Abstract
The pckA gene, encoding the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK), was cloned by PCR amplification from the purple nonsulfur bacterium Rhodopseudomonas palustris No. 7. Sequencing of a 2.5-kb chromosomal SmaI-PstI fragment containing the structural gene revealed an open reading frame encoding 537 amino acids, homologous to known pckA genes. Primer extension analysis identified a transcriptional start site 72 bp upstream of the pckA initiation codon and an upstream sequence similar to ς70 promoters. Studies of a pckA-lacZ gene fusion indicated that when cells were grown in minimal media with various carbon sources, such as succinate, malate, pyruvate, lactate, or ethanol, under both anaerobic light and aerobic dark conditions, the pckA gene was induced in log phase, irrespective of the carbon source. A R. palustris No. 7 PEPCK-deficient strain showed growth characteristics identical to those of the wild-type strain either anaerobically in the light or aerobically in the dark when a C4-dicarboxylic acid, such as succinate or malate, was used as a carbon source. These results indicate that in R. palustris No. 7, an alternative gluconeogenic pathway may exist in addition to PEPCK.
Phosphoenolpyruvate carboxykinase (PEPCK) catalyzes the decarboxylation and phosphorylation of oxaloacetate (OAA) to form phosphoenolpyruvate (PEP) in most organisms. This reaction is the first step in the gluconeogenic pathway, in which citric acid cycle intermediates are converted to hexose. PEPCKs have been generally classified according to their nucleotide specificities: enzymes from bacteria, yeast, and plants mainly use adenosine nucleotides, but enzymes from a variety of eukaryotes and mammals use guanosine or inosine phosphates (24). PEPCK also appears to have an absolute requirement for a divalent metal ion, such as Mn2+, although other divalent ions, such as Mg2+ or Co2+, can substitute for it with reduced activity (44).
In Escherichia coli, besides the PEPCK reaction, there is another pathway to form PEP from C4-dicarboxylic acid which involves the NAD- and NADP-dependent malic enzymes (MAEA and MAEB) and phosphoenolpyruvate synthase (PPS). Strains deficient in all three enzymes cannot grow on C4-dicarboxylic acids as a sole carbon source, whereas strains retaining one of these enzymes are still able to grow (14). In addition, a PEPCK and PPS double mutant also cannot grow on C4-dicarboxylic acids due to the lack of PEP (11).
In Rhizobium species, a PEPCK-deficient mutant fails to grow on minimal medium with succinate or other citric acid cycle intermediates but can grow on glucose and glycerol as a sole carbon source, suggesting that PEPCK functions here as a key gluconeogenic enzyme (30).
Conversely, PEPCK also functions in the anaplerotic pathway of some organisms, such as Anaerobiospirillum succiniciproducens (37), Alcaligenes eutrophus (39), Ruminococcus flavefaciens (40), Dipetalonema viteae (4), and Trypanosoma cruzi (6), where it forms OAA from PEP, which in turn enters the citric acid cycle. Thus, depending on the organism, PEPCK may have different physiological roles as a gluconeogenic or an anaplerotic enzyme.
Purple nonsulfur bacteria (PNSB) are metabolically versatile organisms which are able to grow either aerobically in the dark or anaerobically in the light using hydrogen or organic compounds as electron donors and subsequently fixing CO2 (16). PNSB provide useful tools for the study of carbon metabolism, including CO2 fixation; however, carbon metabolism under both aerobic dark and anaerobic light conditions and the regulation of these reactions are still unclear in these bacteria.
The PNSB Rhodobacter capsulatus and Rhodobacter sphaeroides can utilize sugars, such as sucrose, fructose, and glucose, or glycerol as carbon sources (2), and a phosphoenolpyruvate-sugar phosphotransferase system which functions in the transport and phosphorylation of sugars has been already reported in R. capsulatus (46). However, Rhodopseudomonas palustris No. 7, which is also a PNSB, can use sugars or glycerol only poorly as the sole carbon source (2). This could suggest that gluconeogenesis is important in the supply of sugars for cellular metabolism in R. palustris No. 7.
Furthermore, the analysis of a phosphoenolpyruvate carboxylase (PEPC)-deficient mutant of R. palustris No. 7 indicates that other anaplerotic enzymes besides PEPC may exist (18). It was reported that PEPCK must function in vivo to produce PEP in R. sphaeroides (32). Because the anaplerotic reaction shows great variety in various PNSB (20), PEPCK still ought to be considered a candidate as an anaplerotic enzyme alternative to PEPC in R. palustris No. 7. These observations prompted us to clarify the role of PEPCK as a gluconeogenic or anaplerotic enzyme.
Here we report cloning, sequencing, transcriptional mapping, and expression studies of the pckA gene from the PNSB R. palustris No. 7. We also analyze the physiological role of PEPCK in this strain by constructing pckA-deficient mutants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+ lacIqlacZΔM15] | Takara |
| 1321 | pckA maeA maeB | 14 |
| SM10 λpir | thi-1 thr leu tonA lacY supE recA::RP4-2Tc::Mu, Kmr λpir | 27 |
| P2392 | e14− (McrA−) hsdR514 supE44 supF58 lacY1 or Δ(lacIZY)6 galK2 galT22 metB1 trpR55 (P2 lysogen) | Stratagene |
| R. palustris | ||
| No. 7 | Wild type | 10 |
| ppc mutant | ppc::Kmr | 18 |
| pckA mutant | pckA::Kmr | This work |
| pckA ppc mutant | pckA::Tcrppc::Kmr | This work |
| Plasmids | ||
| pUC118 | Apr; α-lac/MCS M13 ori | Takara |
| pGEM-T | Apr; α-lac/MCS; PCR cloning vector | Promega |
| pMC1871 | Tcr; lacZ fusion vector | Pharmacia |
| pUC4K | Apr Kmr; source of Kmr cartridge | Pharmacia |
| pGP704 | Apr; oriR6K, mobRP4 | 27 |
| pMG102 | Kmr; pHSG298 derivative; E. coli-Rhodopseudomonas sp. shuttle vector | 17 |
| pMG200 | Apr; pUC118 with a 2.5-kb SmaI-PstI fragment containing the R. palustris No. 7 pckA gene | This work |
| pMG201 | Kmr; pMG102 with a 2.5-kb XbaI PCR-amplified fragment containing the R. palustris No. 7 pckA gene | This work |
| pMG202 | Kmr; pMG102 with a 5.0-kb SalI fragment containing the pckA-lacZ fusion | This work |
| pMG300 | Apr Gmr; 1.75-kb EcoRI fragment of Gmr cartridge inserted into pGP704 EcoRI site | 18 |
| pMG312 | Apr Kmr Gmr; 2.8-kb KpnI pckA::Kmr fragment cloned into pMG300 KpnI site | This work |
Chemicals.
All chemicals were of the highest available purity and were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Sigma (St. Louis, Mo.).
Culture conditions.
E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium (36). R. palustris No. 7 was cultivated aerobically in the dark at 35°C in van Niel’s complex medium (45) or in synthetic minimal medium (PMM) without yeast extract (10) and supplemented with a given 20 mM carbon source. Anaerobic growth in the light was at 35°C in synthetic medium containing 20 mM carbon source. When appropriate, media were supplemented with antibiotics. Final concentrations for E. coli were (per milliliter) 50 μg of ampicillin, 50 μg of kanamycin, 20 μg of tetracycline, and 10 μg of gentamicin; for R. palustris No. 7, the final concentrations were (per milliliter) 300 μg of gentamicin, 100 μg of streptomycin, and 200 μg of kanamycin.
DNA manipulations.
Plasmid DNA was isolated by the alkaline lysis procedure (36). Restriction endonucleases and T4 ligase were obtained from Takara (Kyoto, Japan) and used according to the manufacturer’s instructions. E. coli strains were transformed by the CaCl2 method (36). R. palustris No. 7 was transformed by electroporation (7). Restriction fragments were isolated, when required, from agarose gels (1% [wt/vol]) with the Prep-a-gene matrix (Bio-Rad, Richmond, Calif.) according to the manufacturer’s instructions.
PCR.
All PCRs were carried out in a Perkin-Elmer (Foster City, Calif.) DNA thermal cycler model 480 according to the manufacturer’s instructions. For amplification of the pckA gene of R. palustris No. 7, degenerate primers were synthesized. Primer 1 (5′-TCTCGGATCCYTIATIGGIGAYGAYGARCA-3′ [I, inosine; R, A or G; Y, C or T]) and primer 2 (5′-TCTCGGATCCGGYTCIGTIAYICCIYKYTCIGTICCIGC-3′ [K, G or T]) were made for the amino acid sequences of a highly conserved domain of PEPCK proteins, LIGDDEH and AGTE(R/Q/K)G(I/V)T, respectively. PCR fragments were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The PCR fragments were cloned in the pGEM-T vector (Promega Biotech, Madison, Wis.).
DNA sequencing.
pGEM-T clones were sequenced by the dideoxy chain termination method as described by Sanger et al. (38) with a model 373A DNA sequencer (Applied Biosystems–Perkin-Elmer, Foster City, Calif.). Suitable λFIXII clones were identified by hybridization with probes derived from pGEM-T clones. These clones were the sources of restriction fragments, which were then cloned into pUC118 and sequenced as above. The nucleotide sequences of both strands were determined. DNA sequence data were analyzed with the INHERIT programs (Applied Biosystems–Perkin-Elmer) and the Genetyx programs (Software Development, Tokyo, Japan).
RNA isolation and primer extension.
Total RNA was extracted from R. palustris No. 7 cells as previously described (18). Primers were synthesized with 5′ ends at positions 620, 640, and 660, complementary to the mRNA sequence within the structural gene, which begins at sequence position 585. Figure 1A shows the experimental design. The primers were labelled with [γ-32P]ATP (36). Primer extension reactions were assembled as described in Babst et al. (1). Primer extension products were separated on 6% DNA sequencing gels, and the sequence position was determined by alignment with an [α-32P]dATP-labelled DNA sequencing ladder obtained from supercoiled pMG200 DNA, containing the pckA gene, by the dideoxy method (U.S. Biochemicals Sequenase protocol) and with the same primer. Sequencing reactions were performed in the presence of 5% dimethyl sulfoxide. The gels were cut in half, dried, and exposed in two separate Fujix Image cassettes. Autoradiography was performed with a Fujix BAS2000 image analyzer system (Fuji, Tokyo, Japan).
FIG. 1.
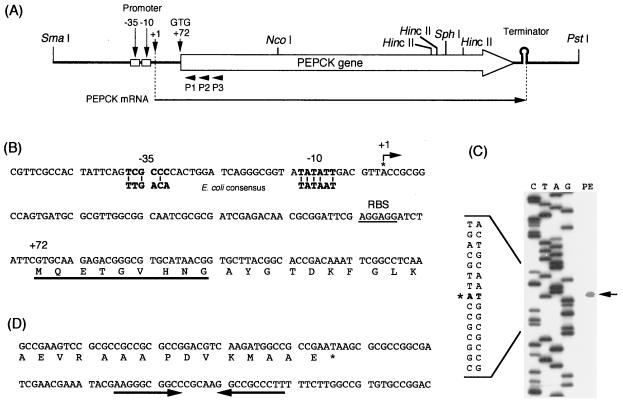
Structural and transcriptional analysis of the pckA gene. (A) Schematic physical map of the 2.5-kb SmaI-PstI fragment containing the R. palustris pckA gene. The pckA gene is indicated by the open arrow. Relevant restriction sites, gene boundaries, the approximate start point of the pckA mRNA, the putative promoter, and the transcriptional terminator-like stem-loop structure are shown above the map. The primers (P1, P2, and P3) used in the primer extension analysis are indicated by the arrowheads below the map. The pckA mRNA is shown by the arrow below the map. (B) Sequence features around the start point of transcription. The DNA sequence around and including the pckA mRNA 5′ end is presented. Regions at the canonical promoter distances of −35 and −10 are indicated by boldface lettering, with the E. coli ς70 recognition consensus below. Conserved bases in the two sequences are indicated by vertical bars. A bent arrow marked +1 shows the start point of the pckA transcript. The putative ribosome binding site is underlined and marked (RBS). The NH2-terminal amino acids whose sequence was determined in this study are indicated by bold underlining. The first base of the start codon is indicated at position +72, and the single-letter amino acid code for the amino terminus of the PEPCK protein is presented below the sequence. (C) Sequencing gel analysis of primer extension (the primer 5′ end is at position 620). High-resolution electrophoresis of 5′-labelled cDNA products is shown aligned with a sequencing ladder generated from DNA from the same region, using the same primer. Dideoxy sequencing marker lanes C, T, A, and G are indicated, and cDNA synthesis of the RNA sample is shown in lane PE. The migration position of the single primer extension product is indicated with an arrow. A portion of the DNA sequence is shown on the left, and the asterisk indicates the putative transcriptional start site. (D) DNA sequence of the carboxyl-terminal region of the pckA gene. The carboxyl-terminal part of the DNA sequence of the pckA gene is translated into amino acids. An inverted repeat which has the characteristics of a rho-independent terminator is denoted by opposing arrows below the sequence.
Construction of pckA-lacZ fusion.
β-Galactosidase was expressed as a hybrid pckA′-′lacZ fusion protein, using the pckA transcriptional and translational signals. The protein fusion was constructed by inserting the 1.9-kb SmaI-HincII fragment (Fig. 1A) encompassing the pckA promoter region into the pMC1871 SmaI site. Pale blue E. coli transformants harbored the recombinant plasmid with the insert in the correct orientation. Subsequently, the pckA-lacZ fusion was subcloned as a 5.0-kb SalI fragment into the SalI site of shuttle vector pMG102 (17). The resulting plasmid, pMG202, was introduced into R. palustris No. 7 by electroporation. Transformants appeared as blue clones on van Niel’s agar plates containing 200 μg of kanamycin/ml and 40 μg of X-Gal (5-bromo-4-chloro-β-d-galactopyranoside)/ml.
Construction of a pckA insertionally inactivated strain.
A 1.3-kb HincII kanamycin cassette from pUC4K was inserted into the unique CpoI site within the pckA gene coding region carried on pMG200, which had been blunt ended with a Takara DNA-blunting kit according to the manufacturer’s instructions. The 2.8-kb KpnI PCR-amplified pckA::Km fragment from this construct was further subcloned into the KpnI site of pMG300, which is a gentamicin derivative of the suicide vector pGP704 (18), yielding pMG312. This plasmid can replicate only in a strain that produces the R6K-specified π protein and was therefore unable to replicate in R. palustris No. 7, which does not produce π. Plasmid pMG312 was transferred from E. coli SM10 λpir, which can produce π, to R. palustris No. 7 by conjugation. Diparental matings between E. coli SM10 λpir(pMG312) and R. palustris No. 7 were performed by using a filter mating technique as described before (18). Both the absence of vector sequences and the presence of pckA::Km cassette sequence were confirmed for several candidates by Southern hybridization analysis. Similarly, a R. palustris No. 7 pckA ppc double mutant was constructed by introducing a copy of the pckA gene disrupted by a tetracycline resistance cassette into a ppc mutant (18).
Enzyme assays.
A crude enzyme extract was prepared as follows. Exponentially growing cells (25 ml) were harvested by centrifugation (10 min; 8,000 × g; 4°C) and washed with STE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA). The cells were resuspended in 1 ml of the same buffer solution and disrupted by sonication (Astrason model XL2020) in an ice water bath for three 2-min periods, interrupted by 2-min cooling intervals. Cell debris was removed by centrifugation (20 min; 10,000 × g; 4°C), and the supernatant was used as a crude extract for enzyme assays.
Protein concentrations were measured with a Bio-Rad protein assay kit in either the standard or detergent-compatible format as appropriate, with bovine serum albumin as a reference standard.
PEPC, PEPCK, MAEA, and MAEB activities were determined as described previously (18, 19).
Pyruvate orthophosphate dikinase (PPDK) and PPS activities were measured as described by Østerås et al. (29), except that imidazole (pH 6.6) was substituted for Tris-HCl (pH 8.5) to obtain optimum enzyme activities.
β-Galactosidase assays were performed according to the method of Miller (26). A β-galactosidase unit is defined as 1 nmol of o-nitrophenyl-β-d-galactoside hydrolyzed per min. The data represent averages of at least three experiments.
Nucleotide sequence accession number.
The sequence data reported here have been deposited in the DDBJ-EMBL-GenBank database under accession no. AB015618.
RESULTS
Cloning of R. palustris No. 7 pckA gene.
Based on the observation that amino acid sequences of known PEPCK proteins share several highly conserved domains, two oligonucleotide primers were designed in order to amplify a portion of the pckA gene from R. palustris No. 7. The degenerate oligonucleotide primers (primer 1 and primer 2 [see Materials and Methods]) were synthesized and used in a PCR with R. palustris No. 7 chromosomal DNA as a template. The 0.4-kb PCR product was subsequently cloned into the pGEM-T vector and sequenced. The resulting amino acid sequence appeared to be greater than 58% homologous to parts of the pckA gene products of Rhizobium meliloti (28), Saccharomyces cerevisiae (21), Staphylococcus aureus (41), E. coli (25), and T. cruzi (22).
The PCR fragment was labelled with α-32P and used as a probe to screen a λFixII R. palustris No. 7 library (18). Screening of 104 plaques yielded 13 positive clones. Restriction analysis demonstrated that all of the positive clones had overlapping inserts and contained a common 3.5-kb PstI fragment which hybridized to the PCR fragment (data not shown). Southern blotting of R. palustris No. 7 PstI-digested chromosomal DNA probed with the same labelled PCR fragment DNA demonstrated that the 3.5-kb PstI fragment had not undergone any major deletions or rearrangements during cloning (data not shown) and was present in only a single copy.
Sequencing of the pckA gene from R. palustris No. 7.
A 2.5-kb SmaI-PstI fragment derived from the 3.5-kb PstI fragment was isolated and subcloned into plasmid pUC118 (Fig. 1A). The nucleotide sequence of this fragment and the deduced amino acid sequence showed homology to known pckA genes and polypeptides.
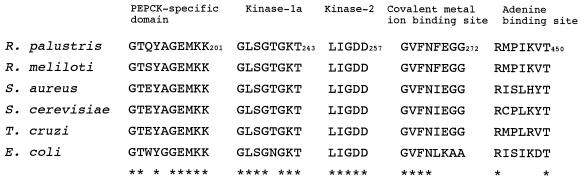
The coding region containing the R. palustris No. 7 pckA gene consists of an open reading frame of 1,614 nucleotides, corresponding to 537 amino acids. The NH2-terminal amino acid sequence of the purified PEPCK protein from R. palustris was determined to be MQETGVHNGAYG. This result corresponds to the deduced NH2-terminal amino acid sequence determined by DNA sequence analysis, and GTG at position 585 was identified as the start codon of the pckA gene (Fig. 1B). At 9 bp upstream of the start codon there is a potential ribosome binding site, AGGAGG (Fig. 1B). An inverted repeat downstream of the stop codon (TAA), centered at position 2237, showed a G+C-rich stem-loop structure followed by five T residues (Fig. 1D), the characteristics of a rho-independent terminator (34). The G+C content of the R. palustris No. 7 pckA coding region was high (64.6%), and the third positions of pckA gene codons showed an extreme preference (89.0%) for G or C, as expected in genes of PNSB (16). The deduced amino acid sequence is 68.2, 50.4, 46.2, 46.0, and 44.3% identical to that of the pckA gene products from R. meliloti, S. aureus, S. cerevisiae, T. cruzi, and E. coli, respectively. Comparative amino acid sequence alignment of ATP-dependent PEPCKs shows that all of the putative functional regions are conserved among this group of enzymes (Fig. 2). The PEPCK-specific domain (22), which is conserved in all ATP- and GTP-dependent PEPCKs analyzed to date, is also found in the R. palustris No. 7 enzyme (residues 192 to 201). In addition, kinase-1a (phosphate and Mg2+ binding site), kinase-2 (Mg2+ binding site) (23), and the covalent metal ion binding sites (22) are essentially identical among all ATP-dependent PEPCKs. On the other hand, more variability was present in the adenine binding site (43), although the key residues, arginine and threonine (R444 and T450 in R. palustris No. 7 PEPCK), which in E. coli interact with the adenine base of ATP, are identical among these sequences.
FIG. 2.
Selected portion of an amino acid sequence alignment of ATP-dependent PEPCKs. Identical residues are marked with asterisks below the sequence alignment.
Gene expression in E. coli and R. palustris No. 7.
The functionality of the R. palustris No. 7 pckA gene was tested by complementation of the E. coli pckA maeA maeB mutant strain 1321, which cannot grow on minimum medium containing succinate as a sole carbon source (14). Plasmid pMG200, which contains the R. palustris No. 7 pckA gene, was able to restore growth of an E. coli pckA maeA maeB mutant on minimum medium containing 0.4% succinate. This observation indicates that the R. palustris No. 7 PEPCK can function as a gluconeogenic enzyme in E. coli and suggests that the pckA promoter might function in E. coli.
To investigate the possibility of multicopy enhancement of PEPCK activity in E. coli and R. palustris No. 7, PEPCK assays were performed with crude extracts from E. coli and R. palustris No. 7 cells bearing either pMG201, which contains the R. palustris No. 7 pckA gene inserted into the shuttle vector pMG102, or vector alone. Plasmid pMG102 has ColE1 and Rhodopseudomonas sp. origins and has a copy number of about five molecules per cell in R. palustris No. 7 (17). Both E. coli and R. palustris No. 7 harboring pMG201 exhibited a PEPCK activity four times greater than that of the control strain, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of pMG201-bearing cells showed enhanced production of a polypeptide of approximately 60 kDa in both strains (data not shown), in agreement with the protein size predicted by sequence analysis.
Identification of the transcription start site.
In order to identify the pckA promoter, the transcriptional start site of the pckA gene was determined by primer extension with 20-mer oligonucleotides (P1, P2, and P3) having 5′ ends at positions 620, 640, and 660, complementary to the coding region. Template RNA was extracted from a R. palustris No. 7 culture in late exponential phase. One major extension product was observed with all three primers, corresponding to a 5′ end at +72 bases upstream of translation initiation codon. The result with the primer at position 620 is shown in Fig. 1C.
Putative −10 and −35 promoter sequences, similar to the E. coli ς70 promoter consensus (15) separated by a typical 18-nucleotide interregion spacing, were identified upstream of the transcriptional start point (Fig. 1B). This result could explain the multicopy enhancement of PEPCK activity observed with plasmid pMG201 in the E. coli host.
Gene expression depends on growth phase.
To facilitate study of the regulation of expression of the pckA gene, we constructed plasmid pMG202, in which lacZ expression is controlled by the transcriptional and translational regulatory signals of the pckA gene. The effect of growth conditions on pckA expression was studied by measuring β-galactosidase levels in R. palustris No. 7 harboring pMG202.
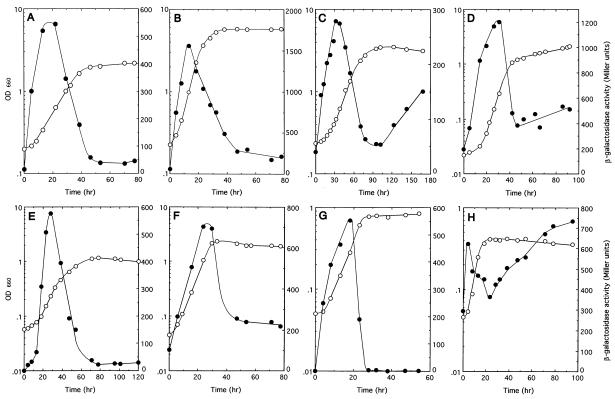
When cells were grown in PMM with gluconeogenic carbon sources, such as succinate (Fig. 3A and E) or malate (data not shown), under either anaerobic light or aerobic dark conditions, pckA expression was strongly induced during log phase and was 10- to 20-fold reduced at the onset of stationary phase. In cells growing with pyruvate (Fig. 3B and F) or lactate (data not shown), either anaerobically in the light or aerobically in the dark, log-phase inductions were also observed; however the magnitude of induction was lower for growth on pyruvate. Under anaerobic light conditions with ethanol and CO2 (Fig. 3C), which is a CO2-fixing condition in R. palustris No. 7 (2), or aerobic dark conditions with ethanol (Fig. 3G), pckA expression was also specifically induced at log phase. In addition, expression gradually increased again during stationary phase under anaerobic light conditions whereas no stationary-phase expression was observed under aerobic dark conditions. These data suggest that log-phase induction of expression of the R. palustris pckA gene does not depend on the carbon source.
FIG. 3.
Relation between growth and pckA-lacZ fusion expression in different media. The solid symbols indicate β-galactosidase activity, and the open symbols represent the culture density. R. palustris No. 7 containing pMG202 was grown under anaerobic light conditions in PMM with succinate (A), pyruvate (B), or ethanol and CO2 (C) or in van Niel’s medium (D) and under aerobic dark conditions in PMM with succinate (E), pyruvate (F), or ethanol (G) or in van Niel’s medium (H). The final concentration of each carbon source was 50 mM.
Regulation of pckA gene expression was also studied during growth in van Niel’s complex medium. Cells grown in van Niel’s medium under anaerobic light (Fig. 3D) and aerobic dark (Fig. 3H) conditions showed the same log-phase induction of the pckA gene as in minimal media with other carbon sources, whereas in cells grown aerobically in the dark, pckA expression was reduced by half at the onset of stationary phase and gradually increased again during stationary phase. Cells grown anaerobically in the light with ethanol and CO2 showed a weaker stationary-phase induction.
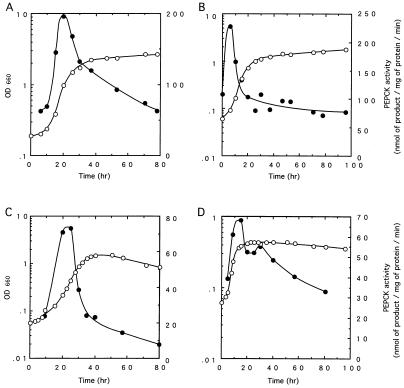
To confirm that the growth phase induction observed represented induction of the pckA gene, we measured PEPCK activity levels in wild-type R. palustris. When cells were grown in PMM with pyruvate under either anaerobic light (Fig. 4A) or aerobic dark (Fig. 4C) conditions, log-phase inductions of PEPCK were observed with magnitudes similar to the β-galactosidase levels of the pckA-lacZ fusion, and the temporal expression patterns of PEPCK and β-galactosidase fusion protein activities corresponded well.
FIG. 4.
Relation between growth and PEPCK activity under different conditions. The solid symbols indicate PEPCK activity, and the open symbols represent the culture density. R. palustris No. 7 was grown under anaerobic light conditions in PMM with pyruvate (A) or in van Niel’s medium (B) and under aerobic dark conditions in PMM with pyruvate (C) or in van Niel’s medium (D). The final concentration of pyruvate was 50 mM.
Cells grown in van Niel’s medium either anaerobically in the light (Fig. 4B) or aerobically in the dark (Fig. 4D) exhibited the same log-phase induction of PEPCK seen for the pckA-lacZ fusion experiments. However, in cells grown aerobically in the dark, PEPCK activity was gradually reduced during stationary phase, and a stationary-phase induction which corresponded to β-galactosidase levels in the pckA-lacZ fusion was not observed. These differences are considerable and suggest that PEPCK enzyme may be differentially unstable due to proteases expressed during stationary phase. High-level expression of the β-galactosidase fusion during stationary phase might represent increased transcription to compensate for PEPCK protein degradation. However, we have not examined PEPCK protein turnover under different growth conditions.
Thus, the log-phase induction of the pckA-lacZ fusion resembles the induction of the PEPCK enzyme seen in wild-type R. palustris and suggests that PEPCK is regulated at the transcriptional level.
In addition, cells grown anaerobically in the light with pyruvate (Fig. 3B) showed three- to fivefold-higher activities than those grown with succinate (Fig. 3A) and ethanol-CO2 (Fig. 3C), respectively. Furthermore, pyruvate-grown cells attained a higher final cell density (optical density at 660 nm [OD660], 5.8) and showed a faster doubling time (5 h) than cells grown on succinate (final OD660, 2.2; doubling time, 11 h) and ethanol-CO2 (final OD660, 3.5; doubling time, 14 h), suggesting that pckA gene expression may also depend on the cell growth rate.
Comparison of growth of a pckA mutant and a pckA ppc double mutant and alternative gluconeogenic enzyme activities.
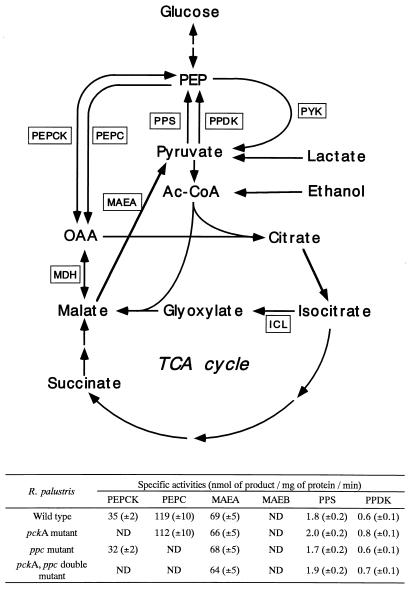
To study the role of PEPCK in R. palustris No. 7 carbon metabolism, we constructed pckA mutants and pckA ppc double mutants. Four clones were identified as pckA mutants that had lost the vector mediating gentamicin resistance (out of 700 Kmr transconjugants). Similarly, 15 clones were isolated as pckA ppc double mutants from 800 Tcr transconjugants. The integration of an inactivated copy of the pckA gene via a double-crossover event was confirmed by Southern hybridization experiments (data not shown). The PEPCK-specific activities in cell extracts of one of each of the recombinant strains and the parental strain were determined (Fig. 5). The recombinant strains showed no detectable PEPCK activity, indicating that the pckA gene was inactivated.
FIG. 5.
Metabolic pathways in R. palustris No. 7 and specific activities of gluconeogenic and anaplerotic enzymes in different strains. PYK, pyruvate kinase; MDH, malate dehydrogenase; ICL, isocitrate lyase; Ac-CoA, acetyl-coenzyme A; ND, not detected. The open boxes indicate relevant enzymes. The values in the table are means ± standard deviations from at least three independent determinations.
The effect of PEPCK deficiency on the growth of R. palustris No. 7 was analyzed. We compared the growth of the pckA mutant strain with that of the wild-type R. palustris No. 7 in synthetic PMM under both aerobic dark and anaerobic light conditions with various carbon sources. When the cells were grown with succinate, malate, pyruvate, lactate, or ethanol, either anaerobically in the light or aerobically in the dark, the growth of the pckA mutant showed no significant difference from that of the wild-type strain (data not shown). We also measured MAEA-, MAEB-, PPS-, and PPDK-specific activities in both the wild-type and pckA mutant strains (Fig. 5). A schematic representation of the metabolic pathways and enzymic reactions also referred to in this paper is shown in Fig. 5. Although MAEB activity was undetectable, both strains show similarly high levels of MAEA activity, which can synthesize pyruvate from malate. Moreover, both PPS and PPDK activities were detected in both strains. Combined activities of MAEA, PPS, and PPDK, therefore, could contribute to the growth of the pckA mutant. This data and the comparative growth experiments indicate that R. palustris, like E. coli, has alternative gluconeogenic pathways and that PEPCK is dispensable as a gluconeogenic enzyme in this organism.
In order to determine if PEPCK played a role as an anaplerotic enzyme, we also compared the growth of the pckA ppc double-mutant strain with those of the wild type, the ppc mutant, and the pckA mutant in PMM under both aerobic dark and anaerobic light conditions with pyruvate. In cells grown with pyruvate aerobically in the dark, a R. palustris ppc mutant showed a doubling time slower than that of the wild-type strain, as described before (18), while the ppc mutant and pckA ppc double mutant showed identical growth. These data imply that PEPC may play an important role in the anaplerotic pathway but that PEPCK does not function similarly and that other anaplerotic enzymes besides PEPC may exist (Fig. 6A).
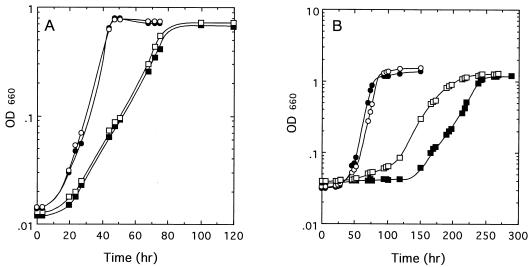
FIG. 6.
Comparative growth of R. palustris No. 7 strains. The wild type (open circles), a pckA mutant (solid circles), a ppc mutant (open squares), and a pckA ppc double mutant (solid squares) were grown in PMM with pyruvate under both aerobic dark (A) and anaerobic light (B) conditions.
When grown anaerobically in the light, the ppc mutant differs from the parental strain in exhibiting slower growth, as previously described (18). However, six independent pckA ppc double mutants showed longer lag times before starting to grow and slower growth than the ppc mutant, while the wild type and the pckA mutant showed only a negligible difference in growth (Fig. 6B). Cells grown with lactate also showed the same growth characteristics compared with pyruvate-grown cells (data not shown). With other carbon sources, such as succinate, malate, or ethanol and CO2, these mutants showed no significant difference from the wild-type strain (data not shown). These results suggest that PEPCK can function weakly as an anaplerotic enzyme in the absence of PEPC under anaerobic light conditions and that residual anaplerotic enzyme activities enable a R. palustris No. 7 pckA ppc double mutant to survive.
DISCUSSION
We have identified the R. palustris No. 7 pckA gene, determined the transcriptional start site, analyzed some basic characteristics of pckA expression, and characterized the growth of pckA-deficient mutants in order to clarify the physiological role of PEPCK.
Expression analyses with a pckA-lacZ fusion and PEPCK assay revealed a strong log-phase induction of the pckA gene in R. palustris No. 7 when cells were grown in minimal media with various carbon sources under both anaerobic light and aerobic dark conditions (Fig. 3). Such a log-phase induction has not been found for other pckA genes characterized to date.
In E. coli, stationary-phase induction of the pckA gene, which requires cyclic AMP, is observed at the onset of stationary phase in LB medium and is repressed by glucose (12). In succinate-grown cells, the level of pckA expression is quite high in all phases of growth (12).
In R. meliloti, the pckA gene was also strongly induced at the onset of stationary phase in LB medium (28). Gluconeogenic carbon sources, like succinate and arabinose, which are metabolized via the trichloroacetic acid cycle in R. meliloti, induced R. meliloti pckA expression, whereas no expression was observed to occur in cells growing on glycolytic carbon sources, like glucose and sucrose. Glucose and sucrose in succinate minimal medium reduced the level of the R. meliloti pckA expression by half (28).
The log-phase induction observed for the R. palustris pckA gene is not found in E. coli and R. meliloti. R. palustris also does not show an induction which depends on gluconeogenic carbon sources as seen in E. coli and R. meliloti. Regulation of the R. palustris pckA gene clearly differs from that in E. coli and R. meliloti. One explanation is that R. palustris can use sugar or glycerol only poorly as a sole carbon source, and therefore PEPCK induction is critical for gluconeogenesis and consequent synthesis of carbohydrate and cell constituents in log phase, irrespective of the carbon source.
Recently, at least four R. meliloti mutations were identified, mapping to different chromosomal locations, which alter the regulation of pckA gene expression such that the pckA gene can be expressed in media containing noninducing carbon sources like glucose and lactate (31). The pckR gene was isolated by complementation of one of these mutations, and the nucleotide sequence of this region revealed that PckR is homologous to the GalR-LacI family of transcriptional regulators (31). These data suggest that the expression of the R. meliloti pckA gene is governed by multiple regulatory controls. In addition, in E. coli it has been reported that the regulatory protein Cra (also known as FruR), which is also a member of the GalR-LacI family, regulates pckA gene expression (35). The R. palustris No. 7 pckA gene may also be controlled by multiple regulatory systems, which may differ for log-phase induction.
Comparative growth experiments indicated that the R. palustris No. 7 wild-type strain and pckA-deficient mutant showed identical growth when cells were grown with a C4-dicarboxylic acid, like succinate or malate, under both anaerobic light and aerobic dark conditions. Furthermore, MAEA, PPDK, and PPS activities were detected in both strains (Fig. 5), despite reports that the photosynthetic bacteria Rhodospirillum rubrum, Chromatium species, and Chlorobium thiosulfatophilum have PPDK activity but no PPS activity (3). The presence of PPS or PPDK enzymes, which synthesize PEP from pyruvate, in R. palustris No. 7 is supported by data showing that a R. palustris ppc mutant grown with pyruvate exhibited a doubling time slower than that of the wild type (18). The low level of these activities may be due to instability, as was seen for other PPDK and PPS enzymes (5, 8). In R. palustris, PPDK and PPS activities could be detected in the pH range of 8.5 to 9.0, unlike the Propionibacterium shermanii (8) and R. meliloti enzymes (29), which showed optimum enzyme activity in the standard pH range of 6.5 to 7.0.
These data suggest that R. palustris No. 7 may have at least two routes for the synthesis of PEP from C4-dicarboxylic acid—catalysis by PEPCK or by the combined activities of MAEA and PPDK and/or PPS—and that the gluconeogenic reaction can function without PEPCK, as is seen in E. coli. For further understanding of the gluconeogenic pathways in R. palustris No. 7, it will be necessary to construct and analyze a PEPCK- and MAEA-deficient mutant and a PEPCK-, PPS-, and PPDK-deficient mutant.
On the other hand, growth experiments with a R. palustris No. 7 pckA ppc double mutant revealed that PEPCK has a limited function as an anaplerotic enzyme in the absence of PEPC under anaerobic light conditions (Fig. 6).
In some organisms, PEPCK can have a significant role as an anaplerotic enzyme (see the introduction). It has been reported that A. succiniciproducens produces a high yield of succinate subsequent to CO2 fixation when cultured under conditions of pH 6.2 and high CO2 concentration (37). Under these conditions, PEPCK activity increased to significant levels, suggesting that PEPCK plays a key role in succinate production by fixing CO2 to form OAA (37). Furthermore, in A. eutrophus, PEPCK was identified as the sole C3-carboxylating enzyme; pyruvate- and other PEP-dependent CO2-fixing enzyme activities were not detected in this organism (39).
Apart from PEPC and PEPCK, additional anaplerotic enzymes in R. palustris are not defined. Possible candidates include pyruvate carboxylase (PC) and glyoxylate cycle enzymes, including isocitrate lyase and malate synthase. Although PC activity was undetectable in R. palustris No. 7, as described previously (18), this could be due to the instability of the enzyme under our assay conditions, as seen for enzymes from Aspergillus niger, Arthrobacter globiformis, and Corynebacterium glutamicum (9, 13, 33). The glyoxylate cycle has been described in R. palustris No. 7, and isocitrate lyase activity was found to be induced in ethanol- and acetate-grown cells (42). Further studies are necessary to clarify whether PC or the glyoxylate cycle can provide an additional anaplerotic function in R. palustris in the presence and/or absence of PEPC. Given our poor understanding of the R. palustris No. 7 carbon metabolic pathways under both aerobic dark and anaerobic light conditions, it is clear that there is a lot to be learned about the regulatory networks that govern the anaplerotic and gluconeogenic pathways, including PEPC and PEPCK, under both conditions.
ACKNOWLEDGMENTS
We are especially grateful to Takaaki Fujii (Chiba University) for the gift of purified PEPCK enzyme and for his valuable discussions. We thank Elliot Juni (University of Michigan) for providing the E. coli pckA maeA maeB mutant strain 1321.
This work was supported by a grant from the New Energy and Industrial Technology Development Organization.
REFERENCES
- 1.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 2.Brenner V, Inui M, Nunoura N, Momma K, Yukawa H. Studies on CO2 fixation in PNSB: utilization of waste as the additional source of carbon for CO2 fixation by PNSB. In: Inui T, Anpo M, Izui K, Yanagida S, Yamaguchi T, editors. Advances in chemical conversions for mitigating carbon dioxide. Studies in surface science and catalysis. Vol. 114. Amsterdam, The Netherlands: Elsevier Science B. V.; 1998. pp. 593–596. [Google Scholar]
- 3.Buchanan B B. Orthophosphate requirement for the formation of phosphoenolpyruvate from pyruvate by enzyme preparations from photosynthetic bacteria. J Bacteriol. 1974;119:1066–1068. doi: 10.1128/jb.119.3.1066-1068.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie D A, Powell J W, Stables J N, Watt R A. A nuclear magnetic resonance study of the role of phosphoenol pyruvate carboxykinase (PEPCK) in the glucose metabolism of Dipetalonema viteae. Mol Biochem Parasitol. 1987;24:125–130. doi: 10.1016/0166-6851(87)90098-3. [DOI] [PubMed] [Google Scholar]
- 5.Cooper R A, Kornberg H L. Phosphoenolpyruvate synthetase and pyruvate, phosphate dikinase. In: Boyer P D, editor. The enzymes. 3rd ed. New York, N.Y: Academic Press; 1974. pp. 631–649. [Google Scholar]
- 6.Cymeryng C, Cazzulo J J, Cannata J J B. Phosphoenolpyruvate carboxykinase from Trypanosoma cruzi: purification and physicochemical and kinetic properties. Mol Biochem Parasitol. 1995;73:91–101. doi: 10.1016/0166-6851(95)00099-m. [DOI] [PubMed] [Google Scholar]
- 7.Donohue T J, Kaplan S. Genetic techniques in Rhodospirillaceae. Methods Enzymol. 1991;204:459–485. doi: 10.1016/0076-6879(91)04024-i. [DOI] [PubMed] [Google Scholar]
- 8.Evans H J, Wood H G. Purification and properties of pyruvate phosphate dikinase from propionic acid bacteria. Biochemistry. 1971;10:721–729. doi: 10.1021/bi00781a001. [DOI] [PubMed] [Google Scholar]
- 9.Feir H A, Suzuki I. Pyruvate carboxylase of Aspergillus niger: kinetic study of a biotin-containing carboxylase. Can J Biochem. 1969;47:697–710. doi: 10.1139/o69-107. [DOI] [PubMed] [Google Scholar]
- 10.Fujii T, Nakazawa A, Sumi N, Tani H, Ando A, Yabuki M. Utilization of alcohols by Rhodopseudomonas sp. No. 7 isolated from n-propanol-enrichment cultures. Agric Biol Chem. 1983;47:2747–2753. [Google Scholar]
- 11.Goldie A H, Sanwal B D. Genetic and physiological characterization of Escherichia coli mutants deficient in phosphoenolpyruvate carboxykinase activity. J Bacteriol. 1980;141:1115–1121. doi: 10.1128/jb.141.3.1115-1121.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldie H. Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J Bacteriol. 1984;159:832–836. doi: 10.1128/jb.159.3.832-836.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurr J A, Jones K M. Purification and characterization of pyruvate carboxylase from Arthrobacter globiformis. Arch Biochem Biophys. 1977;179:444–455. doi: 10.1016/0003-9861(77)90132-1. [DOI] [PubMed] [Google Scholar]
- 14.Hansen E J, Juni E. Isolation of mutants of Escherichia coli lacking NAD- and NADP-linked malic enzyme activities. Biochem Biophys Res Commun. 1975;65:559–566. doi: 10.1016/s0006-291x(75)80183-5. [DOI] [PubMed] [Google Scholar]
- 15.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imhoff J F. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Advances in photosynthesis. Amsterdam, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 17.Inui, M. Unpublished data.
- 18.Inui M, Dumay V, Zahn K, Yamagata H, Yukawa H. Structural and functional analysis of the phosphoenolpyruvate carboxylase gene from the purple nonsulfur bacterium Rhodopseudomonas palustris No. 7. J Bacteriol. 1997;179:4942–4945. doi: 10.1128/jb.179.15.4942-4945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai S, Suzuki H, Yamamoto K, Inui M, Yukawa H, Kumagai H. Purification and characterization of a malic enzyme from the ruminal bacterium Streptococcus bovis ATCC 15352 and cloning and sequencing of its gene. Appl Environ Microbiol. 1996;62:2692–2700. doi: 10.1128/aem.62.8.2692-2700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondratieva E N. Interrelation between modes of carbon assimilation and energy production in phototrophic purple and green bacteria. Int Rev Biochem. 1979;21:117–175. [Google Scholar]
- 21.Krautwurst H, Encinas M V, Marcus F, Latshaw S P, Kemp R G, Frey P A, Cardemil E. Saccharomyces cerevisiae phosphoenolpyruvate carboxykinase: revised amino acid sequence, site-directed mutagenesis, and microenvironment characteristics of cysteine 365 and 458. Biochemistry. 1995;34:6382–6388. doi: 10.1021/bi00019a017. [DOI] [PubMed] [Google Scholar]
- 22.Linss J, Goldenberg S, Urbina J A, Amzel L M. Cloning and characterization of the gene encoding ATP-dependent phospho-enol-pyruvate carboxykinase in Trypanosoma cruzi: comparison of primary and predicted secondary structure with host GTP-dependent enzyme. Gene. 1993;136:69–77. doi: 10.1016/0378-1119(93)90449-d. . (Erratum, 145:157, 1994.) [DOI] [PubMed] [Google Scholar]
- 23.Matte A, Goldie H, Sweet R M, Delbaere L T J. Crystal structure of Escherichia coli phosphoenolpyruvate carboxykinase: a new structural family with the P-loop nucleoside triphosphate hydrolase fold. J Mol Biol. 1996;256:126–143. doi: 10.1006/jmbi.1996.0072. [DOI] [PubMed] [Google Scholar]
- 24.Matte A, Tari L W, Goldie H, Delbaere L T J. Structure and mechanism of phosphoenolpyruvate carboxykinase. J Biol Chem. 1997;272:8105–8108. doi: 10.1074/jbc.272.13.8105. [DOI] [PubMed] [Google Scholar]
- 25.Medina V, Pontarollo R, Glaeske D, Tabel H, Goldie H. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzyme from Trypanosoma brucei and Saccharomyces cerevisiae. J Bacteriol. 1990;172:7151–7156. doi: 10.1128/jb.172.12.7151-7156.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 27.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Østerås M, Driscoll B T, Finan T M. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J Bacteriol. 1995;177:1452–1460. doi: 10.1128/jb.177.6.1452-1460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Østerås M, Driscoll B T, Finan T M. Increased pyruvate orthophosphate dikinase activity results in an alternative gluconeogenic pathway in Rhizobium (Sinorhizobium) meliloti. Microbiology. 1997;143:1639–1648. doi: 10.1099/00221287-143-5-1639. [DOI] [PubMed] [Google Scholar]
- 30.Østerås M, Finan T M, Stanley J. Site-directed mutagenesis and DNA sequence of pckA of Rhizobium NGR234, encoding phosphoenolpyruvate carboxykinase: gluconeogenesis and host-dependent symbiotic phenotype. Mol Gen Genet. 1991;230:257–269. doi: 10.1007/BF00290676. [DOI] [PubMed] [Google Scholar]
- 31.Østerås M, O’Brien S A P, Finan T M. Genetic analysis of mutations affecting pckA regulation in Rhizobium (Sinorhizobium) meliloti. Genetics. 1997;147:1521–1531. doi: 10.1093/genetics/147.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne J, Morris J G. Pyruvate carboxylase in Rhodopseudomonas sphaeroides. J Gen Microbiol. 1969;59:97–101. doi: 10.1099/00221287-59-1-97. [DOI] [PubMed] [Google Scholar]
- 33.Peter-Wendisch P G, Wendisch V F, Paul S, Eikmanns B J, Sahm H. Pyruvate carboxylase as an anaplerotic enzyme in Corynebacterium glutamicum. Microbiology. 1997;143:1095–1103. doi: 10.1099/00221287-143-4-1095. [DOI] [PubMed] [Google Scholar]
- 34.Platt T, Bear D G. Role of RNA polymerase, rho factor, and ribosomes in transcription termination. In: Beckwith J, Davies J, Gallant J A, editors. Gene function in prokaryotes. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 123–161. [Google Scholar]
- 35.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Samuelov N S, Lamed R, Lowe S, Zeikus J G. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl Environ Microbiol. 1991;57:3013–3019. doi: 10.1128/aem.57.10.3013-3019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schobert P, Bowien B. Unusual C3 and C4 metabolism in the chemoautotroph Alcaligenes eutrophus. J Bacteriol. 1984;159:167–172. doi: 10.1128/jb.159.1.167-172.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schocke L, Weimer P J. Purification and characterization of phosphoenolpyruvate carboxykinase from the anaerobic ruminal bacterium Ruminococcus flavefaciens. Arch Microbiol. 1997;167:289–294. doi: 10.1007/s002030050446. [DOI] [PubMed] [Google Scholar]
- 41.Scovill W H, Schreier H J, Bayles K W. Identification and characterization of the pckA gene from Staphylococcus aureus. J Bacteriol. 1996;178:3362–3364. doi: 10.1128/jb.178.11.3362-3364.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tahama H, Shinoyama H, Fujii T. Reaction mechanisms of isocitrate lyase from Rhodopseudomonas sp. No. 7. Biosci Biotech Biochem. 1993;57:140–141. doi: 10.1271/bbb.57.140. [DOI] [PubMed] [Google Scholar]
- 43.Tari L W, Matte A, Pugazhenthi U, Goldie H, Delbaere L T J. Snapshot of an enzyme reaction intermediate in the structure of the ATP-Mg2+-oxalate ternary complex of Escherichia coli PEP carboxykinase. Nat Struct Biol. 1996;3:355–363. doi: 10.1038/nsb0496-355. [DOI] [PubMed] [Google Scholar]
- 44.Utter M F, Kolenbrander H M. Formation of oxalacetate by CO2 fixation on phosphoenolpyruvate. In: Boyer P, editor. The enzymes. Vol. 6. New York, N.Y: Academic Press; 1972. pp. 117–168. [Google Scholar]
- 45.van Niel C B. The culture, general physiology, and classification of the non-sulfur purple and brown bacteria. Bacteriol Rev. 1944;8:1–118. doi: 10.1128/br.8.1.1-118.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L-F, Tomich J M, Saier M H., Jr Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J Mol Biol. 1990;213:687–703. doi: 10.1016/S0022-2836(05)80256-6. [DOI] [PubMed] [Google Scholar]