Abstract
Intestinal homeostasis is key to the maintenance of good health. The small intestine plays important roles in absorption, digestion, hormonal and immune functions. Crypt base columnar (CBC) stem cells residing at the bottom of crypts are nurtured by Paneth cells, and together create the stem cell niche, the foundation of intestinal homeostasis. CBC stem cells replicate to replenish their number, or differentiate into a variety of epithelial cells with specialized functions. Notch signaling is a cell-cell signaling pathway that regulates both the proliferation and differentiation of CBC stem cells. NOTCH1 and NOTCH2 stimulated by canonical Notch ligands DLL1 and DLL4 mediate Notch signaling in the intestine that, in concert with other signaling pathways including the WNT and BMP pathways, determines cell fates. Importantly, interactions between Notch receptors and canonical Notch ligands are regulated by O-glycans linked to Ser/Thr in epidermal growth factor-like (EGF) repeats of the Notch receptor extracellular domain (NECD). The O-glycans attached to NECD are key regulators of the strength of Notch signaling. Imbalances in Notch signaling result in altered cell fate decisions and may lead to cancer in the intestine. In this review, we summarize the impacts of mutations in Notch pathway members on intestinal development and homeostasis, with a focus on the glycosyltransferases that transfer O-glycans to EGF repeats of NOTCH1, NOTCH2, DLL1 and DLL4.
Introduction
The intestine functions in the absorption and digestion of nutrients, and in endocrine and immune responses. The small intestine has renewal capability and is believed to be replaced almost entirely every 3–5 days in mice (1). Finger-like projections towards the lumen of the small intestine are called ‘villi’ and the invaginating regions in between are called ‘crypts’ (2). Intestinal epithelium consists of various cell types with unique functions. Multipotent stem cells expressing Lgr5 and Olfm4 reside at the bottom of crypts and are termed Crypt Base Columnar (CBC) stem cells (3). CBC stem cells self-renew or differentiate into several epithelial cell types generated via the absorptive lineage to produce enterocytes, or via the secretory lineage to produce antimicrobial peptide-secreting Paneth cells, hormone-secreting enteroendocrine cells, mucus-secreting goblet cells, and Tuft cells (4, 5). CBC stem cells initially give rise to intermediate progenitor cells which reside in the transit amplifying [TA] compartment until they differentiate into a specific epithelial cell type and migrate towards either the villus or the crypt. CBC stem cells also generate Bmi1-expressing cells at the +4 cell position from the most distal crypt cell (6). Bmi1+ cells are the principle reserve stem cell that can generate Lgr5+ CBC stem cells in response to stresses such as injury or inflammation (7–9). Notch signaling regulates the balance between Bmi1+ and Lgr5+ cell populations (7). However, cells of almost every differentiated type in the intestinal epithelium have been shown to function as reserve stem cells that give rise to Lgr5+ CBC stem cells following stress (8, 9). In addition to Notch signaling, several other signaling pathways, including WNT, BMP, Hedgehog, EGF and mTORC1 regulate proliferation and differentiation of intestinal epithelium (10–14). These signaling pathways may function between intestinal epithelial cells of the crypt and villus themselves, and between mesenchymal cells, fibroblasts, telocytes or immune cells in surrounding tissue. Thus, the signaling niche supporting replication and differentiation of the intestinal epithelium is extremely complex (8), and poorly understood in both mouse and human. A recent spatio-temporal, single-cell RNA-seq study proposes cell-cell signaling complexities using a technique called Clump-seq (15), a beginning to unraveling functional pathways.
Notch signaling is a highly-conserved, cell-cell signaling pathway necessary for embryonic development and the maintenance of many tissues in the adult (16, 17). Defects in Notch signaling lead to developmental diseases, severe pathologies in adults, and initiation or progression of several cancers. Mammals have four Notch receptors (NOTCH1-NOTCH4) and five canonical Notch ligands (DLL1, DLL3, DLL4, JAG1, JAG2), all of which are single-pass, transmembrane proteins (18, 19). The extracellular domain of Notch receptors (NECD) contains 29–36 EGF repeats that carry O-glycans, followed by three cysteine-rich Lin12/Notch repeats (LNR), and two heterodimeric regions, which form the negative regulatory region. The Notch intracellular domain (NICD) contains an RBP-Jκ-associated module (RAM), and ankyrin (ANK) repeats, both needed for interactions with the transcriptional repressor RBP-Jκ. Nuclear localization signals (NLS) and a transcription transactivating domain (TAD) lie between the ANK repeats and a C-terminal PEST domain. Notch signaling is initiated by Notch ligands on neighboring cells binding to Notch receptors. The Notch/ligand interaction induces conformational changes in NECD that expose a site above the membrane for cleavage by ADAM10. (The related metalloprotease ADAM17 is not required for Notch signaling in mouse intestine (20)). Endocytosis of the released NECD into the ligand-expressing cell is followed by cleavage within the Notch transmembrane domain by γ-secretase. The released NICD goes to the nucleus where it forms a complex with CBF1-Supressor of Hairless-LAG-1 (CSL, also known as RBP-Jκ), Mastermind-like (MAML), and other transcription factors, to stimulate the transcription of many target genes (21). Several Notch target genes are transcriptional repressors, including the hairy and enhancer-of-split 1 (Hes1) family of basic helix-loop-helix transcription factors (22, 23).
Notch signaling in intestinal epithelial cells
Each of the four Notch receptors and all five canonical Notch ligands are expressed in the small intestine (24, 25). At embryonic day E13.5, all five ligands and four Notch receptors are expressed in mesenchyme, while by E18.5 expression of ligands and Notch receptors has become scattered in different parts of intestine. At postnatal day P25, DLL1, DLL4, JAG1, JAG2, NOTCH1 and NOTCH2 are predominantly expressed within individual cells of the crypt while NOTCH3 and NOTCH4 are expressed in cells of the villus and the vasculature, respectively (24). In mouse intestinal organoids, JAG1, DLL1 and DLL4 are expressed in Paneth cells, DLL1 and DLL4 are expressed in goblet cells, while Notch receptors are activated in Lgr5+ CBC stem cells (26). The glycosyltransferases that modify specific EGF repeats and regulate Notch signaling are also restricted in expression, as described below (26).
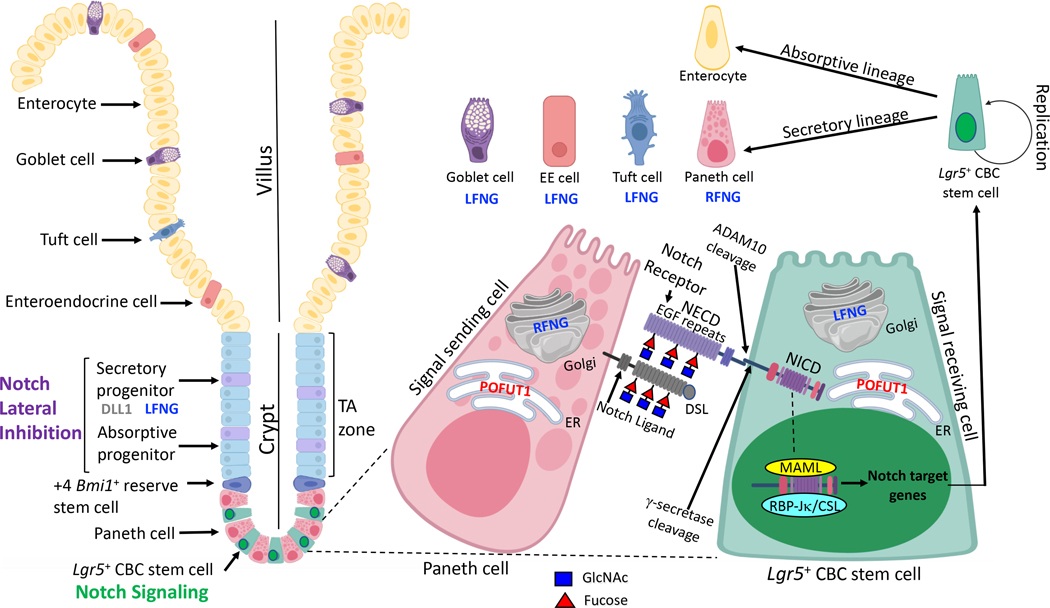
Cell fate decisions controlled by the Notch pathway in the intestine are summarized in Fig. 1. In the crypt, Notch receptors in CBC stem cells are activated by DLL1, DLL4 and JAG1 Notch ligands in Paneth cells. Absorptive or secretory progenitor cells in the TA region derive from CBC stem cells. Notch signaling stimulated by DLL1 in secretory lineage progenitors maintains the balance between absorptive and secretory progenitor cells by a Notch signaling mechanism termed lateral inhibition. The absorptive progenitors become enterocytes while secretory progenitors become goblet, enteroendocrine, Tuft or Paneth cells. Notch signaling strength is regulated by O-glycans attached to Ser/Thr in EGF repeats of the ECD of Notch receptors. Deletion of glycosyltransferase-encoding genes Pofut1, Lfng or Rfng has been shown to alter the balance of intestinal cell fates (7, 26, 27). Fig. 2 summarizes these findings and highlights outstanding questions concerning the interplay between different types of O-glycans on Notch receptors and Notch ligands. There are three classes of O-glycan named for the first sugar attached to an EGF repeat - O-fucose, O-GlcNAc and O-glucose. To date, only O-fucose glycans, initiated by the action of POFUT1 that transfers fucose from GDP-fucose in the endoplasmic reticulum (ER) to an EGF repeat, have been investigated in the intestine. The O-fucose on an EGF repeat becomes a substrate for Golgi enzymes Lunatic, Manic or Radical Fringe (LFNG, MFNG, RFNG), each of which transfers GlcNAc from UDP-GlcNAc to the O-fucose on an EGF repeat. The GlcNAc is often extended by the addition of galactose and sialic acid, by respective glycosyltransferases in the Golgi. O-glucose glycans and O-GlcNAc glycans are initiated in the same manner by their respective enzymes, POGLUT1 and EOGT (Fig. 2). Synergism between the three classes of O-glycans and their constituent sugars in regulating Notch signaling in the intestine is under investigation (28, 29).
Figure 1. Notch signaling in intestinal epithelium.
Lgr5+ CBC stem cells expressing NOTCH1 and NOTCH2 are present at the bottom of crypts interspersed with Paneth cells. Paneth cells express Notch ligands DDL1, DLL4 and JAG1 which stimulate Notch signaling in CBC stem cells. O-glycans attached to EGF repeats on the ECD of Notch receptors regulate receptor and ligand binding. Notch ligands also have EGF repeats modified by O-glycans, and bind Notch receptors via their N-terminal Delta-Serrate-LAG-2 (DSL) domain. POFUT1 initiates formation of O-fucose glycans by adding O-fucose to the consensus site present in numerous EGF repeats of NECD. O-fucose is extended by the addition of GlcNAc by LFNG or RFNG. LFNG is expressed in CBC stem cells, at increased levels in progenitors of the TA zone, and by goblet cells, enteroendocrine (EE) and Tuft cells. RFNG is expressed by Paneth cells. Interactions between Notch receptor O-glycans and Notch ligands determine the strength of Notch signaling, which regulates the replication and differentiation of CBC stem cells. Secretory lineage progenitors expressing DLL1 and LFNG induce lateral inhibition of enterocyte progenitors in the TA zone to control the balance of cell fate decisions and differentiated cells of the villus. The figure was created in part with Biorender.com.
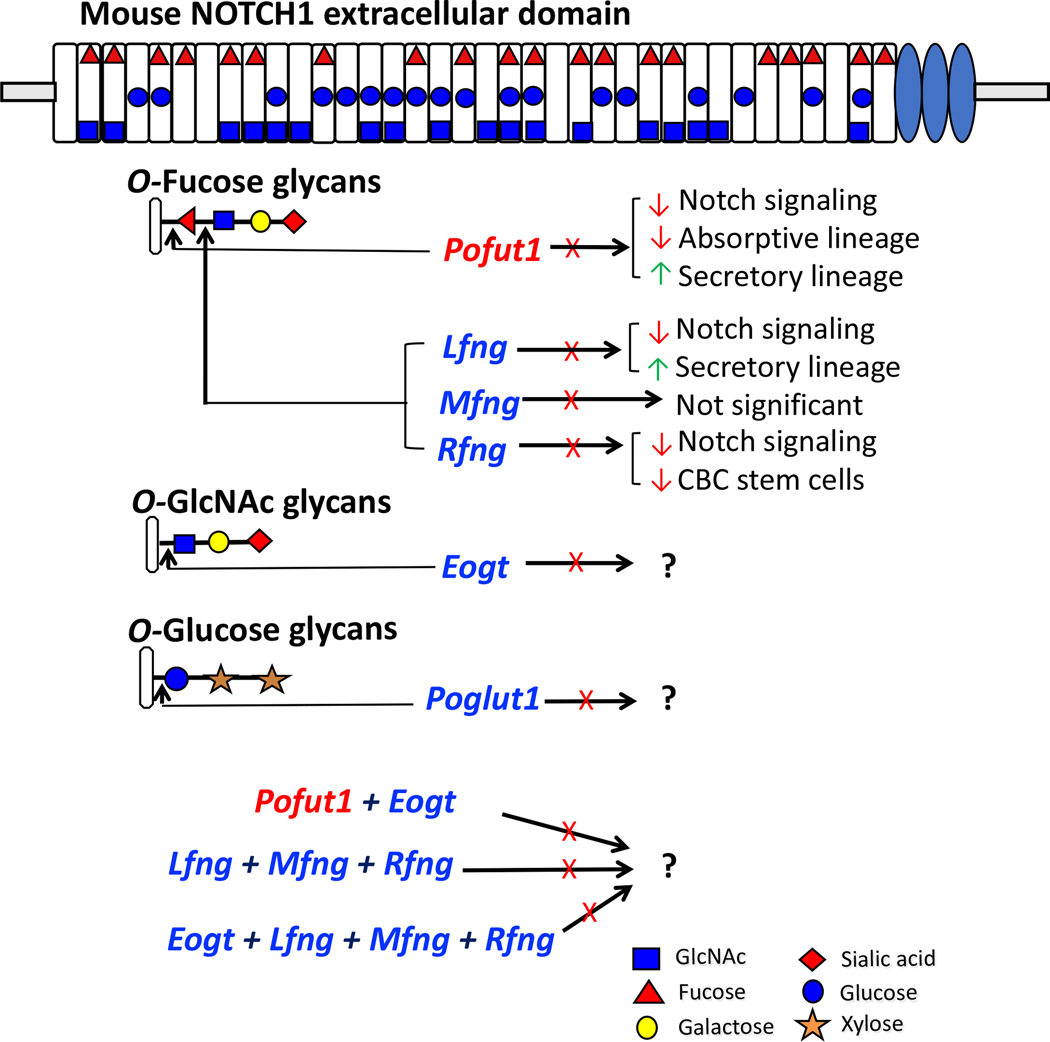
Figure 2. Roles of O-glycans in Notch signaling in small intestine.
Mouse NOTCH1 ECD consists of 36 EGF repeats which are modified by various O-glycans signified by the symbol for the initiating sugar. Each O-glycan is initiated at a consensus site in relevant EGF repeats by a specific glycosyltransferase as shown. The EGF repeats shown to be modified by a particular O-glycan are based on recent reviews for O-fucose and O-glucose (60) glycans, and for O-GlcNAc glycans (39). Further extension of the initial sugar by other glycosyltransferases leads to the formation of an O-glycan. The diagram summarizes the consequences of deleting different glycosyltransferase genes in the intestine (X, gene deleted; red arrow, decrease; green arrow, increase). Proposed or ongoing experiments investigating intestinal consequences in compound mutant mice are also presented.
Since Notch signaling is required for embryogenesis, conditional deletion must often be used to investigate roles for Notch signaling in intestine. The Vil1Cre/1000 strain is highly specific for deletion in intestinal epithelium, although it may also delete in testicular germ cells (30). In this review, names of different mouse strains expressing VillCre and floxed allele(s) are the same as used in the original publication. Deletion of Notch1 or Notch2 by VillCre, compared to deletion of both together, revealed functional redundancy (31). Following tamoxifen injection, adult Villin-CreERT2;N1F/F mice exhibit downregulation of Lgr5, Hes1 and Bmi1, and upregulation of Atoh1/Math1, leading to depletion of CBC stem cells and increased production of Paneth, goblet, and enteroendocrine cells of the secretory lineage. The intestinal phenotype displayed by combined deletion of Notch1 and Notch2 in Villin-CreERT2; N1F/F/N2F/F mice is more severe, and essentially identical to the phenotype induced by treatment with γ-secretase inhibitors (32, 33) [Table 1]. Both Notch1 and Notch2 are required for crypt regeneration post-irradiation (34). Transgenic mice constitutively overexpressing NICD also exhibit altered cell fates in intestine (35, 36). Up-regulation of Hes1 and concomitant downregulation of Math1 and Ngn3 transcripts were observed in mice with NICD overexpression, concomitantly with reduced differentiation of goblet and enteroendocrine cells, and increased CBC stem cell proliferation. Similarly, increased production of Lgr5-expressing stem cells was found in intestinal organoids overexpressing NICD (7).
Table 1.
Notch pathway mutant mouse models
| Experimental model | Phenotype | Gene expression of Notch target genes | References | |
|---|---|---|---|---|
| Downregulation | Upregulation | |||
| Villin-CreERT2; N1F/F | Increased goblet, enteroendocrine and Paneth cells. Decreased LGR5+ and EdU+ cells. Loss of Olfm4+ cells. |
Hes1, Lgr5 and Olfm4 | Math1, Ngn3, Spdef, Muc2, ChgA, Mmp7, Dll1 and Dll4 | (34) |
| Villin-CreERT2; N2F/F | Unaffected goblet cells | Not reported | No changes in Atoh1, ngn3, Muc2, ChgA, Mmp7, Dll1 and Dll4 | (34) |
|
Notch1/Notch2lox/lox-vil-Cre-ERT2 Villin-CreERT2; N1F/F/N2F/F (N1Δ/Δ;N2Δ/Δ) |
Highly increased goblet cells. Reduced Hes1+ cells. |
Hes1 | Muc2, Atoh1, Spdef, ChgA, Mmp7, Dll1 and Dll4 | (31) (34) |
| Dll1flox/flox vil-Cre-ERT2 | Increased goblet cells | Not reported | Not reported | (37) |
| Dll4flox/flox vil-Cre-ERT2 | Unaffected goblet cells | Not reported | Not reported | (37) |
| Dll1-Dll4vil-Cre-ERT2 | Highly increased goblet cells. Decreased Olfm4+ cells. Loss of Lgr5+, Ascl2+, Hes1+ Ki67+ cells. Inhibited NICD1 cleavage. |
Not reported | Math1 | (37) |
|
Villin-Cre;Adam10f/f
Villin-CreERT2;Adam10f/f or organoids from different ADAM10 mutants |
Increased goblet, Paneth, enteroendocrine and cleaved caspase-3+ cells. Decreased enterocytes, BrdU+ and Ki67+ cells. |
Hes1, HeyL and Olfm4 | Tff3, Muc2, Cryptdins, Mmp7, ChgA, Atoh1, Gfi1, Spdef, Ngn3 and Sox9 | (20) |
| γ-secretase inhibitors DBZ/DAPT treatment to mice models or mouse intestinal organoid | Inhibited NICD1 expression. Increased goblet, Paneth enteroendocrine, Tuft, SOX9+ and MATH1+ cells. Decreased Ki67+ and HES1+ cells. |
Olfm4 and Ascl2 | Math1, Lysozyme, Mmp7, EphB3, Ngn3, Spdef, Gfi1, Tff3 and Prom1. | (7, 31–33, 64, 65, 78) |
| RBP-Jfloxed/floxed/P450-Cre Rbpjlox/lox-vil-Cre-ERT2 RBP-Jvil-Cre-ERT2 Rbpjfl/fl:villin-creER RbpjF/F:Villin-CreERT2 |
Increased goblet cells and MATH1+ cells. Reduced HES1+ cells. Loss of Lgr5+, Olfm4+, Acl2+, Hes6+ and Ki67+ cells. |
Hes1 | Math1 | (32) (31) (37) (64) (34) |
| Vil-Cre;Rosa26DN-MAML/+ | Increased goblet, Paneth and enteroendocrine cells. Decreased HES1+ and Ki67+ cells. | Not reported | Not reported | (20) |
| Embryonic Hes1 KO Hes1-KO pups Villin-Cre;Hes1flox/– Hes1fl/fl Cre-ERT2 Hes1fl/flVillin-Cre |
Increased goblet, Paneth, cryptdin 4+ and cleaved caspase-3+ cells. Decreases Ki67+ cells. |
Reg3g, Saa1 and Reg3b | Math1, Lysozyme, Cryptdin 1, 4 and 5 Gast, Gcg, Smst, Cck, Sct, Tph, Gip, ChgA, Atoh1, Isl1, Nkx2–2, Pax6, Pax4, Dll1, Dll3, Hes3, Hes5, Tbp, Atoh5 and Neurod1. | (66) (67) (23) (69) (69) |
| Hes5 −/− | Increased goblet cells | Not reported | Muc2, Dll1 and Fbw7β | (68) |
| Villin-Cre;Hes1 flox/ ; Hes3 –/ ;Hes5 –/– | Increased goblet, Paneth, enteroendocrine and cleaved caspase-3+ cell. Decreased Ki67+ cells. |
Not reported | Not reported | (23) |
|
Math1β-Gal/-
Math1Δintestine/Lacz Math1LoxP/LoxP/AHCre Atoh1flox/flox:Fabp-Cre Atoh1fl/fl;villin-creER Lrig1CreERT2/+;Atoh1fl/fl |
Decreased goblet, enteroendocrine and Paneth cells. | Lack of cryptdins | Not reported | (72) (76) (65) (78) (64) (75) |
The name of each experimental mouse model is written as in the original article. The phenotype observed was based on histology, immunohistochemistry, Western analysis and flow cytometry. KO, knockout.
Inducible deletion of Dll1 in Dll1flox/floxvil-Cre-ERT2 adult mice causes increased production of goblet cells in the intestine, but no changes in the expression of Paneth or enteroendocrine cells (37). Conditional deletion of Dll4 or Jag1 in intestine does not affect secretory lineage differentiation (37). Thus, removal of an individual ligand is not sufficient to recapitulate the altered cell fates observed when Notch signaling is inhibited by γ-secretase inhibitors, or in mice conditionally lacking both Notch1 and Notch2 (31–34). Inducible deletion of both Dll1 and Jag1 had the same effect on stem cell proliferation and differentiation as deletion of Dll1 alone (37). Nevertheless, treatment of intestinal organoids with JAG1 induced the appearance of NICD, upregulation of Hes1 and Hes5 transcripts, and an increase in CBC stem cells, suggesting a Notch signaling role for JAG1 in intestine (7). Conditional deletion of both Dll1 and Dll4 causes massive formation of goblet cells and loss of Lgr5-, Olfm4- and Acsl2-expressing CBC stem cells (37). NICD1 was not detected in Dll1-Dll4vil-Cre-ERT2 intestine which exhibits a loss of Hes1-expressing cells, and elevated expression of Math1 transcripts (37). Mice with conditional deletion of both Notch1 and Notch2 are viable for at least 12 days (31, 34), but Dll1-Dll4vil-Cre-ERT2 mice exhibit a rapid weight loss, cachexia, poor grooming and die 4–6 days after the last dose of tamoxifen (37). Taken together, these findings suggest that DLL1 is the main physiological ligand supporting Notch signaling in mouse intestine but, DLL4 is also necessary for optimal Notch signaling (37) [Table 1].
O-Fucose glycans and Notch signaling in the intestine
Notch receptors and canonical Notch ligands are modified by O-glycans attached to most of the EGF repeats in their ECD (Fig. 2). The O-glycans regulate Notch ligand binding, and also may promote intracellular trafficking and stable expression at the cell surface (38–40). To investigate roles for O-fucose glycans in intestinal Notch signaling, the gene encoding protein O-fucosyltransferase 1 (Pofut1) was conditionally deleted using Villin-Cre (27). POFUT1 resides in the ER and transfers fucose from GDP-fucose to Ser (S) or Thr (T) in the consensus sequence C2XXXXS/TC3 in an EGF repeat (41). Pofut1 is ubiquitously expressed. Autosomal dominant mutations in POFUT1 cause Dowling-Degos disease 2 (DDD2), characterized by reticulate hyperpigmentation (42). Mice lacking Pofut1 die at ∼E9.5 with severe developmental abnormalities typical of embryos lacking different members of the Notch signaling pathway (43, 44). X-ray crystals of NOTCH1 and DLL4 fragments in complex show that O-fucose on EGF12 of NOTCH1 interacts directly with amino acids of DLL4 (45), and O-fucose on NOTCH1 EGF8 and EGF12 interact with the ligand binding domain of JAG1 (46). The biological importance of O-fucose on EGF12 was revealed in mice with a Notch1 point mutation which precludes the addition of O-fucose to EFG12. Depending on genetic background, homozygous mutants exhibit defective T and B cell development (47), or die at ∼E11.5 (48). Villin-Cre-mediated Pofut1 conditional deletion (cKO) showed over-production of Paneth, goblet and enteroendocrine cells at the expense of absorptive enterocytes in intestinal epithelium, with accompanying crypt hyperplasia (27). The expression of Hes1 was reduced, whereas Math1 expression was increased. Pofut1 cKO intestine exhibited T-lymphocyte infiltration, a Th1/Th17 immune response, bacterial translocation to mesenteric lymph nodes, altered species of mucus-associated gut microbiota, chronic intestinal inflammation, and formation of dysplastic foci, with occasional progression of tumors in small intestine as well as colon (27). Thus, lack of O-fucose transferred by POFUT1 leads to reduced Notch signaling, and altered cell fate decisions, as well as inflammation and adenoma in intestine. However, Pofut1F/F:Villin-Cre mice do not die shortly after birth like those conditionally deleted for both Dll1 and Dll4 (37). This may be because other O-glycans on Notch receptors and ligands support Notch signaling in the absence of O-fucose glycans, as discussed below. Lgr5-EGFP-creER/Pofut1flox/flox mice were used to investigate whether Notch signaling regulates interconversion between Bmi1+ reserve stem cells and Lgr5+ CBC stem cells. A decreased ratio of Bmi1+ to Lgr5+ cells was observed following tamoxifen treatment (7). This correlated with loss of Notch signaling following Pofut1 deletion. No NICD1 cleavage was seen in the intestine of Lgr5-EGFP-creER/Pofut1flox/flox mice, while expression of NICD1 via an Lgr5-EGFP-CreERT2/Rosa26-YFP-NICD transgene increased the Bmi1+to Lgr5+ cells ratio [Table 2].
Table 2.
Glycosyltransferase mutant mouse models
| Experimental model | Phenotype | Gene expression of Notch target genes | References | |
|---|---|---|---|---|
| Downregulation | Upregulation | |||
| LGR5-EGFP-creER/ POFUT-1flox/flox
Pofut1F/F :Villin-Cre |
Reduced ratio of BMI1+/LGR5+ cellsIncreased goblet, Paneth and enteroendocrine cells. Decreased HES1. Increased MATH1. Inhibited NICD1 and NICD2 cleavage. |
Hes1 | Notch3, Dll4, Math1, Insm1, Neurod1, Neurog3, Gfi1 and markers of goblet, Paneth and enteroendocrine cells. | (7) (27) |
|
Lfng null Lfng KD organoids |
Increased goblet cells. Inhibited NICD1 cleavage. Reduced NOTCH1-Fc binding. Decreased DLL1 and DLL4 expression. |
Hes1 | Atoh1 | (26) |
|
Rfng null Rfng KD organoids |
Depletion of CBCs. Reduced HES1 and HEY1. Reduced NOTCH1-Fc binding. Decreased DLL1 and DLL4 expression. |
Lgr5 | Not reported | (26) |
|
Mfng null Mfng KD organoids |
No significant phenotypes observed. | No change in Lgr5, Notch1, Hes1, Hes5, Dll1, Dll4 and Jag1. | Not reported | (26) |
The name of each experimental mouse model is written as in the original article. The phenotype observed was based on histology, immunohistochemistry, Western analysis and flow cytometry. KD, knockdown.
Extension of O-fucose on Notch receptors
O-fucose on an EGF repeat provides a substrate for Fringe glycosyltransferases to transfer GlcNAc from UDP-GlcNAc for the generation of a GlcNAcβ1,3Fuc O-glycan. This O-fucose glycan may be further extended by the addition of galactose and sialic acid, catalyzed by respective glycosyltransferases (49). Fringe was first reported as an essential gene required for wing formation in Drosophila (50), and subsequently shown to be a GlcNAc-transferase (51, 52). The three Fringe homologs LFNG, MFNG and RFNG, are expressed in intestine (24, 26). Mice globally deleted for Mfng do not exhibit significant changes in intestinal epithelium, but global deletion of Lfng or Rfng causes intestinal pathologies consistent with reduced Notch signaling (26).
Paneth cells in intestinal crypts support Notch signaling in adjacent CBC stem cells by expressing canonical Notch ligands (26, 53). Intestinal organoids in which Rfng is knocked down have fewer Lgr5-GFP+ stem cells. Similarly, crypts from Rfng null mice are depleted for Lgr5-GFP+ stem cells. Binding of NOTCH1-Fc was reduced on knockdown of Rfng in intestinal organoids, consistent with reduced DLL1 and DLL4 cell surface levels. Notch signaling and Notch targets HES1 and HEY1 were also reduced (26). Thus, Rfng promotes cell surface expression of DLL1 and DLL4 in Paneth cells, and enhances Notch signaling in CBC stem cells. Reduced cell surface expression of Notch ligands may reflect altered trafficking to, or stability at, the cell surface. A potentially direct, functional effect of associated O-fucose glycans on canonical Notch ligands was not investigated. Evidence that O-fucose glycans on a Notch ligand may have a functional effect is from mutational analysis of Dll3 to eliminate its O-fucose glycans (54). O-fucosylation-deficient Dll3 could not fully rescue somitogenesis in mice lacking Dll3 (54).
Lfng is most highly expressed by early progenitors of secretory cells in the transit amplifying (TA) zone in the upper crypt region, and in goblet, enteroendocrine and Tuft cells, throughout villi (26). Lineage tracing experiments show that Dll1 is also expressed in the earliest progenitors of secretory cells. Dll1GFP progenitors derive from Lgr5+ stem cells and may convert to Lgr5+ stem cells in cultured organoids, and after irradiation of the intestine in vivo (55). Lfng knockdown in intestinal organoids leads to less cell surface expression of DLL1 and DLL4, and decreased NOTCH1-Fc binding to ligand-expressing goblet cells. Thus, LFNG also appears to play a supporting role in ligand expression at the surface, and to thereby promote Notch signaling in neighboring cells, especially in the lower crypt (26). Lfng knockdown in Lgr5-GFP+ CBC stem cells reduces their colony-forming efficiency, suggesting that Lfng promotes Notch signaling in stem cells. Stem cells isolated from Lfng null mice exhibit low expression of Hes1 and increased expression of Math1 indicating reduced Notch signaling. Lfng null intestine exhibits accelerated secretory lineage differentiation with large numbers of Paneth, goblet and enteroendocrine cells (26) [Table 2]. Thus, in the intestine, both LFNG and RFNG act in signal-sending cells to promote cell surface expression of Notch ligands, and LFNG acts in CBC stem cells to promote Notch signaling and proliferation. Therefore, both Fringes function in regulating ligand-induced activation of Notch in CBC stem cells in the base of the crypt and also in Notch-controlled lateral inhibition in the TA zone, which maintains the balance of absorptive to secretory progenitors that give rise to differentiated cells of the villus. The high expression of Lfng in goblet, Tuft and enteroendocrine cells compared to CBC stem cells (26) is intriguing, and in need of further functional investigations.
O-GlcNAc glycans of Notch receptors
O-GlcNAc on NOTCH was first discovered on EGF20 of Drosophila Notch (56). Subsequent studies identified an ER-resident, EGF-domain-specific glycosyltransferase (EOGT) that catalyzes the transfer of GlcNAc from UDP-GlcNAc to EGF repeats in the consensus site C5XXXX(T/S)GXXC6 (57, 58). Mammalian O-GlcNAc can be further elongated by galactose and sialic acid in mammals (59). Defects in EOGT cause Adams-Oliver Syndrome 4 (AOS4), a rare, autosomal recessive disease (60). Eogt null mice are healthy and fertile (61). However, Notch signaling is impaired during post-natal retinal angiogenesis (61). Eogt deficiency decreases binding of Delta but not Jagged Notch ligands to certain cell types (61).
Roles for EOGT in intestinal epithelium have not been reported to date. In recent studies we have found that global deletion of Eogt caused only minor changes in morphology and gene expression in small intestine (28, 29). However, experiments to investigate whether O-GlcNAc glycans and EOGT might promote residual Notch signaling in the absence of POFUT1 revealed that both EOGT and POFUT1 contributed to Notch signaling. Global deletion of Eogt in combination with Pofut1F/F:Villin-Cre led to much more severe Notch signaling defects than previously observed in Pofut1F/F:Villin-Cre mice (29). Ongoing studies are investigating differential roles for canonical Notch ligands in interacting with the altered Notch receptors in mice lacking both Eogt and intestinal Pofut1. In other experiments, mice lacking all Fringe activities in intestinal epithelium, either via global knockout or conditionally in LfngF/FMfng−/−Rfng−/−:Villin-Cre mice exhibited a defective Notch signaling phenotype, albeit with some features different from the Pofut1:Villin-Cre phenotype. Surprisingly, deletion of Eogt along with Mfng and Rfng globally and Lfng conditionally in Eogt−/−LfngF/FMfng−/−Rfng−/−:Villin-Cre mice had the effect of rescuing many aspects of the Notch-defective phenotype observed in triple Fng knockout intestinal epithelium (28). Roles for Notch ligands and Notch target genes in facilitating this rescue effect are under investigation.
O-glucose glycans of Notch receptors
Roles for the O-glucose glycans (Fig. 2) in Notch signaling in intestine have not been reported. O-glucose is transferred to EGF repeats in the ER by POGLUT1 at Ser in the C1XSXP/AC2 consensus site, and the glucose may be extended by the addition of two xylose residues (62). Mutations in POGLUT1 cause Dowling Degos Disease 4 (DDD4) or Limb girdle muscular dystrophy R21 (LGMDR21) in humans (60). Cells lacking Poglut1 have reduced Notch signaling but are not defective in Notch ligand binding. Deletion of Poglut1 in the mouse is embryonic lethal (63).
Notch signaling effectors downstream of Notch ligand binding
Villin-CreERT2;Adam10f/f mice (20) lose body weight rapidly and die 7–9 days after the first dose of tamoxifen. Both P0 null and conditional mutants exhibit altered intestinal morphology with reduced cell proliferation, increased apoptosis and increased numbers of Paneth, goblet and enteroendocrine cells at the expense of enterocytes. Expression of Notch targets Hes1 and HeyL is reduced, while expression of Math1, Ngn3, Spdef, Gfi1 and Sox9 is upregulated. Similarly, organoids generated from Adam10 cKO mice exhibit decreased cell proliferation and a reduction in stem cells (20).
Several investigations of the effects of γ-secretase inhibitor dibenzazepine (DBZ) revealed conversion of proliferative cells in intestinal crypts into goblet, Paneth and enteroendocrine cells (33, 64, 65). Treatment of mice or intestinal organoids by another γ-secretase small molecule inhibitor DAPT also leads to increased production of secretory pathway cells and a decreased ratio of Bmi1+to Lgr5+ CBC stem cells (7, 33). Notch target genes are also dysregulated [Table 1]. Due to the well-established effects of γ-secretase inhibitors on Notch signaling, they are routinely used as a positive control to compare with results of other Notch pathway member deletions.
Conditional deletion of Rbp-Jκ leads to downregulation of Hes1 and Hes6 and upregulation of Math1, complete replacement of TA zone cells into post-mitotic goblet cells, and increased numbers of Paneth and enteroendocrine cells, indicative of enhanced differentiation to secretory cells (31, 32, 37, 64). Loss of Notch signaling upon Rbp-Jκ deletion results in loss of proliferative Ki67+ and Lgr5-, Olfm4-, Ascl2-expressing CBC stem cells (32, 37). The intestinal phenotype of Rbp-Jκ cKO mice is similar to that of Notch1/Notch2 and Dll1/Dll4 double cKO mice (31, 32, 34, 37). Intestine-specific expression of dominant-negative Maml (dn-Maml) also reduces Hes1 expression and cell proliferation, and causes large increases in Paneth, goblet and enteroendocrine cells in newborn pups (20) [Table 1].
Downstream effectors of Notch signaling are members of the hairy and enhancer-of-split family basic helix-loop-helix transcription factors including HES1, HES3, HES5, HES7 and related HEY proteins including HEY1, HEY2 and HEYL (22). Small intestine from Hes1 null embryos at E18 displays increased expression of Atoh1 and Atoh5 transcripts, consistent with increased production of enteroendocrine and goblet cells in the duodenum at E19 (66). Similarly, newborn Hes1 null pups show enhanced mRNA levels of Paneth cell markers cryptdins 1, 4 and 5, and a slight increase in mRNA of lysozyme, compared to controls (67). Conditional deletion of Hes1 at P2.5 leads to increased production of goblet, enteroendocrine and cleaved caspase-3+ cells, but these phenotypes disappear when mice reach ∼2 months of age (23). Interestingly, gene expression of Hes3 and Hes5 are upregulated in the intestine of Hes1 mutants at E18, possibly to compensate for the deficiency of Hes1 (66). Consistent with this interpretation, Villin-Cre;Hes1F/-Hes3−/−Hes5−/− mice show a marked increase in the production of goblet, Paneth, and enteroendocrine cells, increased apoptosis and decreased Ki67+ proliferating cells at 2 month, in contrast to mice lacking only Hes1 (23). Hes5 deletion in the intestine results in upregulation of E3 ligase Fbw7β which destabilizes NICD1, inhibits lateral inhibition and leads to an increased number of goblet cells (68). Hes1 deletion in adult mice causes upregulation of Math1 which leads to excessive production of Paneth & endocrine cells, and mucus hyperplasia with microbial dysbiosis and inflammation (69). Math1 null pups die just after birth due to failure of the respiratory system and defects in many essential cell lineages (70, 71). Intestine of Math1β-Gal/- at E18.5 shows a lack of goblet cells and enteroendocrine cells, and also loss of cryptdins in small intestine (72). Single cell RNA-seq indicated that Atoh1/Math1 regulates the process by which CBC stem cells give rise to secretory cells (73–75). Thus, Atoh1F/F:Fabp-Cre mice expressing the Fabpl4X AT−132Cre transgene (76) or β-naphthoflavone-induced Math1LoxP/LoxP:AHCre mice expressing Cre attached to the Cyp1a promoter (AH) (77), show drastic losses of goblet cells, Paneth cells and enteroendocrine cells in small intestine. This phenotype was not altered by treatment with the γ-secretase inhibitor DBZ which causes a massive formation of all secretory cells and loss of Ki67-expressing cells (65, 78). Atoh1/Math1 behaves as a tumor suppressor in colorectal cancer (79, 80). Hence, mice with Math1 deleted in intestine exhibit increased tumor formation in both adenomatosis polyposis coli (APC)min and azoxymethane mouse models (78, 80) [Table 1].
Conclusions
Roles for many members of the Notch signaling pathway have been identified in small intestine. Importantly, different functions of the various members have been revealed by individual and combined gene deletions, and by overexpression studies. Interestingly, the O-glycans on specific EGF repeats, the focus of this review, are required on the ECD of Notch receptors and DLL ligands for optimal Notch signaling in intestine. Loss of O-fucose glycans in Pofut1:Villin-Cre mice primarily reduces Notch signaling in CBC stem cells, with many effects on cell fate decisions, whereas loss of Rfng reduces cell surface expression of DLL ligands in Paneth cells, and thus the induction of Notch signaling. Loss of Lfng affects Notch signaling in CBC stem cells, and Notch ligand cell surface expression in ligand-expressing cells. Pofut1 deletion leads to mucosal inflammation, over-production of mucus, and altered microbiota, culminating in the formation of dysplastic foci with occasional tumor formation (27). In fact, increased expression of POFUT1 is a marker of tumor progression in colorectal cancer in humans (81, 82), and activation of NOTCH1 signaling appears to be a driver of colorectal cancer (83). γ-Secretase inhibitors that inhibit Notch signaling cause intestinal toxicity which may be avoided by using a new Notch inhibitor 6-alkynyl fucose (6AF), that prevents ligand-induced Notch activation of both NOTCH1 and NOTCH2 (84).
Perspectives.
Understanding the consequences of Notch signaling defects for intestinal development and maintenance, as well as roles for Notch signaling in colorectal cancer, are of critical importance to human health.
Identifying different aspects of Notch signaling that regulate intestinal differentiation and homeostasis, and that are dysregulated in cancer, is a current focus. For example, loss of O-glycans transferred by POFUT1, LFNG or RFNG in the intestinal epithelium leads to different defects in Notch signaling, and provide insights into how the Notch signaling pathway is regulated in the intestine.
Understanding synergies between different O-glycans in regulating Notch receptor-ligand interactions, their various effects on the strength of Notch signaling, and the consequences for cell fate decisions, will be important in identifying potential pathologies in human intestine arising from congenital or spontaneous mutations in the genes encoding Notch pathway members. Finding non-toxic inhibitors of Notch signaling to use in the treatment of colorectal cancer will also be important.
Acknowledgements
This work was supported by National Institutes of Health grant RO1 GM106417 to PS.
Abbreviations
- ADAM
a disintegrin and metalloprotease
- ANK
Ankyrin repeats
- AOS4
Adams-Oliver; Syndrome 4
- Ascl2
Achaete-scute homolog 2
- BMP
Bone Morphogenetic Proteins
- C2 domain
module at the N-terminus of Notch ligands
- CBC
Crypt base columnar
- CSL
CBF-1 suppressor of Hairless-LAG-1
- DBZ
dibenzazepine
- DDD
Dowling-Degos disease
- DLL
Delta like
- DSL
Delta-Serrate-LAG-2 domain
- EGF
epidermal growth factor-like
- EE
enteroendocrine
- EOGT
EFG-domain-specific glycosyltransferase
- ER
endoplasmic reticulum
- Gfi1
Growth factor independence 1
- GFP
Green fluorescent protein
- GXYLT
glucoside xylosyltransferase
- Hes
hairy and enhancer split
- IRES
An internal ribosome entry site
- JAG1
Jagged 1
- LFNG
Lunatic fringe
- LGMD
limb-girdle muscular dystrophy
- Lgr5
Leucine-rich repeat-containing G-protein coupled receptor 5
- LNRs
Lin12/Notch repeats
- MAML
mastermind-like
- Math1
Mouse atonal homolog 1
- MFNG
Manic fringe
- mTORC1
Mammalian target of rapamycin complex 1
- NECD
Notch extracellular domain
- Neu5Gc
N-Glycolylneuraminic acid
- Ngn3
Neurogenin 3
- NICD
Notch intracellular domain
- NRR
Negative regulatory region
- O-GlcNAc
O-linked GlcNAc or O-linked β-N-acetylglucosamine
- Olfm4
Olfactomedin-4 precursor
- POFUT1
Protein O-fucosyltransferase1
- POGLUT1
Protein O-glucosyltransferase
- RAM
RBP-Jκ associated module
- RBP-J κ
Recombination signal binding protein for immunoglobulin kappa J region
- RFNG
Radical fringe
- Ser
Serine
- Sox9
SRY-Box Transcription Factor 9
- Spdef
SAM pointed domain-containing Ets transcription factor
- Thr
Threonine
- 6AF
6-alkynyl fucose
Footnotes
Competing interests
Neither Pamela Stanley nor Mohd Nauman have competing interests associated with this manuscript.
References
- 1.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141(4):537–61. [DOI] [PubMed] [Google Scholar]
- 2.Hosoyamada Y, Sakai T. Structural and mechanical architecture of the intestinal villi and crypts in the rat intestine: integrative reevaluation from ultrastructural analysis. Anatomy and embryology. 2005;210(1):1–12. [DOI] [PubMed] [Google Scholar]
- 3.Schuijers J, van der Flier LG, van Es J, Clevers H. Robust cre-mediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem cell reports. 2014;3(2):234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker N Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. [DOI] [PubMed] [Google Scholar]
- 5.Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nature Reviews Gastroenterology & Hepatology. 2019;16(1):19–34. [DOI] [PubMed] [Google Scholar]
- 6.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan T, Than EB, Bu PC, Tung KL, Chen KY, Augenlicht L, et al. Notch signalling regulates asymmetric division and inter-conversion between lgr5 and bmi1 expressing intestinal stem cells. Scientific Reports. 2016;6(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurokawa K, Hayakawa Y, Koike K. Plasticity of Intestinal Epithelium: Stem Cell Niches and Regulatory Signals. Int J Mol Sci. 2020;22(1):357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bankaitis ED, Ha A, Kuo CJ, Magness ST. Reserve Stem Cells in Intestinal Homeostasis and Injury. Gastroenterology. 2018;155(5):1348–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He XC, Zhang J, Tong W-G, Tawfik O, Ross J, Scoville DH, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt–β-catenin signaling. Nature genetics. 2004;36(10):1117–21. [DOI] [PubMed] [Google Scholar]
- 11.Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132(2):279–89. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A, Sekiya S, Gunshima E, Fujii S, Taniguchi H. EGF signaling activates proliferation and blocks apoptosis of mouse and human intestinal stem/progenitor cells in long-term monolayer cell culture. Laboratory investigation. 2010;90(10):1425–36. [DOI] [PubMed] [Google Scholar]
- 13.Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, Costa M, et al. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell reports. 2015;11(1):33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Rychahou P, Wang Q, Weiss HL, Evers BM. TSC2/mTORC1 signaling controls Paneth and goblet cell differentiation in the intestinal epithelium. Cell death & disease. 2015;6(2):e1631-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manco R, Averbukh I, Porat Z, Bahar Halpern K, Amit I, Itzkovitz S. Clump sequencing exposes the spatial expression programs of intestinal secretory cells. Nat Commun. 2021;12(1):3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiological reviews. 2017;97(4):1235–94. [DOI] [PubMed] [Google Scholar]
- 17.Meisel CT, Porcheri C, Mitsiadis TA. Cancer Stem Cells, Quo Vadis? The notch signaling pathway in tumor initiation and progression. Cells. 2020;9(8):1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovall RA, Gebelein B, Sprinzak D, Kopan R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev Cell. 2017;41(3):228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai YH, VanDussen KL, Sawey ET, Wade AW, Kasper C, Rakshit S, et al. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147(4):822–34 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–35. [DOI] [PubMed] [Google Scholar]
- 22.Fischer A, Gessler M. Delta–Notch—and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic acids research. 2007;35(14):4583–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueo T, Imayoshi I, Kobayashi T, Ohtsuka T, Seno H, Nakase H, et al. The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139(6):1071–82. [DOI] [PubMed] [Google Scholar]
- 24.Schroder N, Gossler A. Expression of Notch pathway components in fetal and adult mouse small intestine. Gene Expr Patterns. 2002;2(3–4):247–50. [DOI] [PubMed] [Google Scholar]
- 25.Sander GR, Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004;52(4):509–16. [DOI] [PubMed] [Google Scholar]
- 26.Murthy PKL, Srinivasan T, Bochter MS, Xi R, Varanko AK, Tung KL, et al. Radical and lunatic fringes modulate notch ligands to support mammalian intestinal homeostasis. Elife. 2018;7:e35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guilmeau S, Flandez M, Bancroft L, Sellers RS, Tear B, Stanley P, et al. Intestinal deletion of Pofut1 in the mouse inactivates notch signaling and causes enterocolitis. Gastroenterology. 2008;135(3):849–60, 60 e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nauman M, Stanley P. Glycans that regulate Notch signaling in small intestine. Glycobiology. 2021;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nauman M, Varshney S, Stanley P. Regulation of Notch Signaling by O-Glycans in the Intestine. Glycobiology. 2020;30(12):1082. [Google Scholar]
- 30.Rutlin M, Rastelli D, Kuo WT, Estep JA, Louis A, Riccomagno MM, et al. The Villin1 Gene Promoter Drives Cre Recombinase Expression in Extraintestinal Tissues. Cell Mol Gastroenterol Hepatol. 2020;10(4):864–7 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riccio O, Van Gijn ME, Bezdek AC, Pellegrinet L, Van Es JH, Zimber‐Strobl U, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO reports. 2008;9(4):377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–63. [DOI] [PubMed] [Google Scholar]
- 33.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139(3):488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carulli AJ, Keeley TM, Demitrack ES, Chung J, Maillard I, Samuelson LC. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev Biol. 2015;402(1):98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435(7044):964–8. [DOI] [PubMed] [Google Scholar]
- 36.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102(35):12443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, et al. Dll1-and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140(4):1230–40. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varshney S, Stanley P. Multiple roles for O-glycans in Notch signalling. FEBS Lett. 2018;592(23):3819–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa M, Tashima Y, Sakaguchi Y, Takeuchi H, Okajima T. Contribution of extracellular O-GlcNAc to the stability of folded epidermal growth factor-like domains and Notch1 trafficking. Biochem Biophys Res Commun. 2020;526(1):184–90. [DOI] [PubMed] [Google Scholar]
- 40.Pandey A, Niknejad N, Jafar-Nejad H. Multifaceted regulation of Notch signaling by glycosylation. Glycobiology. 2021;31(1):8–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holdener BC, Haltiwanger RS. Protein O-fucosylation: structure and function. Curr Opin Struct Biol. 2019;56:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephan C, Kurban M, Abbas O. Dowling-Degos disease: a review. Int J Dermatol. 2020;60(8):944–50. [DOI] [PubMed] [Google Scholar]
- 43.Shi SL, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okamura Y, Saga Y. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech Dev. 2008;125(8):663–73. [DOI] [PubMed] [Google Scholar]
- 45.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347(6224):847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luca VC, Kim BC, Ge C, Kakuda S, Wu D, Roein-Peikar M, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355(6331):1320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge C, Stanley P. The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development. Proc Natl Acad Sci U S A. 2008;105(5):1539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varshney S, Wei HX, Batista F, Nauman M, Sundaram S, Siminovitch K, et al. A modifier in the 129S2/SvPasCrl genome is responsible for the viability of Notch1[12f/12f] mice. Bmc Developmental Biology. 2019;19(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struct Biol. 2011;21(5):583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irvine KD, Wieschaus E. Fringe, a Boundary-Specific Signaling Molecule, Mediates Interactions between Dorsal and Ventral Cells during Drosophila Wing Development. Cell. 1994;79(4):595–606. [DOI] [PubMed] [Google Scholar]
- 51.Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–75. [DOI] [PubMed] [Google Scholar]
- 52.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406(6794):411–5. [DOI] [PubMed] [Google Scholar]
- 53.Mei XL, Gu M, Li MY. Plasticity of Paneth cells and their ability to regulate intestinal stem cells. Stem Cell Research & Therapy. 2020;11(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serth K, Schuster-Gossler K, Kremmer E, Hansen B, Marohn-Kohn B, Gossler A. O-fucosylation of DLL3 is required for its function during somitogenesis. PLoS One. 2015;10(4):e0123776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Es JH, Sato T, Van De Wetering M, Lyubimova A, Nee ANY, Gregorieff A, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature cell biology. 2012;14(10):1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, et al. O-Linked N-Acetylglucosamine Is Present on the Extracellular Domain of Notch Receptors. Journal of Biological Chemistry. 2008;283(51):35486–95. [DOI] [PubMed] [Google Scholar]
- 57.Sakaidani Y, Nomura T, Matsuura A, Ito M, Suzuki E, Murakami K, et al. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat Commun. 2011;2(1):583. [DOI] [PubMed] [Google Scholar]
- 58.Muller R, Jenny A, Stanley P. The EGF repeat-specific O-GlcNAc-transferase Eogt interacts with notch signaling and pyrimidine metabolism pathways in Drosophila. PLoS One. 2013;8(5):e62835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogawa M, Senoo Y, Ikeda K, Takeuchi H, Okajima T. Structural Divergence in O-GlcNAc Glycans Displayed on Epidermal Growth Factor-like Repeats of Mammalian Notch1. Molecules. 2018;23(7):1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsumoto K, Luther KB, Haltiwanger RS. Diseases related to Notch glycosylation. Mol Aspects Med. 2020:100938. [DOI] [PubMed] [Google Scholar]
- 61.Sawaguchi S, Varshney S, Ogawa M, Sakaidani Y, Yagi H, Takeshita K, et al. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. Elife. 2017;6:e24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harvey BM, Rana NA, Moss H, Leonardi J, Jafar-Nejad H, Haltiwanger RS. Mapping Sites of O-Glycosylation and Fringe Elongation on Drosophila Notch. J Biol Chem. 2016;291(31):16348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138(10):1925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim TH, Li F, Ferreiro-Neira I, Ho LL, Luyten A, Nalapareddy K, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506(7489):511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, et al. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24(1):36–44. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki K, Fukui H, Kayahara T, Sawada M, Seno H, Hiai H, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochemical and Biophysical Research Communications. 2005;328(1):348–52. [DOI] [PubMed] [Google Scholar]
- 68.Sancho R, Blake SM, Tendeng C, Clurman BE, Lewis J, Behrens A. Fbw7 Repression by Hes5 Creates a Feedback Loop That Modulates Notch-Mediated Intestinal and Neural Stem Cell Fate Decisions. Plos Biology. 2013;11(6):e1001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo XK, Ou J, Liang S, Zhou X, Hu X. Epithelial Hes1 maintains gut homeostasis by preventing microbial dysbiosis. Mucosal Immunol. 2018;11(3):716–26. [DOI] [PubMed] [Google Scholar]
- 70.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390(6656):169–72. [DOI] [PubMed] [Google Scholar]
- 71.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284(5421):1837–41. [DOI] [PubMed] [Google Scholar]
- 72.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294(5549):2155–8. [DOI] [PubMed] [Google Scholar]
- 73.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5. [DOI] [PubMed] [Google Scholar]
- 74.Bottcher A, Buttner M, Tritschler S, Sterr M, Aliluev A, Oppenlander L, et al. Non-canonical Wnt/PCP signalling regulates intestinal stem cell lineage priming towards enteroendocrine and Paneth cell fates. Nat Cell Biol. 2021;23(1):23–31. [DOI] [PubMed] [Google Scholar]
- 75.Herring CA, Banerjee A, McKinley ET, Simmons AJ, Ping J, Roland JT, et al. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst. 2018;6(1):37–51 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132(7):2478–88. [DOI] [PubMed] [Google Scholar]
- 77.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136(5):903–12. [DOI] [PubMed] [Google Scholar]
- 78.Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of Notch/γ-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139(3):918–28. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leow CC, Romero MS, Ross S, Polakis P, Gao WQ. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 2004;64(17):6050–7. [DOI] [PubMed] [Google Scholar]
- 80.Bossuyt W, Kazanjian A, De Geest N, Van Kelst S, De Hertogh G, Geboes K, et al. Atonal homolog 1 Is a Tumor Suppressor Gene. Plos Biology. 2009;7(2):311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komor MA, de Wit M, van den Berg J, Martens de Kemp SR, Delis-van Diemen PM, Bolijn AS, et al. Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int J Cancer. 2020;146(7):1979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lv Y, Xie B, Bai B, Shan L, Zheng W, Huang X, et al. Weighted gene coexpression analysis indicates that PLAGL2 and POFUT1 are related to the differential features of proximal and distal colorectal cancer. Oncol Rep. 2019;42(6):2473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tyagi A, Sharma AK, Damodaran C. A Review on Notch Signaling and Colorectal Cancer. Cells. 2020;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schneider M, Kumar V, Nordstrom LU, Feng L, Takeuchi H, Hao HL, et al. Inhibition of Delta-induced Notch signaling using fucose analogs. Nature Chemical Biology. 2018;14(1):65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]