelhardt W. Protein import into cyanelles and complex plastids. Plant Mol Biol. 1998;38:247–263. [PubMed] [Google Scholar]
- 26.Siewert G, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J Biol Chem. 1968;243:783–790. [PubMed] [Google Scholar]
- 27.Struve W G, Sinha R K, Neuhaus F C. On the initial stage in peptidoglycan synthesis. Phospho-N-acetylmuramyl-pentapeptide translocase (uridine monophosphate) Biochemistry. 1966;5:82–93. doi: 10.1021/bi00865a012. [DOI] [PubMed] [Google Scholar]
- 28.van Heijenoort Y, Gómez M, Derrien M, Ayala J, van Heijenoort J. Membrane intermediates in the peptidoglycan metabolism of Escherichia coli: possible roles of PBP 1b and PBP 3. J Bacteriol. 1992;174:3549–3557. doi: 10.1128/jb.174.11.3549-3557.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheat R W. Analysis of hexosamines in bacterial polysaccharides by chromatographic procedures. Methods Enzymol. 1966;8:60–78. [Google Scholar]
- 30.Work E. Reaction of ninhydrin in acid solution with straight chain amino acids containing two amino groups and its application to the estimation of αɛ-diaminopimelic acid. Biochem J. 1957;67:416–423. doi: 10.1042/bj0670416. [DOI] [PMC free article] [PubMed] [Google Scholar]