Abstract
Bordetella pertussis lipopolysaccharide (LPS) contains a single 2-keto-3-deoxy-d-manno-octulosonic acid (Kdo) residue, whereas LPS from Escherichia coli contains at least two. Here we report that B. pertussis waaA encodes an enzyme capable of transferring only a single Kdo during the biosynthesis of LPS and that this activity is sufficient to complement an E. coli waaA mutation.
Lipopolysaccharide (LPS) is the main constituent of the gram-negative bacterial surface and consists of several domains. All LPS molecules have a lipid A structure that is usually a polyacylated and phosphorylated disaccharide of glucosamine (18) linked to a carbohydrate domain, usually via the eight-carbon sugar 2-keto-3-deoxy-d-manno-octulosonic acid (Kdo). In Escherichia coli, two Kdo molecules are attached in succession to the lipid A precursor lipid IVA before lipid A biosynthesis is completed by the addition of the secondary acyl chains (4, 7, 11). The smallest LPS structure possible in viable E. coli appears to be Kdo2-lipid A. Mutants defective in the biosynthesis of Kdo precursors or the transfer of Kdo to lipid IVA have been isolated only as conditional mutants that die at nonpermissive temperatures (2, 15, 19, 20).
The waaA gene (previously designated kdtA in E. coli and gseA in Chlamydia trachomatis) encodes the enzyme that catalyzes the transfer of Kdo to lipid IVA (3, 4, 11). E. coli WaaA (WaaAEc) consists of 425 amino acids and adds two Kdo residues to lipid IVA without significant accumulation of a monoglycosylated intermediate (4). C. trachomatis WaaA (WaaACt) consists of 431 amino acids and transfers three or more Kdo residues (3). Haemophilus influenzae WaaA (WaaAHi) consists of 427 amino acids (12) and transfers only one Kdo (23). The structural basis for the ability of Kdo transferases to incorporate different numbers of Kdo residues is still unknown. Sequence homology is found for the entire lengths of the different WaaA proteins. Thus, the presence or absence of additional catalytic domains cannot account for the functional differences in Kdo transferase activities.
Bordetella pertussis, the main causative agent of whooping cough in humans (10), possesses a somewhat unusual LPS molecule which contains a single Kdo linking the core to lipid A (14). B. pertussis waaA (waaABp) has been preliminarily identified by its product’s amino acid sequence homology with other known WaaA proteins (1). It is downstream of, and probably in an operon with, waaC, which encodes an enzyme that is thought to catalyze the transfer of heptose to Kdo. B. pertussis WaaA (WaaABp) is 428 amino acids long and has 32, 39, and 24% amino acid identity with WaaAHi, WaaAEc, and WaaACt, respectively.
Based upon structural analysis of B. pertussis LPS, WaaABp has been predicted to be monofunctional. In this study, we confirm enzymologically that this is indeed the case and moreover show that, despite the fact that only a single Kdo is transferred, waaABp can complement a mutation in E. coli waaA (waaAEc).
Bacterial strains and plasmids.
E. coli MC1061 (9) is the parental strain of E. coli CJB26 and E. coli NEB1 (2), in which chromosomal waaA::kan insertion mutations are covered by waaAEc on plasmid pJSC2 (containing a temperature-sensitive origin of replication [2]) and by waaACt on pKEM1 (3), respectively. E. coli X711 (5) is a gmhA strain that synthesizes Re-chemotype LPS. E. coli strains were grown on Luria-Bertani (LB) medium at 28, 37, and 42°C. Antibiotics were added in the following final concentrations: kanamycin, 20 μg/ml; tetracycline, 30 μg/ml; streptomycin, 200 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 10 to 50 μg/ml. H. influenzae 722 (21) and B. pertussis BP536 (1) were grown as described previously.
E. coli N2.8C was generated by replacing pKEM1 in NEB1 with pSK2.8C, which was constructed as follows. A 2,794-bp SacI-BglII fragment from the B. pertussis wlb locus containing the intergenic wlb-waaC promoter region, waaC, waaA, and baf was cloned into the SacI-BamHI site in pBluescript SK(+) (Stratagene, Cambridge, United Kingdom) to make pSK2.8. To generate pSK2.8C, the chloramphenicol resistance gene (cam) originally from pACYC184 was used. pACYC184 was digested with AsuII and NheI, the fragment containing cam was purified, and the sticky ends were filled by using Klenow enzyme. This fragment was then cloned into the EcoRV site of pBluescript II. Subsequently, the cam gene was excised with SmaI and HindIII, the sticky end of this fragment was filled by using Klenow enzyme, and the fragment was then cloned into the ScaI site of pSK2.8. The resultant pSK2.8C thus has an inactivated ampicillin resistance gene and a functional cam gene.
Complementation of a waaC mutation in Salmonella by waaCBp.
To confirm that the waaC-waaA region in pSK2.8C is functional in complementation experiments, pSK2.8C was transformed into the Salmonella typhimurium waaC mutant SA1377, which produces LPS of the Re chemotype. LPS from the resultant transformants expressed O antigen, as shown by the restoration of sensitivity to bacteriophage P22 and by silver-stained tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown). These data indicate that B. pertussis waaC (waaCBp) is transcribed from pSK2.8C and is active in complementing the S. typhimurium waaC mutation and that the downstream waaA gene is also probably transcribed from this plasmid.
Complementation of waaAEc by waaABp.
To study complementation of waaAEc::kan by waaABp, plasmid pSK2.8C was transformed (22) into NEB1 and transformants were selected on LB containing chloramphenicol. The maintenance of chloramphenicol selection throughout a series of subcultures was expected to select for pSK2.8C with concomitant loss of pKEM1 due to incompatibility, since both plasmids belong to the same incompatibility group (16). After a series of such subcultures, seven ampicillin-sensitive, chloramphenicol-resistant colonies were obtained, indicating the loss of pKEM1 and the retention of pSK2.8C. One of these isolates was designated N2.8C. Many rigorous checks, including Southern hybridizations, PCR, and transformation experiments (data not shown), were performed, and all confirmed that the only plasmid in N2.8C was pSK2.8C. This exhaustive series of experiments strongly indicates that pKEM1 had been replaced by pSK2.8C in E. coli N2.8C. This also indicates that the waaA mutation in E. coli CJB26 can be complemented by waaABp.
Kdo transferase activity of N2.8C.
To ascertain whether the WaaABp protein present in N2.8C was monofunctional, Kdo transferase activity in cell extracts was assayed by using thin-layer chromatography as described previously (7) with minor modifications.
Cultures of N2.8C (typically two 125-ml portions) were grown to late-log phase (optical density at 600 nm [OD600] was ∼1.0 to 1.5) at 28°C and harvested by centrifugation at 1,900 × g for 10 min at 4°C. The cell pellet was then washed with 50 ml of 30 mM HEPES (pH 7.5) containing 2.5 mM EDTA and 1 mM EGTA. The washed cell pellet was resuspended in ∼2.5 ml of 30 mM HEPES (pH 7.5) containing 1 mM EDTA and 1 mM EGTA. The cells were ruptured by two passages through an ice-cold French pressure cell (SLM Instruments, Urbana, Ill.) at 18,000 lb/in2. The unbroken cells were removed by centrifugation at 1,900 × g for 10 min at 4°C, and the supernatant was used in the enzyme assays. All extracts were stored in aliquots at −80°C. H. influenzae membrane suspensions were prepared as previously described, as was the recombinant E. coli Kdo transferase (24). Protein concentrations were determined with the bicinchoninic assay (Pierce Chemical Company) with bovine serum albumin as the standard.
The substrates for the reaction, lipid IVA (17) and [4′-32P]lipid IVA (6), were isolated as previously described. To generate CMP-Kdo in situ, reaction mixtures (10 to 20 μl) contained 50 mM HEPES (pH 7.5), 2 mM Kdo, 0.1% Triton X-100, 50 μM [4′-32P]lipid IVA (3,000 to 6,000 cpm/nmol), 5 mM CTP, 10 mM MgCl2, and 1.8 mU of partially purified CMP-Kdo synthase. Kdo transferase assays were initiated by the addition of enzyme or bacterial cell extract and incubated at 30°C. The reactions were terminated by spotting 5 μl of the mixtures onto a thin-layer silica plate. The plate was then air dried and developed in chloroform–pyridine–88% formic acid–H2O (30:70:16:10, vol/vol). The solvent was evaporated with a hot air stream, and the plate was exposed to a PhosphorImager screen for 12 to 16 h. The extent of conversion of [4′-32P]lipid IVA to the products of interest was quantified by using a Molecular Dynamics PhosphorImager equipped with the ImageQuant program.
Extracts prepared from strain N2.8C and B. pertussis are capable of converting [4′-32P]lipid IVA to a new, more slowly migrating species (Fig. 1, lanes 4 and 5) that comigrates with the Kdo-lipid IVA produced by H. influenzae membranes (Fig. 1, lane 3). The reaction product generated by N2.8C extracts also migrates faster than the Kdo2-lipid IVA formed by the E. coli Kdo transferase under the same conditions (Fig. 1, lane 2). The data show that WaaABp can utilize CMP-Kdo in the same fashion as other Kdo transferases tested previously (WaaAEc, WaaACt, and WaaAHi) (3, 4, 8, 23) and that WaaABp is monofunctional, transferring only a single Kdo residue to lipid IVA. Only the monofunctional, Kdo transferase activity is present in N2.8C, and this activity alone is sufficient to support growth of E. coli.
FIG. 1.
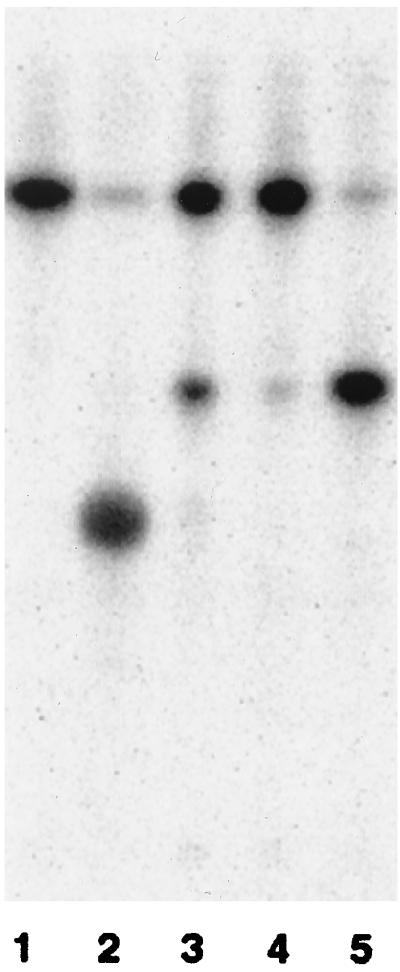
Thin-layer chromatography showing the conversion of [4′-32P]lipid IVA to Kdo-[4′-32P]lipid IVA by N2.8C extracts. The reactions were initiated with enzyme or bacterial cell extract and were terminated after 10 min. Reactions initiated with the following enzyme sources are shown: a nonenzymatic control (lane 1), purified E. coli Kdo transferase (8 μU) (lane 2), membranes from H. influenzae 722 (0.5 mg/ml) (lane 3), extract from wild-type B. pertussis BP536 (lane 4), and extract from N2.8C (lane 5).
Analysis of the LPS phenotype.
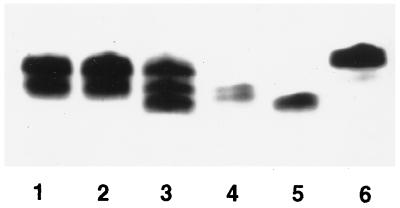
To investigate the effect on E. coli of relying on WaaABp for Kdo transfer, the LPSs of N2.8C, CJB26, MC1061, and NEB1 were extracted and analyzed by silver-stained tricine-SDS-PAGE. This revealed that N2.8C synthesizes LPS which is distinct from that of the other strains (Fig. 2). LPSs from MC1061 and CJB26 were identical, as expected, and produced two bands in the gel (Fig. 2, lanes 1 and 2). NEB1 also produced these bands as well as a third, faster-migrating band (Fig. 2, lane 3). N2.8C LPS appears as two bands, very close together, between the two lower bands observed in NEB1 (Fig. 2, lane 4) and migrating slower than Re-chemotype LPS produced by X711 (5) (Fig. 2, lane 5). If N2.8C produces LPS with a structure consisting of Kdo-lipid A, this LPS might be expected to migrate faster than that of X711. These results suggest that this is not the case and that the Kdo-lipid A is further substituted, given that the core oligosaccharide usually extends from the inner Kdo in E. coli. On the other hand, the observed extension might be a consequence of B. pertussis waaC, encoding heptosyltransferase I, being introduced together with waaABp on pSK2.8C. It is possible that WaaCBp is competing with WaaCEc to add heptose to the Kdo-lipid A being synthesized in N2.8C, given that Kdo-lipid A is a less adequate substrate for WaaCEc than Kdo2-lipid A (13). A full understanding of the N2.8C LPS will require a definitive structural analysis.
FIG. 2.
Silver-stained tricine-SDS-PAGE of LPSs from E. coli MC1061 (lane 1), CJB26 (waaAEc; lane 2), NEB1 (waaACt; lane 3), N2.8C (waaABp; lane 4), and X711 (gmhA mutant of E. coli, Re-LPS; lane 5) and wild-type B. pertussis BP536 (lane 6).
Temperature-sensitive growth of N2.8C.
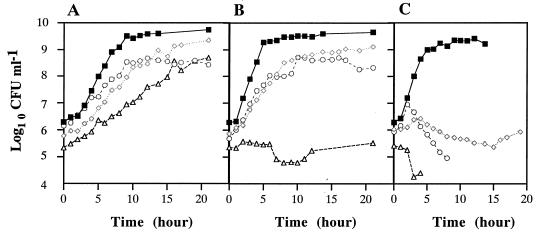
The effect of synthesizing LPS containing one, two, or three Kdo residues on the growth of E. coli was investigated by determining the growth rates of N2.8C, MC1061, CJB26, and NEB1 at three different temperatures: 28, 37, and 42°C (Fig. 3).
FIG. 3.
Growth curves of MC1061 (■), CJB26 (◊), NEB1 (○), and N2.8C (▵). Bacteria were grown at 28 (A), 37 (B), and 42°C (C). An aliquot was removed from the culture at each time point and used to determine CFU by plating on LB agar and incubating at 28°C until bacterial colonies were visible.
MC1061 was grown from a single colony in LB broth containing streptomycin at 28°C with shaking at 160 rpm to an OD600 of 1. CJB26, NEB1, and N2.8C were grown in LB medium containing streptomycin, tetracycline, and kanamycin. Sufficient culture was used to inoculate 100 ml of prewarmed LB medium supplemented with streptomycin, to an OD600 of ∼0.001 (5 × 105 cells/ml), and was incubated at 28, 37, or 42°C with shaking at 160 rpm. The growth rate was determined by performing viability counts every hour.
MC1061 grew exponentially at all three temperatures, reaching a maximum cell density of 6 × 109 CFU/ml. CJB26 and NEB1 grew slower than MC1061 at 28 and 37°C and grew poorly or not at all at 42°C. This result differs from data previously obtained where NEB1 grew at elevated temperatures, albeit less well than MC1061 (2). The reason for this difference is unknown, but differences in the protocols for performing the respective growth curves may play a role. For example, in the previous study, the growth curve was started with a culture at an OD600 of 0.2, whereas in this study, the growth curve was started with a culture at an OD600 of 0.001. Also, in the previous study, the cultures were intermittently back-diluted, whereas in this study, no back-dilution was performed and the growth curve was derived simply from sampling aliquots at different times and performing viability counts.
N2.8C grew slowly but steadily at 28°C, reaching a cell density of 5 × 108 CFU/ml after 27 h, which is equivalent to the level of NEB1 attained. However, at 37°C growth was dramatically reduced, while at 42°C no viable N2.8C cells were recovered after 5 h (Fig. 3). This suggests that N2.8C may be accumulating LPS precursors that are toxic at high concentrations and so inhibit normal growth. The transfer of heptose or the incorporation of laurate into Kdo-lipid IVA may be limited.
It is interesting that the bacteria with the complemented mutation observed under the electron microscope appear to have an altered morphology (data not shown; pictures available on request). Scanning electron microscopy revealed that the surfaces of NEB1 and N2.8C were rough in comparison to the parental strain MC1061. In addition, the surface of N2.8C appeared to be indented, a trait which was observed neither in the parental strain nor in NEB1. Considerable variations in cell length were also observed for NEB1 and N2.8C cultures. Transmission electron microscopy was performed to visualize the physical state of the cell envelopes. MC1061 and NEB1 had cell envelopes which appeared intact and periplasmic regions with constant widths throughout. The cell envelopes of N2.8C appeared less consistent, showing dissociations of outer and inner membranes.
In conclusion, we have shown that waaABp encodes an enzyme capable of transferring only a single Kdo residue to the growing LPS molecule. In addition, this gene is sufficient to complement an E. coli waaA mutation, but the bacteria with the complemented mutation are not as robust as wild-type E. coli in terms of growth at elevated temperatures. This is reflected in the morphology of the bacteria with the complemented mutation, which appears to indicate that the outer cell membrane is not as well formed as that of wild-type E. coli.
Acknowledgments
This work was funded by The Welcome Trust, UK, project grant 045666. K. A. White and C. R. H. Raetz were supported by National Institutes of Health grant GM51310.
We thank Mary Soane (National Heart and Lung Institute, The Royal Brompton Hospital, Imperial College of Science, Technology and Medicine, London, United Kingdom) and Ian Morris (Department of Biology, Imperial College of Science, Technology and Medicine) for their help with scanning electron microscopy.
REFERENCES
- 1.Allen A, Maskell D. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol Microbiol. 1996;19:37–52. doi: 10.1046/j.1365-2958.1996.354877.x. [DOI] [PubMed] [Google Scholar]
- 2.Belunis C J, Clementz T, Carty S M, Raetz C R. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J Biol Chem. 1995;270:27646–27652. doi: 10.1074/jbc.270.46.27646. [DOI] [PubMed] [Google Scholar]
- 3.Belunis C J, Mdluli K E, Raetz C R, Nano F E. A novel 3-deoxy-d-manno-octulosonic acid transferase from Chlamydia trachomatis required for expression of the genus-specific epitope. J Biol Chem. 1992;267:18702–18707. [PubMed] [Google Scholar]
- 4.Belunis C J, Raetz C R. Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-d-manno-octulosonic acid transferase from Escherichia coli. J Biol Chem. 1992;267:9988–9997. [PubMed] [Google Scholar]
- 5.Brooke J S, Valvano M A. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J Biol Chem. 1996;271:3608–3614. doi: 10.1074/jbc.271.7.3608. [DOI] [PubMed] [Google Scholar]
- 6.Brozek K A, Bulawa C E, Raetz C R. Biosynthesis of lipid A precursors in Escherichia coli. A membrane-bound enzyme that transfers a palmitoyl residue from a glycerophospholipid to lipid X. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 7.Brozek K A, Hosaka K, Robertson A D, Raetz C R. Biosynthesis of lipopolysaccharide in Escherichia coli. Cytoplasmic enzymes that attach 3-deoxy-d-manno-octulosonic acid to lipid A. J Biol Chem. 1989;264:6956–6966. [PubMed] [Google Scholar]
- 8.Brozek K A, Raetz C R. 3-Deoxy-d-manno-octulosonate transferase and late acyltransferases of lipopolysaccharide biosynthesis. Methods Enzymol. 1992;209:476–485. doi: 10.1016/0076-6879(92)09058-b. [DOI] [PubMed] [Google Scholar]
- 9.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 10.Cherry J D. Historical review of pertussis and the classical vaccine. J Infect Dis. 1996;174:S259–S263. doi: 10.1093/infdis/174.supplement_3.s259. [DOI] [PubMed] [Google Scholar]
- 11.Clementz T, Raetz C R. A gene coding for 3-deoxy-d-manno-octulosonic-acid transferase in Escherichia coli. Identification, mapping, cloning, and sequencing. J Biol Chem. 1991;266:9687–9696. [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Kadrmas J L, Raetz C R H. Enzymatic synthesis of lipopolysaccharide in Escherichia coli. Purification and properties of heptosyltransferase I. J Biol Chem. 1998;273:2799–2807. doi: 10.1074/jbc.273.5.2799. [DOI] [PubMed] [Google Scholar]
- 14.Le Dur A, Caroff M, Chaby R, Szabo L. A novel type of endotoxin structure present in Bordetella pertussis. Isolation of two different polysaccharides bound to lipid A. Eur J Biochem. 1978;84:579–589. doi: 10.1111/j.1432-1033.1978.tb12201.x. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann V, Rupprecht E, Osborn M J. Isolation of mutants conditionally blocked in the biosynthesis of the 3-deoxy-d-manno-octulosonic-acid-lipid-A part of lipopolysaccharides derived from Salmonella typhimurium. Eur J Biochem. 1977;76:41–49. doi: 10.1111/j.1432-1033.1977.tb11568.x. [DOI] [PubMed] [Google Scholar]
- 16.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raetz C R, Purcell S, Meyer M V, Qureshi N, Takayama K. Isolation and characterization of eight lipid A precursors from a 3-deoxy-d-manno-octylosonic acid-deficient mutant of Salmonella typhimurium. J Biol Chem. 1985;260:16080–16088. [PubMed] [Google Scholar]
- 18.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 19.Rick P D, Fung L W, Ho C, Osborn M J. Lipid A mutants of Salmonella typhimurium. Purification and characterization of a lipid A precursor produced by a mutant in 3-deoxy-d-mannooctulosonate-8-phosphate synthetase. J Biol Chem. 1977;252:4904–4912. [PubMed] [Google Scholar]
- 20.Rick P D, Osborn M J. Lipid A mutants of Salmonella typhimurium. Characterization of a conditional lethal mutant in 3-deoxy-d-mannooctulosonate-8-phosphate synthetase. J Biol Chem. 1977;252:4895–4903. [PubMed] [Google Scholar]
- 21.Ruan M, Akkoyunlu M, Grubb A, Forsgren A. Protein D of Haemophilus influenzae. A novel bacterial surface protein with affinity for human IgD. J Immunol. 1990;145:3379–3384. [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.White K A, Kaltashov I A, Cotter R J, Raetz C R H. A mono-functional 3-deoxy-d-manno-octulosonic acid (Kdo) transferase and a Kdo kinase in extracts of Haemophilus influenzae. J Biol Chem. 1997;272:16555–16563. doi: 10.1074/jbc.272.26.16555. [DOI] [PubMed] [Google Scholar]
- 24.White K A, Raetz C R H. Characterization of a mono-functional Kdo transferase in extracts of Haemophilus influenzae. FASEB J. 1995;9:A1376. doi: 10.1074/jbc.272.26.16555. [DOI] [PubMed] [Google Scholar]