Abstract
We herein report that Azorhizobium caulinodans PII and GlnK are not necessary for glutamine synthetase (GS) adenylylation whereas both proteins are required for complete GS deadenylylation. The disruption of both glnB and glnK resulted in a high level of GS adenylylation under the condition of nitrogen fixation, leading to ammonium excretion in the free-living state. PII and GlnK also controlled nif gene expression because NifA activated nifH transcription and nitrogenase activity was derepressed in glnB glnK double mutants, but not in wild-type bacteria, grown in the presence of ammonia.
Azorhizobium caulinodans reduces atmospheric nitrogen both in the free-living state and in symbiosis with its host plant, the tropical legume Sesbania rostrata (11). In pure culture, this bacterium grows using molecular nitrogen, whereas during symbiosis, fixed nitrogen is exported from the bacteroid to the plant cell and assimilated by the host. Thus, the coupling between nitrogen fixation and ammonia assimilation that exists in the free-living state must be abolished during symbiosis.
Ammonia assimilation proceeds through the glutamine synthetase (GS)-glutamine oxoglutarate aminotransferase pathway. A. caulinodans has a single GS (encoded by glnA), the activity of which is regulated by adenylylation (10). Two genes with products similar to PII have been characterized in A. caulinodans: glnB, which is cotranscribed with glnA (17); and glnK, which is cotranscribed with amtB, a gene encoding a protein similar to a known ammonium transporter (18). As in Escherichia coli, glnB is constitutively transcribed whereas glnK expression is regulated by ammonia (17, 22). Neither PII nor GlnK is required for nitrogen fixation in the free-living state. glnB mutants are impaired in symbiotic nitrogen fixation (Fix−), whereas glnK mutants are not (Fix+). PII and GlnK have a minor effect on GS adenylylation (17, 18).
Two proteins similar to PII (PII and GlnK) have been identified in several gram-negative bacteria, including Herbaspirillum seropedicae, Azospirillum brasilense, and E. coli (3, 8, 22). These two proteins are not equivalent in H. seropedicae and A. brasilense because glnB single mutants have impaired nitrogen fixation (3, 9). In contrast, E. coli PII and GlnK seem to control GS deadenylylation in the absence of ammonia (2).
We report herein the properties of an A. caulinodans glnB glnK double mutant. In contrast to the glnB and glnK single mutants, GS deadenylylation was strongly impaired during nitrogenase derepression in the double mutant. We also found that the glnB glnK double mutant, but not the wild type, derepressed nitrogenase activity in the presence of ammonia, indicating that PII and GlnK are also involved in the regulation of nitrogen fixation.
Characterization of the growth properties of the glnB glnK double mutant.
A glnB glnK double mutant (strain 57625) was constructed by transferring the glnK interposon mutation (18) into the glnB mutant strain (57620) by conjugation, in order to study the effect of the absence of both proteins.
As previously reported for the glnB mutant, the glnB glnK mutant (strain 57625) grew less well than the wild type and the glnK mutant in liquid minimal medium containing 15 mM ammonia as the sole nitrogen source (17, 18). The generation time of the double mutant strain was 174 min, whereas that of the wild-type strain was 120 min. Maximum optical density (600 nm) for the mutant was 2.4, whereas that for the wild type was 5.5. Both the glnB mutant and the glnB glnK double mutant grew less well than the wild type on solid nitrogen-free medium containing 15 mM ammonia, 1 mM ammonia, 10 mM nitrate, or 10 mM histidine but grew as well as the wild type on 10 mM glutamine. Unlike the glnB or glnK single mutants, the glnB glnK mutant could not use molecular nitrogen for growth.
PII or GlnK was required for GS deadenylylation.
Unadenylylated and total GS activities were measured by the γ-glutamyltransferase assay in the presence and the absence, respectively, of 60 mM Mg2+ (Table 1), on whole cells cultured under nitrogenase-derepressing conditions (17) with and without shock by addition of 0.2% NH4+. As reported for the glnB mutant (57620) (17), total GS activity, which depends on the total amount of enzyme, was higher in the glnB glnK mutant (57625) than in the wild type. This may be due to there being more glnA transcription under the control of the promoter of the aphII gene (which confers kanamycin resistance) inserted in the glnB coding sequence. For all strains tested, there was less or equal amount of unadenylylated (or active) GS after ammonia shock than under nitrogenase-derepressing conditions, suggesting that GS adenylylation does not require PII or GlnK.
TABLE 1.
Effect of ammonia shock on total GS activity and the percentage of unadenylylated (active) GS in A. caulinodans ORS571 and mutant strains
| Strain | GS sp acta,b
|
% active GSb (unadenylylated)
|
||
|---|---|---|---|---|
| N freec | NH4+ shockd | N freec | NH4+ shockd | |
| ORS571 (wild type) | 4.59 ± 0.59 | 2.80 ± 0.58 | 70.8 ± 4.1 | 41.4 ± 6.0 |
| 57620 (glnB mutant) | 32.74 ± 2.02 | 20.14 ± 3.43 | 66.4 ± 3.0 | 12.4 ± 3.2 |
| 57621 (glnK mutant) | 7.43 ± 1.26 | 5.10 ± 1.29 | 70.4 ± 11.3 | 34.2 ± 9.2 |
| 57625 (glnB glnK mutant) | 13.05 ± 2.9 | 11.68 ± 3.16 | 11.3 ± 2.8 | 12.6 ± 4.6 |
| 57625 (glnB glnK mutant)/ pRS1045e | 21.49 ± 2.26 | 13.99 ± 2.30 | 68.2 ± 7.4 | 26.1 ± 6.2 |
| 57625 (glnB glnK mutant)/ pRS1046e | 19.77 ± 3.44 | 13.50 ± 1.89 | 64.2 ± 13.0 | 14.6 ± 1.8 |
Specific activity of GS in pure culture; 1 unit corresponds to 1 μmol of γ-glutamyl hydroxamate · min−1 · mg of protein−1.
Values are the means ± standard deviations from at least three independent experiments.
Cells were cultured for 3 h in nitrogen-free medium under microaerobic conditions.
Cells were cultured as described in footnote c and shocked by incubation with 0.2% NH4+.
The percentages of unadenylylated GS were similar in the wild-type strain and the glnB or glnK single mutants (about 70%) under nitrogenase-derepressing conditions, but the percentage was much lower in the glnB glnK double mutant (11%) (Table 1). It must be mentioned that the percentage of unadenylylated GS was probably underestimated since, under these assay conditions, unadenylylated GS may have a specific transferase activity different from that of adenylylated GS, which may account for the increase of the total activities (10). However, the low level of active GS present was correlated with the impaired growth of the glnB glnK mutant on molecular nitrogen. Both GS activity (Table 1) and growth on molecular dinitrogen were restored in the double mutant strain by expression from plasmids of either glnB (from pRS1045) or glnK (from pRS1046). Therefore, at least one of the proteins is required for GS deadenylylation under nitrogenase-derepressing conditions.
It is unclear why A. caulinodans PII and GlnK are functionally equivalent in GS deadenylylation and not in symbiotic nitrogen fixation. The difference in function may be due to a difference in gene expression during symbiosis. It is also possible that PII and GlnK have activities that differ according to their molecular forms. PII is active as a homotrimer (5, 7), but it is likely that PII/GlnK heterotrimers exist. Thus, PII or GlnK homotrimers may activate GS deadenylylation, and heterotrimers or PII homotrimers may activate symbiotic nitrogen fixation.
The glnB glnK double mutant excreted ammonia.
A. caulinodans, unlike Bradyrhizobium species, can grow by consuming molecular nitrogen in pure culture. In the free-living state, only 10% of fixed nitrogen (NH3) is exported from the cell, the remaining 90% being used for growth (13), whereas Bradyrhizobium cultures export all their fixed nitrogen to the medium (4). The absence or inhibition of GS activity blocks ammonium transport in Klebsiella pneumoniae (16). We tested whether the low level of unadenylylated GS in the glnB glnK double mutant (57625) or the absence of GS in the glnBA mutant (57619) affected ammonium excretion during nitrogen fixation in the free-living state by the indophenol method (6). No NH4+ was detected in the supernatants of cultures of glnB or glnK single mutant strains (57620 and 57621), as was also previously reported for the wild-type strain (13). A large amount of NH4+ was present in the supernatants of cultures of the glnBA mutant and glnB glnK double mutant (310 and 362 μM extracellular NH4+/optical density unit, respectively). Excretion of NH4+ was completely abolished in the glnB glnK mutant by expression from plasmids of either glnB (57625/pRS1045) or glnK (57625/pRS1046). Thus, the absence or inactivation of GS may lead to the accumulation of fixed nitrogen in A. caulinodans cells and ultimately to its excretion into the medium.
The glnB glnK mutant strain expressed nitrogen fixation genes in the presence of ammonia.
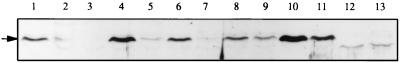
The PII and GlnK proteins control ammonium metabolism, in response to ammonia availability, by regulating GS activity. Ammonia negatively regulates nitrogen fixation genes in A. caulinodans. In particular it affects nifA transcription (15, 21). Thus, we investigated whether PII and GlnK were also involved in the regulation of nifA expression. The A. caulinodans NifA protein has an estimated molecular mass of 66.8 kDa. It was detected by Western blot analysis in whole-cell extracts from wild-type and mutant strains, using Bradyrhizobium japonicum anti-NifA antibodies (19) (Fig. 1). NifA was detected in the wild-type strain and in glnB and glnK single mutants (57620 and 57621) cultivated under nitrogenase-derepressing conditions (Fig. 1, lanes 1, 4, and 6) but not in the presence of NH4+ (lanes 2, 5, and 7) or in the nifA mutant (lane 3) (20). NifA was detected in the presence and absence of ammonia in the glnB glnK double mutant (lanes 8 and 9) and in the nifA mutant containing either the A. caulinodans nifA gene (lanes 10 and 11) or the B. japonicum nifA gene expressed constitutively in the presence of ammonia (lanes 12 and 13).
FIG. 1.
Immunodetection of NifA from A. caulinodans cells incubated under microaerobic conditions either in nitrogen-free medium (lanes 1, 3, 4, 6, 8, 10, and 12) or in the presence of 15 mM ammonia (lanes 2, 5, 7, 9, 11, and 13). Lanes 1 and 2, ORS571 (wild type); lane 3, ORS571A5 (nifA mutant); lanes 4 and 5, 57620 (glnB mutant); lanes 6 and 7, 57621 (glnK mutant); lanes 8 and 9, 57625 (glnB glnK mutant); lanes 10 and 11, ORS571A5/pRS1022 (containing A. caulinodans constitutive nifA [15]); and lanes 12 and 13, ORS571A5/pRJ7556 (containing B. japonicum constitutive nifA [12]).
nifA was expressed in the presence of ammonia in the glnB glnK double mutant strain, indicating that PII and GlnK may inhibit nifA transcription and/or regulate nifA posttranscriptionally under these conditions. This absence of ammonia regulation could be explained by the very low level of active GS, which could lead to a decrease in the glutamine pool and therefore to an increased α-cetoglutarate/glutamine ratio. This could mimic nitrogen fixation conditions, stimulating expression of nif genes, even in the presence of ammonia. However, similar amounts of active GS were found in the glnB mutant strain and glnB glnK double mutant strain in the presence of ammonia, but only the glnB glnK mutant had no ammonia regulation. Thus, PII and GlnK may be involved directly in the repression of NifA synthesis. This control is independent from NtrC, since a translational glnK-lacZ fusion (glnK is the only gene that is strictly under the control of NtrC to have been characterized for A. caulinodans) recombined into the chromosome of either the wild-type strain or the glnB glnK mutant is expressed at low levels in the presence of ammonia (1,500 and 2,000 Miller units/mg of protein, respectively, as contrasted with 15,000 Miller units/mg of protein in the wild-type strain under nitrogen-limiting conditions). Thus, the synthesis of a NifA protein by the glnB glnK mutant in the presence of ammonia cannot be accounted for by a constitutive NtrC activity.
We assessed NifA activity by integrating a translational nifH-lacZY fusion (15) into the chromosomes of the same strains. The activation of nifH transcription correlated with the detection of the NifA polypeptide in all but one case (Table 2). The A. caulinodans NifA protein was detected in the presence of ammonia if it was produced constitutively (Fig. 1). It did not activate A. caulinodans nifH expression, whereas the B. japonicum NifA protein, which is active in the presence of ammonia, did. This may be due to regulation of the activity or differences in the stability of the A. caulinodans NifA protein, in the presence of ammonia.
TABLE 2.
β-Galactosidase activities of the translational nifH-lacZY fusion recombined into the chromosomes of the wild-type and mutant strains of A. caulinodans carrying or not carrying the constitutively expressed nifA from A. caulinodans or B. japonicum
| Strain | β-Galactosidase sp acta
|
||
|---|---|---|---|
| N free, 3% O2b | NH4+, 3% O2c | NH4+, aird | |
| 57721 (wild type) | 14,229.0 ± 3,876.5 | 34.6 ± 32.5 | 10.5 ± 0.81 |
| 57820 (glnB mutant) | 20,123.0 ± 5,480.3 | 22.6 ± 18.3 | 14.6 ± 11.1 |
| 57821 (glnK mutant) | 9,804.6 ± 953.8 | 6.2 ± 5.5 | 4.0 ± 0.3 |
| 57720 (nifA mutant) | 11.5 ± 9.3 | 3.3 ± 2.3 | 9.9 ± 9.9 |
| 57825 (glnB glnK mutant) | 7,661.7 ± 478.8 | 6,535.4 ± 532.1 | 10.7 ± 8.3 |
| 57720 (nifA mutant)/ pRS1022e | 17,169.6 ± 3,463.2 | 91.0 ± 27.3 | 5.5 ± 2.7 |
| 57820 (glnB mutant)/ pRS1022e | 28,319.8 ± 4,652.6 | 305.5 ± 221.8 | 9.7 ± 0.6 |
| 57821 (glnK mutant)/ pRS1022e | 15,791.0 ± 3,111.6 | 798.6 ± 205.1 | 13.4 ± 7.3 |
| 57720 (nifA mutant)/ pRJ7556e | 31,650.4 ± 6,762.7 | 19,878.6 ± 6,333.1 | 175.75 ± 71.5 |
Specific activity of β-galactosidase in pure culture expressed in Miller units · mg of protein−1. Values are the means ± standard deviations from at least three independent experiments.
Cells were cultured for 4 h in nitrogen-free medium under microaerobic conditions.
Cells were cultured for 4 h in minimal medium containing 20 mM NH4+ under microaerobic conditions.
Cells were cultured for 4 h in minimal medium containing 20 mM NH4+ in the presence of air.
PII or GlnK may be required in any case because nifH transcription, under nitrogenase-derepressing conditions, in the glnB glnK mutant (strain 57825) is half that in the wild type (strain 57721), suggesting that PII and GlnK might also have a positive role in nifH transcription in the absence of ammonia. However, transcription levels were similar in the absence and presence of ammonia in the double mutant, suggesting that PII and GlnK are required for nif gene repression by ammonia (Table 2). The absence of either PII (strain 57820) or GlnK (strain 57821) did not lead to activation of nifH transcription by the constitutively expressed NifA, in the presence of ammonia (Table 2).
Two mechanisms have been put forward to account for the regulation of NifA activity in response to ammonia. Arsène et al. suggested that the N-terminal part of the A. brasilense NifA negatively regulates the activating domain, whereas PII maintains NifA in an active form under nitrogenase-derepressing conditions (1). This model is not applicable to A. caulinodans because (i) PII and GlnK are not required for NifA activity under nitrogen-derepressing conditions and (ii) NifA proteins from which the N terminus has been deleted are inactive (data not shown). NifA activity in K. pneumoniae is inhibited by NifL in the presence of excess ammonia. GlnK, whether uridylylated or not, is required to abolish the inhibition of NifA activity by NifL under nitrogen-limiting conditions, but this inhibition was restored in the presence of ammonia, suggesting the existence of another mechanism (14). This model could be applied to A. caulinodans if one postulates the existence of a repressor of NifA activity in the presence of ammonia.
Nitrogenase was active in the presence of ammonia in the glnB glnK mutant strain.
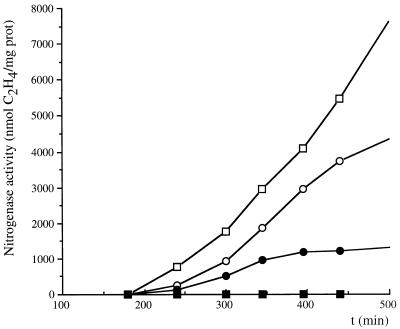
As nifH was expressed in the presence of ammonia in the glnB glnK mutant strain, we investigated whether the nitrogenase was active (17). Nitrogenase activities were similar in the wild-type strain and the glnB glnK mutant strain under nitrogenase-derepressing conditions (Fig. 2). No nitrogenase activity was detected in the wild-type strain in the presence of 10 mM NH4+, whereas nitrogenase activity was detected in the glnB glnK mutant strain (Fig. 2), suggesting that PII and GlnK may also be required for the regulation of nitrogenase activity.
FIG. 2.
Kinetics of nitrogenase activities in ORS571 (wild type) (open squares) or the 57625 strain (glnB glnK mutant) (open circles) under microaerobic conditions in nitrogen-free medium or of the same strains in medium with 10 mM ammonia added to the medium at time 0 (closed squares and closed circles, respectively).
In summary, PII and GlnK are the key elements controlling nitrogen fixation and ammonia assimilation in A. caulinodans. In the presence of ammonia, either protein is involved in the repression of nitrogen fixation, whereas under nitrogen-fixing conditions they stimulate GS deadenylylation. Future work should focus on determining the mechanisms by which these two proteins regulate both processes.
Acknowledgments
N. M.-R. is a recipient of a predoctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche.
We thank C. Elmerich for critical reading of the manuscript, M. de Zamaroczy for discussion, and N. Desnoues for skillful technical help. We also thank H.-M. Fischer for kindly providing a NifA antiserum and plasmid pRJ7556, which constitutively expresses the B. japonicum nifA gene.
REFERENCES
- 1.Arsène F, Kaminski P A, Elmerich C. Modulation of NifA activity by PII in Azospirillum brasilense: evidence for a regulatory role of the NifA N-terminal domain. J Bacteriol. 1996;178:4830–4838. doi: 10.1128/jb.178.16.4830-4838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson M R, Ninfa A J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–447. doi: 10.1046/j.1365-2958.1998.00932.x. [DOI] [PubMed] [Google Scholar]
- 3.Benelli E M, Souza E M, Funayama S, Rigo L U, Pedrosa F O. Evidence for two possible glnB-type genes in Herbaspirillum seropedicae. J Bacteriol. 1997;179:4623–4626. doi: 10.1128/jb.179.14.4623-4626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C M, Dilworth M J. Ammonia assimilation by Rhizobium cultures and bacteroids. J Gen Microbiol. 1975;86:39–48. doi: 10.1099/00221287-86-1-39. [DOI] [PubMed] [Google Scholar]
- 5.Carr P D, Cheah E, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. X-ray structure of the signal transduction protein PII from Escherichia coli at 1.9 angstrom. Acta Crystallogr Sect D. 1996;52:93–104. doi: 10.1107/S0907444995007293. [DOI] [PubMed] [Google Scholar]
- 6.Chaney A L, Marbach E P. Modified reagents for determination of urea and ammonia. Clin Chem. 1962;8:130–132. [PubMed] [Google Scholar]
- 7.Cheah E, Carr P D, Suffolk P M, Vasudevan S G, Dixon N E, Ollis D L. Structure of the Escherichia coli signal transducing protein PII. Structure. 1994;2:981–990. doi: 10.1016/s0969-2126(94)00100-6. [DOI] [PubMed] [Google Scholar]
- 8.de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Zamaroczy M, Paquelin A, Peltre G, Forchhammer K, Elmerich C. Coexistence of two structurally similar but functionally different PII proteins in Azospirillum brasilense. J Bacteriol. 1996;178:4143–4149. doi: 10.1128/jb.178.14.4143-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donald R G K, Ludwig R A. Rhizobium sp. strain ORS571 ammonium assimilation and nitrogen fixation. J Bacteriol. 1984;158:1144–1151. doi: 10.1128/jb.158.3.1144-1151.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfus B L, Elmerich C, Dommergues Y R. Free-living Rhizobium strain able to grow on N2 as the sole nitrogen source. Appl Environ Microbiol. 1983;45:711–713. doi: 10.1128/aem.45.2.711-713.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer H-M, Hennecke H. Direct response of Bradyrhizobium japonicum nifA-mediated nif gene regulation to cellular oxygen status. Mol Gen Genet. 1987;209:621–626. doi: 10.1007/BF00331174. [DOI] [PubMed] [Google Scholar]
- 13.Gebhardt C, Turner G L, Gibson A H, Dreyfus B L, Bergersen F J. Nitrogen-fixing growth in continuous culture of a strain of Rhizobium sp. isolated from stem nodules on Sesbania rostrata. J Gen Microbiol. 1984;130:843–848. [Google Scholar]
- 14.He L, Soupene E, Ninfa A, Kustu S. Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions. J Bacteriol. 1998;180:6661–6667. doi: 10.1128/jb.180.24.6661-6667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaminski P A, Elmerich C. The control of Azorhizobium caulinodans nifA expression by oxygen, ammonia and by the HF-I-like protein, NrfA. Mol Microbiol. 1998;28:603–613. doi: 10.1046/j.1365-2958.1998.00823.x. [DOI] [PubMed] [Google Scholar]
- 16.Kleiner D. Bacterial ammonium transport. FEMS Microbiol Rev. 1985;32:87–100. [Google Scholar]
- 17.Michel-Reydellet N, Desnoues N, Elmerich C, Kaminski P A. Characterization of Azorhizobium caulinodans glnB and glnA genes: involvement of the PII protein in symbiotic nitrogen fixation. J Bacteriol. 1997;179:3580–3587. doi: 10.1128/jb.179.11.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel-Reydellet N, Desnoues N, de Zamaroczy M, Elmerich C, Kaminski P A. Characterisation of the glnK-amtB operon and involvement of AmtB in methylammonium uptake in Azorhizobium caulinodans. Mol Gen Genet. 1998;258:671–677. doi: 10.1007/s004380050781. [DOI] [PubMed] [Google Scholar]
- 19.Morett E, Fischer H M, Hennecke H. Influence of oxygen on DNA binding, positive control, and stability of the Bradyrhizobium japonicum NifA regulatory protein. J Bacteriol. 1991;173:3478–3487. doi: 10.1128/jb.173.11.3478-3487.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawlowski K, Ratet P, Schell J, de Bruijn F. Cloning and characterization of nifA and ntrC genes of the stem nodulating bacterium ORS571, the nitrogen fixing symbiont of Sesbania rostrata: regulation of nitrogen fixation (nif) genes in the free-living versus symbiotic state. Mol Gen Genet. 1987;206:207–219. [Google Scholar]
- 21.Ratet P, Pawlowski K, Schell J, de Bruijn F. The Azorhizobium caulinodans nitrogen-fixation regulatory gene, nifA, is controlled by the cellular nitrogen and oxygen status. Mol Microbiol. 1989;3:825–838. doi: 10.1111/j.1365-2958.1989.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 22.van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]