Abstract
2,4,6-Trinitrophenol (picric acid) and 2,4-dinitrophenol were readily biodegraded by the strain Nocardioides simplex FJ2-1A. Aerobic bacterial degradation of these π-electron-deficient aromatic compounds is initiated by hydrogenation at the aromatic ring. A two-component enzyme system was identified which catalyzes hydride transfer to picric acid and 2,4-dinitrophenol. Enzymatic activity was dependent on NADPH and coenzyme F420. The latter could be replaced by an authentic preparation of coenzyme F420 from Methanobacterium thermoautotrophicum. One of the protein components functions as a NADPH-dependent F420 reductase. A second component is a hydride transferase which transfers hydride from reduced coenzyme F420 to the aromatic system of the nitrophenols. The N-terminal sequence of the F420 reductase showed high homology with an F420-dependent NADP reductase found in archaea. In contrast, no N-terminal similarity to any known protein was found for the hydride-transferring enzyme.
The majority of nitroaromatic compounds in the environment are due to anthropogenic activities. Since nitrogroups can readily be converted into other functional groups, nitroaromatic compounds are important starting materials for the production of aromatic amines, hydrazo- and azo-compounds, isocyanates, benzidin derivatives, and haloaromatic structures. Hence, some of them occur as contaminants in wastewater. Trinitroaromatics are used as explosives and thus have been found as contaminants in ground water at certain military sites and former production facilities (28).
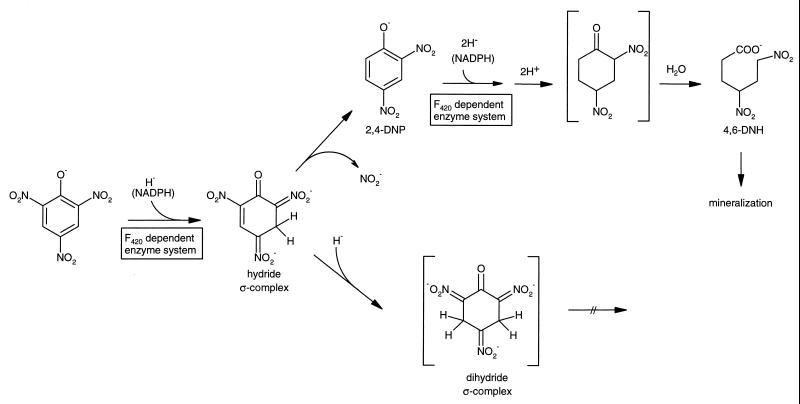
Due to the presence of electron-withdrawing nitro groups as substituents, dinitroaromatic compounds and particular trinitroaromatic compounds like 2,4,6-trinitrotoluene (TNT) and 2,4,6-trinitrophenol (picric acid) are readily susceptible to initial reductive rather than oxidative attack (23, 27). Consequently, initial oxidations by microbial mono- or dioxygenases of aerobic microorganisms are unknown for this class of xenobiotic compounds. Besides specific and unspecific reductions of the nitro groups (26), unusual hydrogenations of the aromatic ring system have been observed for picrate and TNT. Thus, hydride and dihydride complexes have been identified as initial metabolites (23, 27). In addition, the identification of 2,4-dinitrophenol (2,4-DNP) and 4,6-dinitrohexanoate (4,6-DNH) (13, 14, 23, 22) as metabolites of picrate indicates that extensive hydrogenation of the aromatic system (6 H per mol of picrate and 4 H per mol of 2,4-DNP) gives rise to a non-oxygenolytic ring cleavage. As outlined in Fig. 1, transformation of picrate via a hydride ς-complex to 2,4-DNP and 4,6-DNH is considered part of a productive catabolic sequence in Rhodococcus erythropolis HL PM-1, whereas the dihydride complexes of trinitroaromatics are dead-end products (14, 27).
FIG. 1.
Proposed steps in initial degradation of nitroaromatic compounds.
However, information on the origin and transfer of the hydride ion has still been missing. The present paper describes an enzyme system from Nocardioides simplex FJ2-1A that catalyzes hydride transfer from NADPH to picrate and 2,4-DNP. These enzymes of strain FJ2-1A are readily accessible compared to the previously described enzymes of R. erythropolis HL PM-1 (14), which strain is resistant to common cell-disrupting techniques.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
N. simplex FJ2-1A was previously isolated from picric acid containing wastewater and identified by 16S rRNA analysis by Rajan et. al (21). The strain was grown in batch cultures in a 10-liter fermenter (BIO-MAG; Fa. Bio-Chem-Color, Göttingen, Germany) at 30°C, 550 rpm, and 1.2 liters of air min−1 with 50 mM phosphate buffer (pH = 7.1) containing 0.7 mM picrate, 20 mM acetate, 0.5 g of yeast extract liter−1, 0.5 g of proteose peptone liter−1, 0.5 g of Casamino Acids liter−1, and mineral salts. Mineral salts without nitrogen contained 20 mg of Fe(III)-citrate liter−1, 1 g of MgSO4 · 7 H2O liter−1, 50 mg of CaCl2 · 2 H2O liter−1, and 1 ml of trace element solution (19). After consumption of 0.7 mM picrate, we added 0.35 mM picrate to maintain induction. The cells were then harvested by centrifugation immediately after decolorization of the medium. They were frozen in liquid nitrogen and stored at −30°C.
DOC Die-Away test.
The test was performed as described in the Organization for Economic Cooperation and Development (OECD) guideline for testing chemicals (18). Precultures were grown in a medium containing only mineral salts, as described above, and 0.7 mM picrate as the sole source of nitrogen, carbon, and energy. Cells were harvested by centrifugation and were washed twice. The test medium was inoculated to a final concentration of 30 mg of suspended solids liter−1 with an initial concentration of 146 mg liter−1 (0.64 mM) picrate, corresponding to 46 mg of dissolved organic carbon (DOC) liter−1 (blank without additional carbon source). Cells were incubated at a temperature of 20°C. The DOC concentration was monitored over a time period of 28 days. For comparison the same test was performed with unadapted activated sludge from a municipal sewage treatment plant. Benzoate at a concentration of 72 mg liter−1 (0.6 mM), corresponding to 50 mg of DOC liter−1 served as a reference substance.
Enzyme assay.
The activity of the system toward picrate or 2,4-DNP as substrate was routinely assayed under aerobic conditions by photometric determination of the decrease of absorption of NADPH at 340 nm or repeated recording of UV-visible spectra between 200 and 600 nm at 20°C. The test was conducted in 50 mM TRIS-HCl (pH = 7.5) containing 0.2 mM NADPH, 0.05 mM 2,4-DNP or 0.07 mM picrate, F420, and components A and B. Test solutions monitored by high-pressure liquid chromatography (HPLC) analysis contained 50 mM TRIS-HCl (pH = 7.5), 1.6 mM NADPH, 0.4 mM 2,4-DNP or 0.56 mM picrate, 0.04 mg of protein component A ml−1 (see below), 0.05 mg of protein component B containing Q Sepharose fractions ml−1, and nearly 5 nM coenzyme F420. Reactions were started by the addition of substrate (2,4-DNP or picrate). Reactions were stopped by the addition of phosphoric acid (85%), and the samples were frozen in liquid nitrogen and stored at −30°C until being analyzed by HPLC.
The assay for testing the function of component A used a solution containing citric acid-phosphate buffer (pH = 5.5) (9)–0.3 mM NADPH–0.06 mM coenzyme F420 and the partially purified component A from the Q Sepharose step at a protein concentration of 5 μg ml−1. The test was conducted under anaerobic conditions in rubber-stoppered cuvettes with nitrogen as the gas phase and was started by the addition of substrate.
Preparation of cell extract.
Frozen cells were resuspended in 50 mM TRIS-HCl (pH = 7.5) and disrupted by multiple French press treatment at 137 MPa. Cell debris were removed by centrifugation at 100,000 × g and 4°C for 45 min.
Purification of coenzyme F420, the F420-dependent NADPH oxidoreductase, and the hydride transferase.
The cell extract (105 mg of protein) was passed through a Q Sepharose column (1.6 × 10 cm) preequilibrated with basic buffer (50 mM TRIS-HCl [pH = 7.5]). Three of the different fractions were eluted from the column with a linear gradient (300 ml) from 0 to 1 M NaCl in basic buffer at NaCl concentrations of approximately 0.34, 0.42 and 0.5 M, respectively. They were designated components A, B, and C.
The fraction containing component C was heated to 100°C for 15 min. The precipitate was removed by centrifugation. The supernatant was diluted with basic buffer at a ratio of 1:1 and added to a Mono Q column (0.5 × 5 cm) preequilibrated with basic buffer. Component C eluted at an NaCl concentration of 0.42 M.
Ammonium sulfate was added to the fraction containing component A to a final concentration of 1.25 M. The solution was applied to a phenyl Superose HR column (1 × 10 cm) preequilibrated with 1.25 M ammonium sulfate in basic buffer. The protein was eluted with a linear gradient (100 ml) from 1.25 to 0 M ammonium sulfate in basic buffer at a concentration of 0.81 M (NH4)2SO4. Fractions containing component A were pooled. The sample was diluted with basic buffer and concentrated with a microconcentrator (10K; Filtron, Northborough, Mass.). This step was repeated twice to desalt the sample. Then the sample was passed through a Mono Q column (0.5 × 5 cm) preequilibrated with basic buffer. Component A was eluted with a linear gradient (70 ml) from 0 to 1 M NaCl in basic buffer at a concentration of 0.26 M NaCl.
The fraction containing component B was treated the same way as described above for component A. Component B was eluted with 0.69 M (NH4)2SO4 from the phenyl Superose column and with 0.37 M NaCl from the Mono Q column. The protein concentration was estimated according to Bradford (5) or Scopes (25) with bovine serum albumin as the standard. The molecular masses of the protein subunits were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 10% polyacrylamide gel stained with silver. The standard proteins were phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa). The concentration of F420 was calculated from the UV-visible spectrum with an extinction coefficient at 420 nm of 41.4 mM−1 cm−1 (pH = 7.5) (20).
Analysis of protein sequences.
Purified proteins were blotted onto a polyvinylidene difluoride membrane (ProSorb; Applied Biosystems, Weiterstadt, Germany) and were subjected to automatic sequencing (491 protein sequencer; Applied Biosystems). Database searches were performed with BLAST (2).
Chemical reduction of F420.
To obtain reduced coenzyme F420 we used an anaerobic solution of F420 in water, 50 mM TRIS-HCl (pH = 7.5), or 50 mM phosphate buffer (pH = 7.1). Reductants were applied as crystals or as solution. Sodium borohydride, sodium dithionite, zinc dust, and NADPH in the presence of component A served as reductants. Progress in reduction was monitored by UV-visible spectroscopy. The loss of absorption at 420 nm indicated the reduction of coenzyme F420.
Analytical methods.
For quantification of substrates and identification of coenzyme F420 an HPLC system with a Gromsil 120 Oc4 column (125 × 4 mm; particle size, 5 μm) was used. The mobile phase consisted of 80% water, 20% acetonitrile, and 0.26% H3PO4. Concentrations were determined at 210 nm. Identification via UV-visible spectrum was performed with a photodiode array (PDA) detector (UVD 340S; Gynkotek, Germering, Germany). DOC concentration was analyzed with a Beckmann Industrial 915B total organic carbon (TOC) analyzer. Nitrite concentration was determined by HPLC as described previously (23). Fluorescence of compound C was detected with a common UV lamp at 360 nm.
Materials.
All chemicals used were of the highest available purity and were purchased from Aldrich (Steinheim, Germany), Fluka (Neu-Ulm, Germany), Merck (Darmstadt, Germany), and Sigma (Deisenhofen, Germany).
RESULTS
Biodegradability of picrate.
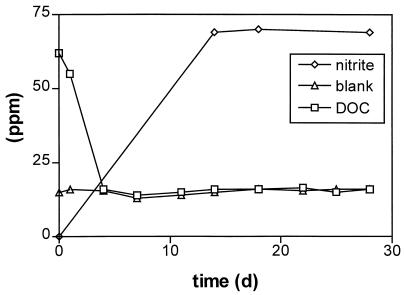
Picrate belongs to those xenobiotic compounds that are generally not degraded in natural populations, although single isolates have been described to degrade this compound. Therefore the biodegradability of picrate was compared by a standard OECD method (test on ready biodegradability) with N. simplex FJ2-1A and unadapted activated sludge. As shown in Fig. 2, biodegradation was monitored over a time period of 28 days. To avoid an accumulation of storage products the inoculum of N. simplex was pregrown with picrate as the sole source of carbon, nitrogen, and energy over three generations. The test sample was inoculated to a dry mass concentration of 30 mg liter−1. After 4 days nearly 100% of the initial carbon concentration was removed. Nitrite elimination amounted to 78% of the theoretically expected value. Presumably the organism assimilated some of the nitrite. In control experiments using activated sludge (30 mg of suspended solids liter−1) or heated inactivated cells of N. simplex, picrate was not degraded under the conditions of the OECD test. This underlines the ability of N. simplex to use the carbon backbone of picrate as the sole source of carbon and energy.
FIG. 2.
Decrease of DOC and release of nitrite by N. simplex in an OECD biodegradation test. The initial concentration of picrate was 146 mg liter−1 or 0.64 mM. The test was inoculated to a final cell density of 30 mg of suspended solids/liter.
Purification of a coenzyme F420-dependent NADPH oxidoreductase and a hydride transferase.
To identify the enzyme responsible for the initial hydrogenation step of picrate and 2,4-DNP, cell extract was fractionated by using an anion exchanger column. This resulted in a complete loss of activity. Combination of distinct fractions restored the activity of picrate and 2,4-DNP turnover. These fractions were designated components A, B, and C. Component C showed a yellowish green color and a bright green fluorescence at an excitation wavelength of 360 nm. In contrast, UV-visible spectra of components A and B displayed no characteristic absorbance.
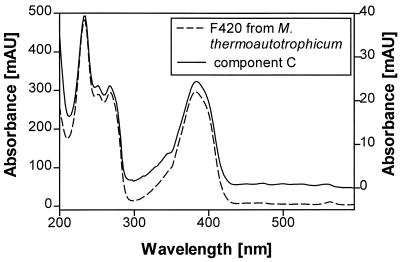
The component C-containing fractions were heated to 100°C for 15 min in order to denature protein if present. This procedure did not affect the activity of this component in the enzyme assay. Further investigations were performed with HPLC-PDA analysis and revealed a sharp band eluting at 1.9 min, which displayed a characteristic UV-visible spectrum (Fig. 3). Comparison of the retention time and UV-visible spectrum with those of an authentic preparation from Methanobacterium thermoautotrophicum identified this component as coenzyme F420. Enzyme tests, in which component C was replaced by the authentic coenzyme F420, revealed the same or even greater activity as in the reconstituted mix of components A, B, and C. Heating of component C-containing fractions or the authentic coenzyme F420 at 100°C for 2 h in 2 M HCl resulted in a total loss of activity. This indicates that hydrolysis products of coenzyme F420 cannot function as a cofactor. In contrast, a hydrolysis product from Streptomyces aureofaciens does act as a catalyst in the reduction of 5a,11a-dehydrochlortetracycline to chlortetracycline (17) or is involved in an enzymatic step leading to the synthesis of propyl proline in lincomycin production by Streptomyces lincolnensis (8).
FIG. 3.
UV-visible spectra recorded during HPLC run of coenzyme F420 of M. thermoautotrophicum (left y axis)/(right y axis) and component C under acidic conditions.
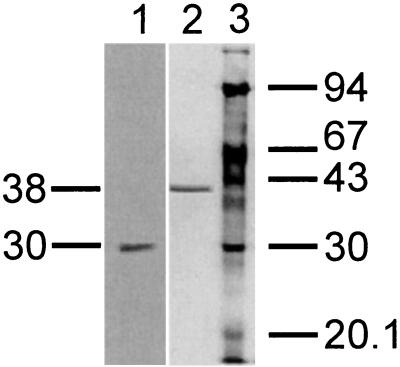
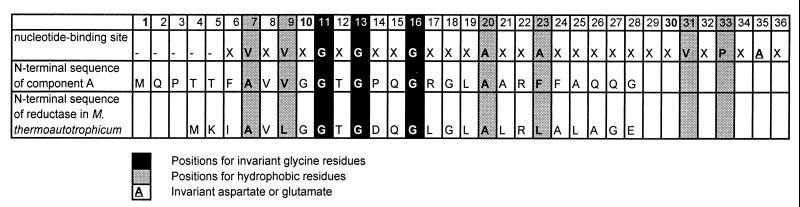
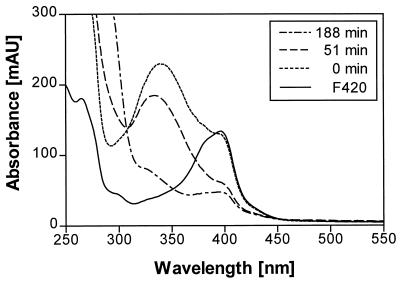
Practically no major activity was observed if one of the three components was missing in the enzymatic test. Obviously, all three components are necessary for activity. Further purification of components A and B yielded small amounts of homogenous proteins. Component A was judged to be homogenous on the basis of SDS-PAGE (Fig. 4). A 10% SDS-PAGE gel revealed a single protein band. The apparent molecular mass was calculated to be 30 kDa. N-terminal amino acid sequencing of the purified component A by Edman degradation revealed the sequence MQPTTFAVVGGTGPQGRGLAARFAQQG (Fig. 5). The characteristic pattern for a nucleotide binding site is found within these 27 amino acid residues. BLAST (2) comparison showed a very high similarity of nearly 66% to an F420-dependent NADP reductase of M. thermoautothrophicum. Component A from N. simplex catalyzes the reduction of coenzyme F420 (Fig. 6). This was also demonstrated for the reference sample F420 from M. thermoautothrophicum (not shown). NADPH is required as hydride donor and could not be substituted by NADH. The assay was carried out at pH = 5.5, which is the pH optimum for F420 reduction by the oxidoreductase of Methanogenium organophilum (4).
FIG. 4.
SDS-PAGE of components A and B. A 10% polyacrylamide gel was used and stained with silver. The molecular mass markers in lane 3 were phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (20.1 kDa), and α-lactalbumin (14.4 kDa). Lane 1, purified enzyme A; lane 2, purified enzyme B.
FIG. 5.
N-terminal sequence of component A compared to the homologous region of the F420-dependent NADP reductase of M. thermoautotrophicum and the nucleotide-binding motif.
FIG. 6.
UV-visible spectrum of coenzyme F420 from N. simplex FJ2-1A in citric acid-phosphate buffer at pH = 5.5 and repeated UV-visible spectra during the reduction of coenzyme F420 with NADPH-dependent F420 reductase from N. simplex after 0, 55, and 188 min in citric acid-phosphate buffer at pH = 5.5.
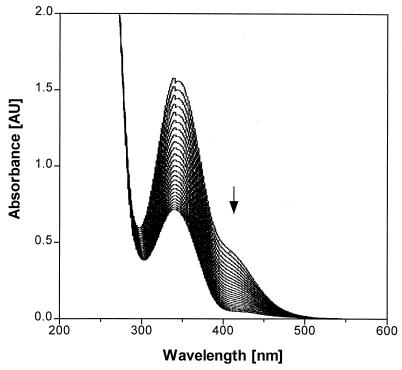
For component B an apparent molecular mass of 38 kDa was determined by 10% SDS-PAGE (Fig. 4). N-terminal amino acid sequencing revealed the sequence MIKGIQLHAWAGGPEMVEFAEIAAQEF. BLAST comparison gave no similarity to known protein sequences. The function of component B was investigated by repeated recording of the UV-visible spectrum (Fig. 7) during an aerobic enzymatic assay using a solution which contained 2,4-DNP, the partially purified components A, B, and C, plus NADPH as described in Materials and Methods. A decrease of absorption in the spectra represents disappearance of both NADPH and 2,4-DNP. The final spectrum corresponds to that for residual NADPH. Complete disappearance of 2,4-DNP was confirmed by HPLC analysis. As described above, component A reduced coenzyme F420 at the expense of NADPH. Addition of component B enabled the system to transfer hydride from coenzyme F420 to the aromatic ring of 2,4-DNP. Reduction of the aromatic ring is indicated by the following observations: (i) loss of UV-visible absorption characteristic for the nitrophenolic chromophore, (ii) detection of 4,6-DNH by HPLC (nonstoichiometric amounts), and (iii) absence of an amino aromatic structure, which was not detected by HPLC and might have been generated via nitro group reduction. Stoichiometry of 4,6-DNH formation cannot be expected because of its instability at pH = 7.5 (t1/2 = 55.6 min). No hydride transfer from coenzyme F420 to 2,4-DNP was observed without component B. Hence, component B is identified as a hydride transferase.
FIG. 7.
Repeated recording of the UV-visible spectrum during an enzymatic assay containing 0.05 mM 2,4-DNP, partially purified components A and B (0.244 mg of protein each), component C in a concentration of 8.3 μM F420, and 0.2 mM NADPH. The spectra were recorded at intervals of 3 min. The arrow indicates the disappearance of the characteristic absorption shoulder of 2,4-DNP. This was confirmed by HPLC analysis.
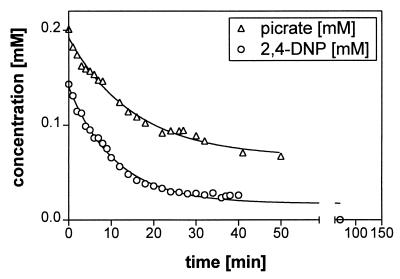
To investigate the enzymatic turnover by HPLC higher concentrations of substrate, cofactor, and partially purified protein were applied. Finally, the reaction was stopped with phosphoric acid. Picrate was tested as a substrate for the hydride transferase because the first step in picrate metabolism is the formation of a hydride ς-complex (21) which subsequently leads to 2,4-DNP (14, 23). As shown in Fig. 8 the initial activities for both substrates were similar. For picrate, this activity was 39 U mg of protein−1 and for 2,4-DNP it was 51 U mg of protein−1. This suggests that the hydride transferase is responsible for the initial reduction of the aromatic ring of 2,4-DNP and picrate.
FIG. 8.
Conversion of picrate and 2,4-DNP by partially purified enzymes of N. simplex. Concentrations were determined by HPLC analysis. The test contained 0.04 mg of component A ml−1, 0.05 mg of component B containing Q Sepharose fractions ml−1, and nearly 5 nM coenzyme F420.
DISCUSSION
During the in vitro studies on the biodegradation of picrate and 2,4-DNP, coenzyme F420 and its reductase, which are typical for methanogenic archaea, were surprisingly isolated from the aerobic bacterium N. simplex. These and a novel hydride transferase were identified as parts of a redox enzyme system which appears to have a key function in the catabolism of picrate and 2,4-DNP.
Picrate is readily biodegraded by N. simplex as shown by the DOC Die-Away test. The fast and complete carbon removal within 4 days strongly indicates that picrate is completely mineralized and utilized as the sole source of carbon and energy. In contrast, under the same standardized conditions picrate is not degraded by activated sludge or by inactivated cells of N. simplex. Hence, the highly efficient catabolic system of N. simplex, as well as that in R. erythropolis HL PM-1, as previously described (14), is unique, and obviously such organisms are not or are only marginally present in unadapted activated sludge.
With partially purified enzymes the system showed activity with 2,4-DNP and picrate as substrates. Activity was dependent on NADPH, which could not be replaced by NADH. From the transformation of picrate 2,4-DNP is transiently accumulated, and 4,6-DNH was identified as a metabolite from both nitrophenols. So it is evident that hydride ions were transferred to the π-electron-deficient system of these nitroaromatic compounds. The initial activities toward picrate and 2,4-DNP were very high but soon leveled off in the course of the reaction. This may be due to a low substrate affinity of the initial hydride transferase or, more likely, to decomposition of coenzyme F420 upon exposure to oxygen and light during the test. Such decomposition of coenzyme F420 from M. thermoautotrophicum by oxygen has been described by Schönheit et al. (24). Also, photodecomposition of coenzyme F420 has been reported for cell extracts from Methanobacterium sp. strain M.o.H. (6).
In order to resolve the hydride transfer from NADPH to picrate and 2,4-DNP into single reaction steps we reduced coenzyme F420 separately. Under anoxic conditions reduction of coenzyme F420 could be achieved with reductants such as sodium borohydride, sodium dithionite, zinc dust, or NADPH in the presence of component A. In the absence of reductant fast reoxidation of coenzyme F420 occurred. An excess of inorganic reductant, however, inevitably gave rise to chemical reduction of the nitrophenols, most likely by attacking the nitrogroups. Thus, enzymatic F420 reduction could not be detached from the hydride-transferring reaction, and an isolated system containing only component B, reduced coenzyme F420, and substrate could not be established. In addition, UV-visible spectroscopic measurements were hampered by the overlapping absorption of 2,4-DNP, picrate, and NADPH in a rather narrow wavelength range, so that the stoichiometry for the hydride transfer to the aromatic system of 2,4-DNP could not be fully defined. UV-visible spectroscopic estimation of NADPH consumption in the enzymatic turnover of 2,4-DNP indicates that the complete reduction of 1 mol of 2,4-DNP required 2 mol of NADPH.
Coenzyme F420 appears in archaea (4, 6, 10, 12, 15, 20) and also in some actinomycetes (11). It functions as a two-electron carrier for several redox reactions. It transfers, for example, hydride to methenyl- and methylenetetrahydromethanopterin during reduction of CO2 by methanogenic archaea. The reduced cofactor is generated by an F420-reducing hydrogenase, by two enzymes that together in vitro can catalyze the reduction of F420 with H2 (1), or by an F420-dependent NADP reductase (4). Up to now a hydrolysis product of F420 in aerobic bacteria has only been detected in S. aureofaciens (17) and S. lincolnensis (8).
Component A from N. simplex catalyzed the reduction of coenzyme F420 in an NADPH-dependent reaction and is therefore identified as an NADPH-dependent F420 reductase. By BLAST comparison (2) the N-terminal amino acid sequence of component A showed high similarity to an F420-dependent NADP reductase occurring in methanogenic archaea such as M. thermoautotrophicum (4), Methanogenium organophilum, and Methanococcus jannaschii and also in nonmethanogenic archaea such as Archaeoglobus fulgidus. Despite the large evolutionary distance between archaea and proteobacteria the oxidoreductase of the N. simplex strain has the same function as the F420-dependent NADP reductases in archaea. According to Chistoserdova et al. (7), this indicates that either the gene encoding this enzyme has been conserved, because these organisms evolved from a common ancestor, or it has been transferred horizontally between more recent ancestors. This consideration implies that the same enzymes in archaea and proteobacteria are involved in the reductive metabolism. Other authors have reported on an F420-dependent NADP reductase that was purified and characterized from S. griseus (11). However, metabolic functions could not be assigned to the oxidoreductase except for its being a ground-state electron carrier and a photosensitizer (11).
For the hydride transferase (component B) no N-terminal similarity to any known protein was found. It seems to be a novel enzyme which is responsible for the hydride transfer to the nitroaromatic ring. Due to the electron-withdrawing effect of the nitro substituents, the π-electron deficiency of the aromatic system favors nucleophilic hydride additions. Such reactions are known for chemical hydride donors like sodium borohydride generating hydride or even dihydride ς-complexes of nitroaromatic compounds (14, 23, 27). More recently, hydride complexes were observed in our lab as microbial metabolites of picrate and TNT. Whereas the hydride ς-complex of picrate is an intermediate of a productive catabolic pathway (see Fig. 1) (23), the hydride ς-complex of TNT (2,4,6-trinitrotoluene) is further reduced to a dihydride ς-complex which proved to be a stable dead-end product (27).
The hydride transferase (component B) of strain FJ2-1A obviously transfers hydride not only to picrate but also to 2,4-DNP. Thus, it appears to have multiple functions in the degradation of picrate via 2,4-DNP. Since 4,6-DNH is formed from 2,4-DNP with the partially purified enzyme, two hydride ions must be transferred. This is supported by estimation of NADPH consumption. In the first step a hydride ς-complex of 2,4-DNP may be generated. Thereafter, a second hydride ion may give rise to a dihydride complex. In the case of the picrate-dihydride complex, acid-catalyzed hydrolytic cleavage and subsequent decarboxylation can generate 1,3,5-trinitropentane (14). A dihydride complex of 2,4-DNP can form 2,4-dinitrocyclohexanone by protonation (shown in brackets in Fig. 1). As described in the literature for 2-nitrocyclohexanone (3, 16) such an activated α-nitroketo group may hydrolyze easily, yielding 6-nitrohexanoate (Fig. 1). Current analytical work with larger amounts of pure enzyme should provide sufficient quantitative data with respect to the stoichiometry of hydride transfer to picrate or 2,4-DNP and the formation of 4,6-DNH.
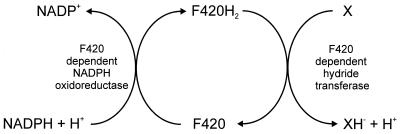
This report describes a novel enzyme system which is responsible for the transfer of hydride ions from NADPH to 2,4-DNP and picrate (Fig. 9). This system consists of three components, the NADPH-dependent F420 reductase, coenzyme F420 as a mediator, and a new hydride transferase. Such an F420-dependent enzyme system seems to be of general importance in picrate and 2,4-DNP metabolism: picrate- or 2,4-DNP-degrading strains from different habitats were all gram positive and exhibited the characteristic blue-green fluorescence exhibited by strain FJ2-1A when examined under the microscope.
FIG. 9.
Tentative scheme of the enzymatic transfer of hydride from NADPH to 2,4-DNP or picrate (X).
ACKNOWLEDGMENTS
We thank K. Thauer (Marburg, Germany) for providing authentic coenzyme F420 from Methanobacterium thermoautotrophicum, DuPont for supplying us with N. simplex FJ2-1A, Rainer Russ for stimulating discussions, Volker Nödinger for protein sequence analysis, and Monica Orendi for carrying out the DOC Die-Away tests.
REFERENCES
- 1.Afting C, Hochheimer A, Thauer R K. Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch Microbiol. 1998;169:206–210. doi: 10.1007/s002030050562. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballini R, Petrini M. Ring cleavage of cyclic 2-nitroketones by KF catalyst: a general synthesis of ω-nitroacids and ω-nitroesters. Synth Commun. 1986;16:1781–1788. [Google Scholar]
- 4.Berk H, Thauer R K. Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch Microbiol. 1997;168:396–402. doi: 10.1007/s002030050514. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman P, Toms-Wood A, Wolfe R S. Isolation and properties of a fluorescent compound, factor420 from Methanobacterium strain M.o.H. J Bacteriol. 1972;112:527–531. doi: 10.1128/jb.112.1.527-531.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 8.Coats J H, Li G P, Kuo M-S T, Yurek D A. Discovery, production, and biological assay of an unusual flavenoid cofactor involved in lincomycin biosynthesis. J Antibiot. 1989;42:472–473. doi: 10.7164/antibiotics.42.472. [DOI] [PubMed] [Google Scholar]
- 9.Dawson R M C, Elliot D C, Elliot W H, Jones K M. Vitamins and coenzymes. In: Dawson R M C, editor. Data for biochemical research. 3rd ed. Oxford, Great Britain: Oxford University Press; 1986. pp. 115–139. [Google Scholar]
- 10.Eirich L D, Vogels G D, Wolfe R S. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978;17:4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 11.Eker A P M, Hessels J K C, Meerwaldt R. Characterization of an 8-hydroxy-5-deazaflavin: NADPH oxidoreductase from Streptomyces griseus. Biochim Biophys Acta. 1989;990:80–86. doi: 10.1016/s0304-4165(89)80015-7. [DOI] [PubMed] [Google Scholar]
- 12.Jaenchen P, Schönheit P, Thauer R K. Studies on the biosynthesis of coenzyme F420 in methanogenic bacteria. Arch Microbiol. 1984;137:362–365. doi: 10.1007/BF00410735. [DOI] [PubMed] [Google Scholar]
- 13.Lenke H, Pieper D H, Bruhn C, Knackmuss H-J. Degradation of 2,4-dinitrophenol by two Rhodococcus erythropolis strains, HL 24-1 and HL 24-2. Appl Environ Microbiol. 1992;58:2928–2932. doi: 10.1128/aem.58.9.2928-2932.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenke H, Knackmuss H-J. Initial hydrogenation during catabolism of picric acid by Rhodococcus erythropolis HL 24-2. Appl Environ Microbiol. 1992;58:2933–2937. doi: 10.1128/aem.58.9.2933-2937.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X-L, White R H. Occurrence of coenzyme F420 and its γ-monoglutamyl derivative in nonmethanogenic archaebacteria. J Bacteriol. 1986;168:444–448. doi: 10.1128/jb.168.1.444-448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matlack A S, Breslow D S. Cleavage of 2-nitrocyclohexanone by base. J Org Chem. 1967;32:1995–1996. [Google Scholar]
- 17.McCormick J R D, Morton G O. Identity of cosynthetic factor 1 of Streptomyces aureofaciens and fragment FO from coenzyme F420 of Methanobacterium species. J Am Chem Soc. 1982;104:4014–4015. [Google Scholar]
- 18.Organization for Economic Cooperation and Development. Guidelines for testing of chemicals. Ready biodegradability: DOC Die-Away test 301A. Paris, France: Organization for Economic Cooperation and Development; 1993. [Google Scholar]
- 19.Pfennig N, Lippert K D. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Microbiol. 1966;55:245–256. [Google Scholar]
- 20.Purwantini E, Mukhopadhyay B, Spencer R W, Daniels L. Effect of temperature on the spectral properties of coenzyme F420 and related compounds. Anal Biochem. 1992;205:342–350. doi: 10.1016/0003-2697(92)90446-e. [DOI] [PubMed] [Google Scholar]
- 21.Rajan J, Valli K, Perkins R E, Sariaslani F S, Barns S M, Reysenbach A-L, Rehm S, Ehringer M, Pace N R. Mineralization of 2,4,6-trinitrophenol (picric acid): characterization and phylogenetic identification of microbial strains. J Ind Microbiol. 1996;16:319–324. doi: 10.1007/BF01570041. [DOI] [PubMed] [Google Scholar]
- 22.Rieger P-G, Knackmuss H-J. Basic knowledge and perspectives on biodegradation of 2,4,6-trinitrotoluene and related nitroaromatic compounds in contaminated soil. In: Spain J C, editor. Biodegradation of nitroaromatic compounds. New York, N.Y: Plenum Press; 1995. [Google Scholar]
- 23.Rieger P-G, Sinnwell V, Preuss A, Franke W, Knackmuss H-J. Hydride-Meisenheimer complex formation and protonation as key reactions of 2,4,6-trinitrophenol biodegradation by Rhodococcus erythropolis. J Bacteriol. 1999;181:1189–1195. doi: 10.1128/jb.181.4.1189-1195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schönheit P, Keweloh H, Thauer R K. Factor F420 degradation during exposure to oxygen. FEMS Microbiol Lett. 1981;12:347–349. [Google Scholar]
- 25.Scopes R K. Measurement of protein by spectrometry at 205 nm. Anal Biochem. 1974;59:277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- 26.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 27.Vorbeck C, Lenke H, Fischer P, Spain J C, Knackmuss H-J. Initial reductive reactions in aerobic microbial metabolism of 2,4,6-trinitrotoluene. Appl Environ Microbiol. 1998;64:246–252. doi: 10.1128/aem.64.1.246-252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyman J F, Serve M P, Hobson D W, Lee L H, Uddin D E. Acute toxicity, distribution, and metabolism of 2,4,6-trinitrophenol (picric acid) in Fischer 344 rats. J Toxicol Environ Health. 1992;37:313–327. doi: 10.1080/15287399209531672. [DOI] [PubMed] [Google Scholar]