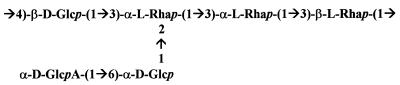Abstract
The type 2 capsule locus of Streptococcus pneumoniae was characterized in Avery’s strain D39, which is the parent strain of the standard transformation recipients currently used in pneumococcal research and is largely used as a virulent strain in studies on the pathogenesis of pneumococcal infections. The capsule locus was sequenced by using a 21.7-kb PCR fragment from the D39 genome as a template. Sequence data analysis showed the presence of 18 open reading frames, 17 of which have the same direction of transcription and all of which are potentially involved in capsule biosynthesis. It was also shown that R36A and R6, which are unencapsulated (rough) derivatives of D39, carry a 7,504-bp deletion involving nine capsule genes.
Streptococcus pneumoniae (pneumococcus) is an important human pathogen that causes such bacteremic infections as pneumonia, bacteremia, and meningitis, resulting in high mortality rates even when treated with antimicrobials (4). The polysaccharide capsule is the major pathogenicity determinant of S. pneumoniae, and its presence is a conditio sine qua non of pneumococcal virulence. The capsular polysaccharide varies from strain to strain, and 90 different capsular serotypes have been recognized (14). Transformation-mediated exchange of capsular genes has long been known to occur in the pneumococcus (5), but only recently has information on the genes involved in capsule biosynthesis begun to accumulate. Nucleotide sequence data is now available for a limited number of types, including 1, 3, 14, 19B, 19F, and 23F (3, 8, 9, 18, 20–22). Capsular transformation has been shown to occur in vivo, and it is believed to play a role both in the spread of drug-resistant clones and in the long-term efficacy of vaccines based on a limited number of serotypes (8, 24).
The type 2 capsular polysaccharide is composed of singly branched hexasaccharide repeating units, each containing one d-glucuronic acid, two d-glucose, and three l-rhamnose residues (Fig. 1) (16). In this work we determined the nucleotide sequences of the genes involved in type 2 polysaccharide biosynthesis in Avery’s strain D39 (5), which is the parent of the standard transformation recipients currently used in pneumococcal research, such as R36A, R6, Rx1, R800, and CP1200 (1, 13, 26–28), and is largely used as a virulent strain in studies on the pathogenesis of pneumococcal infections (6). We also characterized the deletion which occurred in the capsule locus of D39 when the unencapsulated transformable strain R36A was generated (5). Since capsular genes are in a chromosomal locus between genes dexB (11) and aliA (plpA) (1, 25) in all types studied so far (3, 8, 9, 18, 20–22), we proceeded to sequence the DNA between dexB and aliA in the type 2 strain D39.
FIG. 1.
Structure of the repeating unit of the type 2 capsular polysaccharide of S. pneumoniae (16). Glc, glucose; Rha, rhamnose; GlcA, glucuronic acid.
Sequencing and sequence analysis.
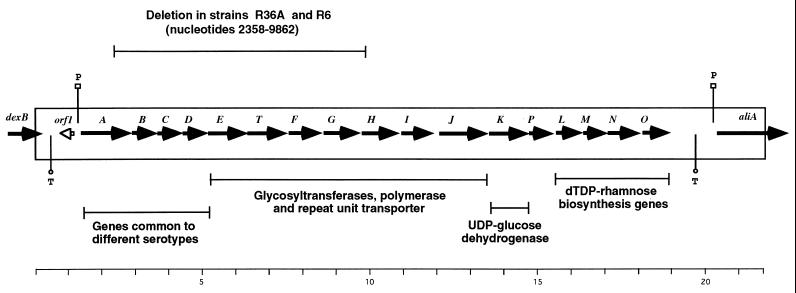
To avoid problems encountered when trying to clone pneumococcal DNA in Escherichia coli (7, 10, 19), the type 2 capsule locus was sequenced by using a 21.7-kb PCR fragment obtained by using primers designed on dexB and aliA as a starting template (Fig. 2). The method for direct sequencing of long PCR fragments from the pneumococcal genome has already been described in detail (15). Gapped BLASTX software (2) was used to translate the sequences of both strands of DNA in all six reading frames and to conduct homology searches of the nucleotide and protein databases available at the National Center for Biotechnology Information. The compilation and analyses of the sequences were carried out with Dnasis version 3.6 software (Hitachi, San Bruno, Calif.) and the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.).
FIG. 2.
Structure of the type 2 capsule locus in S. pneumoniae D39. The 21,709-bp PCR fragment used as a template for sequencing was obtained by using primers FI3 and FI4, designed on dexB and aliA, respectively. FI3, 5′-TCT TAG TTC CAT GGG ATG CTT TCT GTG TG-3′, nucleotides 657 to 685 of dexB (GenBank accession no. Z47210); FI4, 5′-CGC TGA ACT TTT GTA GTT GCT GTC TGG TCA AC-3′, complementary to nucleotides 1301 to 1270 of aliA (plpA) (GenBank accession no. L20556). Sequenced DNA is represented by the rectangle. cps genes are indicated only by their letters, and the ORFs and their direction of transcription are represented by arrows. Putative promoters (P) and transcription terminators (T) are indicated. Scale is in kilobases.
Genetic organization of the type 2 capsule locus.
Sequence data analysis showed the presence of 18 open reading frames (ORFs), 17 of which have the same direction of transcription and all of which are potentially involved in capsule biosynthesis. As for other serotypes, the 17 type 2 capsule genes are apparently arranged in a single transcriptional unit, with a promoter-like sequence located immediately upstream of cps2A (−35, nucleotides 1403 to 1408; −10, nucleotides 1426 to 1431), 100% identical to that proposed for the type 19F capsule operon (12). Stem-loop structures resembling transcription terminators are present downstream of dexB (nucleotides 387 to 444) and downstream of the last capsular gene, csp20 (nucleotides 19742 to 19832). Between the dexB transcription terminator and the capsule operon promoter, there is a 327-bp ORF (orf1) oriented opposite to the cps2 genes (Fig. 2). The orf1 gene product is similar to several transposases of the Synechocystis sp. genome (GenBank accession no. D90915) and is probably part of an insertion sequence present seven times in the type 4 pneumococcal genome (15a). Interestingly, orf1 occupies the same positions in the capsule loci of types 1 (GenBank accession no. Z83335), 4, and 19F (GenBank accession no. AF030367).
The csp2 genes.
Type 2 capsule genes are named according to the nomenclature adopted for types 19F (12, 21) and 23F (GenBank accession no. AF030373). Comparison of sequence data shows that the first four genes, cps2A through -D, have a very high similarity to the corresponding genes present in the capsule loci of other serotypes. The putative functions of these common genes have been already discussed in detail by other authors (11, 12, 17). The central portion of the locus is occupied by seven genes (cps2E, cps2T, and cps2F through -J) encoding five putative glycosyltransferases, a polysaccharide polymerase, and a repeat unit transporter (Table 1). The conversion of glucose to glucuronic acid is probably catalized by the cps2K gene product, since it is homologous to the type 1 (89% similarity) and type 3 (74% similarity) UDP-glucose dehydrogenases. The csp2P gene is unique to the type 2 locus, and its gene product shows similarity to the UDP-galactopyranose mutases of many microorganisms, including E. coli (67% similarity) (23) and Mycobacterium tuberculosis (58% similarity). The four genes involved in dTDP-rhamnose biosynthesis (csp2L through -O) are located at the 3′ end of the locus and show a very strong similarity (up to 99% amino acid identity) to the corresponding genes of types 19F, 23F, and 1 (cryptic genes) (21, 22). The coding sequences of both cps2J and cps2P start with a TTG codon, as previously observed for other pneumococcal cps genes (21).
TABLE 1.
Homologies of type 2 capsule genes with other bacterial genesa
| Type 2 gene | Homologous gene | Origin species or S. pneumoniae type | Proposed function of gene product | Amino acid identity and similarity (%) |
|---|---|---|---|---|
| cps2E | cps23fE | Type 23F | Undecaprenyl-phosphate glucose-1-phosphate transferase | 95, 96 |
| cps14E | Type 14 | Glucosyl-1-phosphate transferase | 60, 79 | |
| cps19fE | Type 19F | Uridine diphosphate glycosyltransferase | 60, 78 | |
| cps2T | cps23fT | Type 23F | Rhamnosyl transferase | 81, 88 |
| cps2F | cps23fU | Type 23F | Galactosyltransferase | 23, 50 |
| cps14J | Type 14 | ss-1,4-galactosyltransferase | 19, 46 | |
| cps14I | Type 14 | ss-1,3-N-acetylglucosaminyltransferase | 18, 43 | |
| cps2G | Sequence encoding protein RPN00103 | Type 4 | Undecaprenyl-phosphate galactosephosphotransferase | 35, 55 |
| icsA | Neisseria meningitidis | Lipopolysaccharide glycosyltransferase | 19, 48 | |
| cps2H | cps23fI | Type 23F | Polysaccharide polymerase | 22, 52 |
| cps14H | Type 14 | Polysaccharide polymerase | 21, 51 | |
| cps19fI | Type 19F | Polysaccharide polymerase | 20, 50 | |
| cps2I | rfaK | N. meningitidis | Alpha 1,2 N-acetylglucosamine transferase | 25, 52 |
| icsA | N. meningitidis | Lipopolysaccharide glycosyltransferase | 24, 52 | |
| cps2J | cps23fJ | Type 23F | Repeat unit transporter | 36, 63 |
| cps14L | Type 14 | Repeat unit transporter | 20, 49 |
Deletion in rough strains R36A and R6.
R36A is an unencapsulated (rough) derivative of D39 obtained by growing D39 in the presence of anti-type 2 rabbit serum for 36 serial passages. This strain is used as a transformation recipient and has never been found to revert to capsule production (5). R6 is a subclone of R36A selected in the 1950s for continued competence and used since in many laboratories (28). Sequence analyses of the cps loci in R36A and R6 showed a 7,504-bp deletion corresponding to nucleotides 2358 through 9862 of the D39 capsule locus (Fig. 2) with a 25-bp insertion of an inverted portion of csp2A (nucleotides 2502 through 2526). The deletion involves the 3′ end of cps2A, the 5′ end of cps2H, and seven other whole genes (Fig. 2).
The data reported here are important not only because they add to the knowledge of the genetic variability within the capsule locus of S. pneumoniae but also because they describe a genetic locus responsible for a crucial phenotype (smooth versus rough) of the most used bacterial strains in pneumococcal research. Since Avery’s time, R36A and its derivatives have been used as recipients in transformation experiments in all laboratories working on the genetics of S. pneumoniae.
Nucleotide sequence accession number.
The nucleotide sequence of the type 2 capsule locus of D39 is assigned GenBank accession no. AF026471, and the deletion mapped in R36A and R6 has been assigned accession no. AF029368.
Acknowledgments
This work was supported in part by grants from GlaxoWellcome Verona, M.U.R.S.T. (60%), and CNR (P. F. Biotecnologie, contract 97.01185.PF49).
We thank Marco Oggioni for helpful advice, James Paton for critically reading the manuscript, and Lorenzo Morelli and Marisa Callegari for gracious hospitality at the CRB sequencing facility (Cremona, Italy).
REFERENCES
- 1.Alloing G, de Philip P, Claverys J P. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J Mol Biol. 1994;241:44–58. doi: 10.1006/jmbi.1994.1472. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrecubieta C, Garcia E, Lopez R. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene. 1995;167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 4.Austrian R. Pneumococcal infections. In: Germanier R, editor. Bacterial vaccines. Orlando, Fla: Academic Press Inc.; 1984. pp. 257–288. [Google Scholar]
- 5.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry A M, Alexander J E, Mitchell T J, Andrew P W, Hansman D, Paton J C. Effect of defined point mutations in the pneumolysin gene on the virulence of Streptococcus pneumoniae. Infect Immun. 1995;63:1969–1974. doi: 10.1128/iai.63.5.1969-1974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J D, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 9.Dillard J P, Vandersea M W, Yother J. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J Exp Med. 1995;181:973–983. doi: 10.1084/jem.181.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillard J P, Yother J. Analysis of Streptococcus pneumoniae sequences cloned into Escherichia coli: effect of promoter strength and transcription terminators. J Bacteriol. 1991;173:5105–5109. doi: 10.1128/jb.173.16.5105-5109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia E, Lopez R. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett. 1997;149:1–10. doi: 10.1111/j.1574-6968.1997.tb10300.x. [DOI] [PubMed] [Google Scholar]
- 12.Guidolin A, Morona J K, Morona R, Hansman D, Paton J C. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun. 1994;62:5384–5396. doi: 10.1128/iai.62.12.5384-5396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrichsen J. Six newly recognized types of Streptococcus pneumoniae. J Clin Microbiol. 1995;33:2759–2762. doi: 10.1128/jcm.33.10.2759-2762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannelli F, Giunti L, Pozzi G. Direct sequencing of long PCR fragments. Mol Biotechnol. 1998;10:183–185. doi: 10.1007/BF02760864. [DOI] [PubMed] [Google Scholar]
- 15a.The Institute for Genomic Research. 16 December 1998, revision date. [Online.] http://www.tigr.org. [25 January 1999, last date accessed.]
- 16.Jansson P E, Lindberg B, Andersson M, Lindquist U, Henrichsen J. The structure of capsular polysaccharide of the pneumococcus type II. Carbohydr Res. 1975;40:69–75. doi: 10.1016/s0008-6215(00)82669-5. [DOI] [PubMed] [Google Scholar]
- 17.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 18.Kolkman M A B, Wakarchuk W, van der Zeijst B A M, van der Zeijst N. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 19.Martin B, Alloing G, Boucraut C, Claverys J P. The difficulty of cloning Streptococcus pneumoniae mal and ami loci in Escherichia coli: toxicity of malX and amiA gene products. Gene. 1989;80:227–238. doi: 10.1016/0378-1119(89)90287-4. [DOI] [PubMed] [Google Scholar]
- 20.Morona J K, Morona R, Paton J C. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 19B. J Bacteriol. 1997;179:4953–4958. doi: 10.1128/jb.179.15.4953-4958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 22.Munoz R, Mollerach M, Lopez R, Garcia E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 23.Nassau P M, Martin S L, Brown R E, Weston A, Monsey D, McNeil M R, Duncan K. Galactofuranose biosynthesis in Escherichia coli K-12: identification and cloning of UDP-galactopyranose mutase. J Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 25.Pearce B J, Naughton A M, Masure H R. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol Microbiol. 1994;12:881–892. doi: 10.1111/j.1365-2958.1994.tb01076.x. [DOI] [PubMed] [Google Scholar]
- 26.Pozzi G, Masala L, Iannelli F, Manganelli R, Håvarstein L S, Piccoli L, Simon D, Morrison D A. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravin A W. Reciprocal capsular transformations of pneumococci. J Bacteriol. 1959;77:296–309. doi: 10.1128/jb.77.3.296-309.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith M D, Guild W R. A plasmid in Streptococcus pneumoniae. J Bacteriol. 1979;137:735–739. doi: 10.1128/jb.137.2.735-739.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.WIT, Computational Biology Group at Argonne National Laboratory. [Online.] http://selkov-7.mcs.anl.gov/WIT2/CGI. [25 January 1999, last date accessed.]