Abstract
The Bacillus subtilis membrane-bound division proteins, DivIB and DivIC, each contain a single transmembrane segment flanked by a short cytoplasmic N-terminal domain and a larger external C-terminal domain. Both proteins become localized at the division site prior to septation. Mutagenesis of both divIB and divIC was performed whereby the sequences encoding the cytoplasmic domains were replaced by the corresponding sequence of the other gene. Finally, the cytoplasmic-plus-transmembrane-encoding domain of each protein was replaced by a totally foreign sequence not involved in division, that encodes the N-terminal-plus-transmembrane domains of the Escherichia coli TolR protein. B. subtilis strains expressing the divIB and divIC hybrids, in the absence of the wild-type gene, were viable when grown under conditions in which the wild-type genes were found previously to be essential. Furthermore, these strains were able to sporulate to near normal levels. Thus, the cytoplasmic and transmembrane segments of DivIB and DivIC do not appear to have any specific functions other than to anchor these proteins correctly in the membrane. The implications of these findings are discussed.
Bacillus subtilis is a rod-shaped bacterium that divides by forming a septum at mid cell, between segregated nucleoids. Under conditions of nutrient starvation, a specialized acentral division event occurs, whereby a nucleoid is actively translocated into a small polar compartment, which eventually becomes a spore. Although many of the genes involved in cell division in B. subtilis and Escherichia coli have been identified, knowledge of the molecular mechanism and control of cell division in bacteria is just starting to emerge (see references 5 and 28 for recent reviews). The highly conserved FtsZ protein has a key role. This cytoplasmic protein can dynamically self-associate into a ring structure at the division site, along the inner edge of the membrane (4). During septation, the ring constricts and then disassembles when division is complete. The peripheral membrane protein FtsA is known to associate with this ring, at least in E. coli (1, 29, 34). Most of the other division proteins are transmembrane proteins exhibiting a simple bitopic topology, with a small N-terminal cytoplasmic domain and a larger external C-terminal domain separated by a single transmembrane segment. B. subtilis contains four such division proteins—PBP2B, DivIB, DivIC, and FtsL. PBP2B is a homolog of E. coli FtsI (35), which is involved specifically in septal peptidoglycan synthesis (23). The functions of the other three remain obscure.
DivIB and DivIC are highly abundant proteins which localize to the division site in B. subtilis (19, 26, 31). While DivIC is essential for division (27), cells can still divide in the absence of DivIB at low temperatures (30°C or below [3]). As the temperature increases, more DivIB is required to maintain the normal rate of cell division (31). It has been suggested that DivIB is involved in stabilizing or promoting the formation of the division complex at high temperatures. DivIB is a 263-residue protein, with a short cytoplasmic domain (31 residues) and a large external domain (210 residues). Several temperature-sensitive filamenting mutants have been found to contain a mutation within the divIB gene. Three of these (ts12, div104, and div105) are clustered near the 3′ end of the gene (17). Thus, at least the extreme C-terminal segment of DivIB is important for its function. In contrast to DivIB, DivIC is quite small (121 residues), with relatively short cytoplasmic (37 residues) and external (60 residues) domains. One temperature-sensitive mutation (div-355) occurs within the coding sequence corresponding to the external domain and causes a frameshift mutation at residue 106 (27).
Although no mutations have been identified within the cytoplasmic or transmembrane domains of DivIB or DivIC, it is possible that these domains have a role other than simply anchoring the proteins within the membrane. It has been proposed previously that the cytoplasmic domain of FtsQ, an E. coli homolog of DivIB, is involved in signal transduction from the cytoplasm to the exterior of the cytoplasmic membrane (6). This could be the case for B. subtilis DivIB and DivIC, reflecting a role in coordinating events occurring in the cytoplasm, such as chromosome replication and partitioning, with an external event such as septal peptidoglycan synthesis. Alternatively, the cytoplasmic domain may associate with cytoplasmic division proteins, such as FtsA or FtsZ, as an important step in the division process. It is known that both DivIB and DivIC require FtsL for localization to the division site (10). The cytoplasmic and transmembrane domains of these bitopic division proteins may facilitate dimerization or complex formation with themselves or each other. Consistent with a specialized role for the cytoplasmic domain of DivIB, there is a high degree of homology in this region between the B. subtilis and Bacillus licheniformis DivIB proteins (16). The cytoplasmic domains are 87% identical, in contrast to 68% identity over the entire sequence; the membrane-spanning segments are 77% identical.
Because the bitopic membrane-bound proteins contain topologically separate domains, alterations in the sequence of one domain should not affect the conformation of the other domains. Using this rationale, mutagenesis studies on the E. coli bitopic transmembrane division proteins (FtsQ, FtsI, FtsL, and FtsN) were carried out, whereby the cytoplasmic and transmembrane domains were either exchanged with an equivalent domain from among the four division proteins or replaced with a sequence from a protein not involved in cell division (9, 14). In summary, these studies showed that, of the cytoplasmic N-terminal and transmembrane domains of E. coli FtsI and FtsL, both are essential for cell division; for FtsQ, only the cytoplasmic domain is essential. Neither of the domains is essential for FtsN function.
We have made similar types of hybrid constructs of both DivIB and DivIC. In the first set of hybrids the cytoplasmic domains of the two proteins were exchanged. Subsequently, more drastic changes were made, such that the entire cytoplasmic-plus-transmembrane domains were replaced with equivalent domains of a totally foreign (E. coli) membrane-bound protein not involved in division. It is shown that retention of only the external domains of DivIB and DivIC is sufficient for both central vegetative and acentral sporulation division. It is also shown directly that the presence of the foreign N-terminal and transmembrane sequences allows proper localization of at least the DivIC hybrid at the division site. It is concluded that both the targeting of these proteins to the division site and their activity reside solely in the external domain, suggesting roles in the regulation of septal peptidoglycan synthesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids are listed in Table 1. Unless otherwise stated, tryptose blood agar base plates supplemented with thymine (20 μg/ml) and appropriate antibiotics were used for B. subtilis colony growth. Chloramphenicol, kanamycin, and spectinomycin were used at 5, 10, and 100 μg/ml, respectively. Growth in liquid media was performed in Penassay or L broth (29a). Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added when needed. E. coli DH5α was used as the host for plasmid cloning.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or construction | Source or reference |
|---|---|---|
| B. subtilis | ||
| 168 (SU5)a | trpC2 | E. Nester |
| SB19 | Prototroph | E. Nester |
| ts355 | trpC2 thyA1 thyB1 div-355 | N. Mendelson |
| SU347 | div-355 | 26 |
| PL171 | amyE::divIC divICΔ::spr | 27 |
| KU608 | metC85::Tn917 trpC2 divIBΔ::cat | 3 |
| SU301a | trpC2 murB:pVK2:divIB cat (Pspac-CBB) | This studyc |
| SU320a | trpC2 murB:pVK3:divIB cat (Pspac-BBB) | This study |
| SU321a | trpC2 divIBΔ::cat | This study |
| SU325a | trpC2 murB:pVK2:divIB cat (Pspac-CBB) orf4:pVK6:orf5 aph (trp a) | This study |
| SU326a | trpC2 murB:pVK3:divIB cat (Pspac-BBB) orf4:pVK6:orf5 aph (trp a) | This studyc |
| SU338b | trpC2 thyA1 thyB1 div-355 amyE::pVK8 cat BCC | This study |
| SU339b | trpC2 thyA1 thyB1 div-355 amyE::pVK9 cat CCC | This study |
| SU386b | trpC2 thyA1 thyB1 amyE::pVK8 cat BCC divICΔ::spr | This study |
| SU387b | trpC2 thyA1 thyB1 amyE::pVK9 cat CCC divICΔ::spr | This study |
| SU388a | trpC2 murB:pVK12:divIB cat (Pspac-RRB) | This study |
| SU389a | trpC2 div-355 amyE::pVK13 cat (Pspac-RRC) | This study |
| SU390a | trpC2 div-355 amyE::pVK14 cat (Pspac-CCC) | This study |
| SU391a | trpC2 amyE::pVK13 cat (Pspac-RRC) divICΔ::spr | This study |
| SU392a | trpC2 amyE::pVK14 cat (Pspac-CCC) divICΔ::spr | This study |
| E. coli | ||
| DH5α | F−endA1 hsdR17 supE44 λ− thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80dlac) Δ(lacZ)M15 | GIBCO BRL |
| Plasmids | ||
| pDH88 | B. subtilis integration vector; Pspac lacI bla cat | 20 |
| pDG364 | B. subtilis amyE integration vector; bla cat | 8 |
| pPY18 | B. subtilis amyE integration vector; Pspac lacI bla cat | 11 |
| pSG122 | bla aph | J. Errington |
| pLH3 | Contains 1.7-kb BamHI-HindIII fragment encoding divIB (3′ end), orf4 and orf5; bla | 18 |
| pLH8 | Contains 0.7-kb mnlI fragment encoding the 3′ end of divIB (encoding residues 27 to 263); bla | 18 |
| pLH10 | Contains 2.7-kb EcoRI-HhaI fragment encoding the entire divIB gene; bla | 15 |
| pVK2 | pDH88 containing 3′ end of murB and Pspac-CBB hybrid for integration into the divIB locus; lacI bla cat | This study |
| pVK3 | pDH88 containing 3′ end of murB and Pspac-BBB for integration into the divIB locus; lacI bla cat | This study |
| pVK6 | pLH3 containing trp a transcriptional terminator between the StuI-EcoRV sites within orf4 and orf5; bla aph | This study |
| pVK8 | pDG364 containing BCC; bla cat | This study |
| pVK9 | pDG364 containing CCC; bla cat | This study |
| pVK12 | pDH88 containing 3′ end of murB and Pspac-RRB hybrid for integration into the divIB locus; lacI bla cat | This study |
| pVK13 | pPY18 containing Pspac-RRC; lacI bla cat | This study |
| pVK14 | pPY18 containing Pspac-CCC; lacI bla cat | This study |
The original 168 strain has been maintained in our collection as SU5. New strains based on SU5 have also been marked.
These strains are based on ts355.
See also reference 31.
General methods.
Competent B. subtilis cells were made according to the method of Anagnostopoulos and Spizizen (2), and transformations were performed by using standard procedures and at the appropriate temperature. amyE mutants were tested on 1% starch plates, using Gram’s iodine stain (8). General DNA manipulation techniques were performed as described by Sambrook et al. (32). Chromosomal DNA was prepared according to the method of Errington (12). Sporulation was performed at 30°C in Schaeffer’s sporulation medium containing 1 mM IPTG and assessed as previously described (31).
Oligonucleotides.
Oligonucleotides are listed below with their collection numbers. 161, GCGGACGAAGCTTGAAGAGATGAAAGGAGAACCG; 162, AATAAATGAAATTAACCGTCTGTACAGCCCTTTGC; 163, AGACGGGTTAATTTCATTTATTAGCTCTTTTTTATTATGGTGC; 164, GGATACGTCTAGATTCGGTCTGC; 167, GAAGACGGATCGATTGAGTGGCTG; 168, CGGCATACTAGCATGCCGTTTGTCG; 214, GCGGACGAAGCTTGAAGAGATGAAAGGAGGACCGTCTTTGAACCCGGGTCAAGACCGAG; 238, AATTCCTAAAAAAAAGCCCGCTCATTAGGCGGGCTAGGCATGCAGCAGG; 239, CCTGCTGCATGCCTAGCCCGCCTAATGAGCGGGCTTTTTTTTAGG; 248, CCGGAATTCAGGATTTACATCTTTTTAGCGG; 249, GACCCGGGTTCAAACCAGACGGTCCTCCTTTC; 250, GGACCGTCTGGTTTGAACCCGGGTCAAGACCGAG; 251, CGAATACAGTTAACCGGCGGTTTGCCTTTTGTTTCCTC; 252, GGCAAACCGCCGGTTAACTGTATTCGGCGCCCTAG; 253, CACCGTCCAAGCTTGGCTACTTGCTCTTCTTCTCCAC; 345, AGCTTGAAGAGATGAAAGGAGGACCGTCTGGTATGGCTCGTGCACGCGGTCGCGGACGTCGCGATCTTAAATCTGAAATCAA; 346, AGGTACAATGTTGATTTCAGATTTAAGATCGCGACGTCCGCGACCGCGTGCACGAGCCATACCAGACGGTCCTCCTTTCATCTCTTCA; 347, CATTGTACCTTTACTTGATGTTTTGTTAGTATTACTTTTAATCTTCATGGCTACAGCTCCAATCATTACTCAATCTGTTGAAGTA; 348, GATCTACTTCAACAGATTGAGTAATGATTGGAGCTGTAGCCATGAAGATTAAAAGTAATACTAACAAAACATCAAGTAA; 349, GCTCGAAGATCTTAAAGAAGAAAAGAAAGAACAGCTTG; 350, GCTCCAATCGATGGCTACTTGCTCTTCTCCAC; 351, GCTCGAAGATCTTAGTAAAGTATCAACAATCTCTGTTAC; and 352, GCAGCTTTATCGATTTTCCCAAACTC.
PCR amplification of divIB, divIC, and hybrid DNA sequences.
PCR was performed with Tth plus DNA polymerase (Biotech International Ltd.) essentially according to the supplier’s instructions. MgCl2 was added to the reaction mixture at various concentrations (range, 1.5 to 5 mM) to optimize product yield. Starting template DNAs (2 ng) were amplified for 15 or 30 cycles (for plasmid or chromosomal template, respectively), using denaturation, annealing, and extension temperatures of 95, 43 to 52, and 72°C, respectively.
Gene splicing by overlap extension (SOE [22]) was used to PCR amplify and fuse the gene sequences of the CBB and BCC hybrids (see Results section for naming of hybrids). The divIB and divIC portions of the hybrids were PCR amplified separately. Due to primer design, the two PCR products contained identical sequences at one end. When added together in a second PCR, the products fused (over a 21- to 26-bp region) and extended, resulting in a full-length hybrid sequence. The second PCR mixture contained the two noncomplementary primers used in the primary reaction to further amplify the hybrid sequence (for 15 cycles). For the BCC hybrid, a third PCR step was needed (see below). The primary PCR products were treated with Klenow (4.2 U/μl) (Boehringer Mannheim) plus deoxynucleoside triphosphates (0.5 mM) for 10 min at 37°C, to remove extra 3′ nucleotides. Klenow-treated PCR products were gel purified prior to the SOE reaction. All cloned PCR products were sequenced to establish their validity.
Construction of SU301 and SU320.
The CBB hybrid sequence was PCR amplified and spliced together as follows. Primers 161 (containing a HindIII site) and 162 were used to amplify divIC (encoding residues 1 to 37) plus the upstream Shine-Dalgarno sequence, from SB19 DNA. Primers 163 and 164 (containing an XbaI site) were used to PCR amplify the divIB sequence (encoding residues 32 to 221) from plasmid pLH8. The two amplified sequences were spliced together and amplified further, using primers 161 and 164. The hybrid was cloned into pDH88 cut with HindIII and XbaI, placing it under control of the Pspac promoter (36). Primers 167 and 168, containing ClaI and SphI restriction sites, respectively, were used to amplify 0.5 kb of upstream divIB sequence (containing the 3′ end of murB). This was inserted between the ClaI and SphI sites of pDH88, together with the CBB sequence, giving plasmid pVK2. The plasmid was linearized with ClaI and used to transform the 168 strain (SU5 in our collection) to chloramphenicol resistance, via a double crossover between murB and the 3′ region of divIB. This allowed Pspac-CBB to replace wild-type divIB, which was confirmed in the resulting strain (SU301) by Southern blotting.
Wild-type divIB (BBB) was PCR amplified with primers 214 (containing a HindIII site) and 164. This gave the divIB sequence (encoding residues 1 to 221) plus an upstream divIC Shine-Dalgarno sequence (as for the CBB construct). This was cloned into the HindIII and XbaI sites of pDH88, followed by insertion into the resulting plasmid of the murB sequence between the ClaI and SphI sites, giving plasmid pVK3. This plasmid was linearized and transformed into 168 as for pVK2. Replacement of divIB by Pspac-BBB was confirmed in the resulting strain (SU320) by Southern blotting.
Construction of SU325 and SU326.
The strong transcriptional terminator trp a (7), from the E. coli trp operon, was synthesized (oligonucleotides 238 and 239) with an EcoRI site at one end and SphI plus StuI sites at the other and placed between the nonessential orf4 and orf5 genes (downstream of divIB) within pLH3. EcoRI linkers were inserted within the EcoRV site of pLH3 to allow directional cloning of trp a between the EcoRI and StuI sites. The aph (kanamycin resistance) gene, cut out of pSG122 with SphI and SmaI, was inserted between the SphI and StuI sites of the cloned terminator. The resulting plasmid (pVK6) was linearized with SacI and used to transform SU301 and SU320 to kanamycin resistance. The aph gene plus trp a DNA was inserted across the orf4-orf5 junction (truncating both genes) by double crossover, giving SU325 and SU326, respectively. The expected chromosomal structures were confirmed by Southern blotting.
Construction of SU338, SU339, SU386, and SU387.
The BCC sequence was amplified as follows. Primers 248 (containing an EcoRI site) and 249 were used to PCR amplify 455 bp of upstream divIC sequence from SB19 DNA. Primers 250 and 251 were used to PCR amplify the divIB sequence (encoding residues 1 to 31) from pLH10. Primers 252 and 253 (containing a HindIII site) were used to PCR amplify the divIC sequence (encoding residues 38 to 125) from SB19 DNA. The three PCR products were joined in two stages. First, the divIB and divIC coding regions were joined and PCR amplified by using primers 250 and 253. Then, the hybrid sequence was joined to the divIC upstream sequence in a final PCR round, using primers 248 and 253. The full-length hybrid was inserted between the EcoRI and HindIII sites of the amyE vector, pDG364, resulting in plasmid pVK8. Wild-type divIC (CCC) was PCR amplified by using primers 248 and 253 and cloned into the EcoRI and HindIII sites of pDG364 to give plasmid pVK9. Both pVK8 and pVK9 were linearized with PstI and inserted into the amyE locus of ts355. Transformants were selected on plates containing chloramphenicol. Correct insertion into the amyE locus in the resulting strains (SU338 and SU339 for BCC and CCC constructs, respectively) was confirmed by Southern blotting and lack of amylase activity. Both SU338 and SU339 were transformed to spectinomycin resistance with PL171 chromosomal DNA, resulting in SU386 and SU387, respectively.
Construction of SU388, SU389, SU390, SU391, and SU392.
The DNA sequence that would encode the cytoplasmic-plus-transmembrane domains of TolR (residues 1 to 47) was constructed from synthetic oligonucleotides. Two DNA sequences were assembled separately (oligonucleotide 345 was annealed to 346, and oligonucleotide 347 was annealed to 348) and then joined together through a 10-bp overlap. Oligonucleotides 346 and 347 were phosphorylated at their 5′ ends to allow ligation. The codons chosen were those present in highly expressed B. subtilis genes (33). Also, the divIC Shine-Dalgarno sequence was incorporated upstream of the coding region. Overhanging HindIII and BglII sites were incorporated at the ends. Sequences encoding the C termini of DivIB and DivIC were PCR amplified by using primer 349 with 350 for divIC and primer 351 with 352 for divIB. Primers 349 and 351 contained BglII sites, while primers 350 and 352 contained ClaI sites. The tolR sequence was joined to the PCR-amplified divIB and divIC sequences at the BglII site (giving RRB and RRC, respectively) and cloned into the HindIII and ClaI sites of pVK3 (with the BBB sequence removed) and pPY18 (Pspac amyE vector), respectively, to give plasmids pVK12 and pVK13. As a positive control for RRC, the entire divIC gene (plus the Shine-Dalgarno sequence) was PCR amplified by using primers 161 and 350 and cloned into pPY18 between the HindIII and ClaI sites, resulting in plasmid pVK14. pVK12 was cut with ClaI and transformed into 168 (to give SU388), while pVK13 and pVK14 were cut with PstI and transformed into SU347 (giving SU389 and SU390, respectively). Southern blotting analysis of the three resulting chloramphenicol-resistant strains established that the plasmids had inserted correctly into the chromosome. The div-355 gene of SU389 and SU390 was inactivated with PL171 DNA, resulting in strains SU391 and SU392, respectively.
Western blotting, subcellular fractionation, and proteinase K treatment.
B. subtilis cells were solubilized as previously described (17) and then heated for 5 min at 95°C prior to electrophoresis. Fifteen percent glycine and 10% Tricine sodium dodecyl sulfate-polyacrylamide gels were used for electrophoresis of protein from divIB and divIC hybrid-expressing strains (and controls), respectively. Western transfer and color immunodetection of DivIB and DivIC were performed essentially as described by Katis et al. (26), using a 1 in 5,000 and 1 in 1,000 dilution of anti-DivIB and anti-DivIC polyclonal antisera, respectively. B. subtilis cells were fractionated and treated with proteinase K as previously described (26).
IFM.
DivIC immunofluorescence microscopy (IFM) was performed essentially as described previously (19, 26), except that cells were incubated in 2% bovine serum albumin–0.05% Tween 20–5% whole goat serum (Jackson Immunoresearch) prior to anti-DivIC antibody addition. Also, the cells were incubated in 5% whole goat serum in phosphate-buffered saline for 30 min and washed once with phosphate-buffered saline prior to the incubation with sheep anti-rabbit fluorescein isothiocyanate (FITC)-conjugated antibody. Affinity purified anti-DivIC antibodies were used at a 1 in 100 dilution.
RESULTS
Approach to DivIB and DivIC domain replacement.
In the first hybrids made, the cytoplasmic domain of DivIB was replaced with the corresponding domain of DivIC and vice versa. The second group of hybrids made use of the E. coli TolR protein, involved in colicin uptake. TolR is a bitopic transmembrane protein, with a small positively charged N-terminal cytoplasmic domain and a larger periplasmic C-terminal domain (24, 30). Residues 1 to 47 of TolR, encoding the cytoplasmic and membrane-spanning domain, were fused to the DivIB and DivIC external domains.
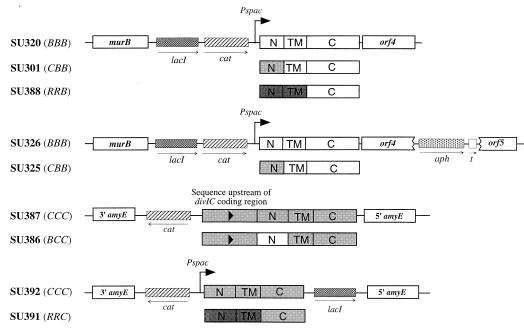
Each of the various hybrid constructs and controls was given a three-letter code. The first letter refers to the gene from which the cytoplasmic domain originated, the second refers to the transmembrane domain, and the third refers to the external domain (B denotes divIB, C denotes divIC, and R denotes tolR). For the gene, the letters are italicized (e.g., CBB); for the protein product they are not italicized and the letters are separated by a slash (e.g., C/B/B). The amino acid sequences of the cytoplasmic and transmembrane domains of the hybrids and controls are shown in Fig. 1 (see Fig. 2 for the structures of the chromosomal regions containing the hybrid genes). Note that an IPTG-inducible Pspac promoter (36) was incorporated in order to control the expression of all hybrid genes and controls, except BCC.
FIG. 1.
N-terminal sequences of hybrids and controls used in this study. The regions replaced in either DivIB or DivIC are lightly shaded. The transmembrane segments are underlined. Residue charges are shown above the sequences. B denotes DivIB, C denotes DivIC, and R denotes TolR, with each pair of domains separated by a slash.
FIG. 2.
Chromosomal structures of hybrid strains and controls used in this study, showing the region of plasmid insertion at either the divIB (136°) or amyE (28°) loci. N, TM, and C denote N-terminal-, transmembrane-, and C-terminal-encoding segments of the hybrid genes, respectively.
Analysis of DivIB hybrids.
DivIB is involved in maintaining the normal rate of cell division (3, 17). Although not essential for division at 30°C or lower, DivIB is necessary at high temperatures (3, 31). Thus, it is possible to replace the wild-type divIB gene at its normal chromosomal location with hybrid genes, and the resulting strains should be viable at low temperatures, even if the hybrid is nonfunctional.
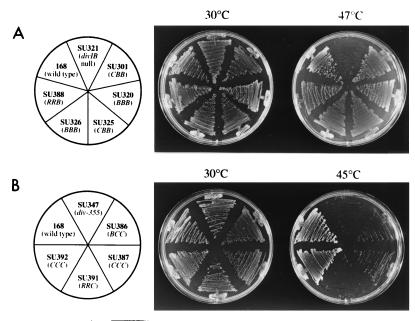
The CBB gene was constructed as described in Materials and Methods and inserted into the B. subtilis chromosome between murB and the C-terminal-encoding divIB sequence. Thus, the wild-type divIB gene was replaced by the IPTG-inducible CBB hybrid, encoding a full-length DivIB C terminus (SU301, Fig. 2). As a control, BBB was inserted into the chromosome in the same way (SU320). Selection of the final transformants was at 25°C on plates containing IPTG. As expected, both SU301 and SU320 (typical of several transformants), as well as a divIB null strain with an isogenic background (SU321, constructed by transformation of wild-type 168 with KU608 DNA), were able to grow at 30°C on plates containing IPTG (Fig. 3). At 47°C, the null strain failed to grow, while both SU301 and SU320 grew well (Fig. 3A). Both new strains produced levels of their respective proteins close to that of DivIB in wild-type 168 (Fig. 4A; compare lanes 1, 3, and 4), although C/B/B production in SU301 was slightly higher (∼1.5 times, assessed semiquantitatively by eye). Cell fractionation and Western blotting experiments showed that the C/B/B protein was membrane bound. Accessibility of the external DivIB domain to proteinase K confirmed that it was oriented in the membrane correctly (data not shown).
FIG. 3.
Viability of hybrid-expressing strains on plates incubated at various temperatures. The strains were streaked out onto tryptose blood agar base with thymine plates supplemented with IPTG (1 mM). (A) divIB hybrid-expressing strains (and controls), grown at either 30°C or 47°C. (B) divIC hybrid-expressing strains (and controls) grown at either 30°C or 45°C. The plates were incubated for either 24 h (30°C) or 12 h (45 or 47°C).
FIG. 4.
Detection of control and hybrid DivIB and DivIC proteins in strains used in this study. Strains were grown in Penassay broth at 30°C (plus IPTG where appropriate) to mid-exponential phase prior to harvesting for Western blotting. Coomassie blue staining of gels with the same loading as used for Western transfer confirmed that equal amounts of protein were loaded in each well (not shown). Both the DivIB and DivIC antisera used recognize only the external C-terminal portion of the proteins. Brackets denote the position of migration of DivIB and DivIC hybrid proteins and controls. Standards shown are expressed in kilodaltons. (A) Western blot of DivIB hybrid strains and controls. Lane 1, wild-type 168; lane 2, SU321 (divIB null); lane 3, SU320 (BBB); lane 4, SU301 (CBB); lane 5, SU326 (BBB + trp a); lane 6, SU325 (CBB + trp a); lane 7, SU388 (RRB). (B) Western blot of DivIC hybrid strains and controls. Lane 1, wild-type 168; lane 2, SU347 (div-355); lane 3, SU387 (CCC); lane 4, SU386 (BCC); lane 5, SU392 (CCC); lane 6, SU391 (RRC).
It was possible that the ability of the C/B/B hybrid to promote division at high temperatures was due to polar effects on the downstream division genes, ftsA and ftsZ. Upregulation of either ftsA or ftsZ, which are normally expressed from immediately upstream promoters in B. subtilis (13), might have aided division at the nonpermissive temperature. To rule out this possibility, a strong transcriptional terminator (trp a) was inserted into the nonessential orf4-orf5 region, between the divIB region and ftsA/Z. Such strains containing CBB and BBB (SU325 and SU326, respectively; see Fig. 2), produced normal levels of their proteins (Fig. 4A, lanes 5 and 6) and, furthermore, were able to grow at 47°C (Fig. 3A). Thus, the ability of the CBB strain to grow and divide at high temperatures was not due to enhanced ftsA or ftsZ expression.
Since the C/B/B hybrid protein functioned normally, the cytoplasmic domain of DivIB most probably does not play an essential specific role, such as in signal transduction. Although there is no sequence homology between the cytoplasmic domains of DivIB and DivIC, it is possible that they share a more general role in common, such as in protein dimerization or septal localization. This was one of the reasons for making a more drastic change by constructing the RRB strain (SU388; see Fig. 2). It should be noted that this construct would contain up to four external TolR residues that extended past the membrane (Fig. 1). The final transformation to obtain SU388 was again performed at 25°C. SU388 grew normally at 47°C (Fig. 3A) and produced a detectable but low level of R/R/B protein (approximately one-third of the wild-type protein level) with a higher mobility than that of the wild-type protein, as expected (Fig. 4A, lane 7). Although the presence and topology of the R/R/B protein in the membrane were not tested, since it was functional, presumably it was correctly positioned. In conclusion, the ability of the C/B/B- and R/R/B-producing strains to grow and divide at high temperatures, in the absence of wild-type divIB, suggests that the cytoplasmic domain and transmembrane segments of DivIB play no essential role other than that achieved by anchoring the protein in the membrane in the correct orientation.
Analysis of DivIC hybrids.
Because DivIC is an essential protein (27) and there was a possibility that the hybrid constructs would be lethal, the hybrid divIC genes were not inserted directly into the divIC locus. Instead, they were placed at the amyE locus in the temperature-sensitive divIC mutant ts355. The hybrids were then tested for their ability to rescue the mutant phenotype at 45°C.
Levin and Losick (27) previously showed that the divIC gene, with 455 bp of upstream DNA, could be placed at the amyE locus (by using the amyE integration vector pDG364) and give rise to viable colonies (at least at 30°C), in the absence of divIC at its normal locus. Using a similar approach, the BCC sequence was fused to the same upstream 455-bp sequence and inserted into the amyE locus of ts355. Of the many transformants obtained for both the final BCC strain and the positive CCC control at 30°C, several were tested for growth at 45°C. Both types grew poorly, while ts355 failed to grow (not shown). The poor growth at high temperatures of the strain producing the normal C/C/C protein made it difficult to assess whether the B/C/C hybrid could complement the DivIC(Ts) protein in ts355 (containing the div-355 mutation). To address this problem, div-355 (in ts355) was inactivated in both the B/C/C- and C/C/C-producing strains, by transformation with PL171 chromosomal DNA to spectinomycin resistance. PL171 contains the divIC gene at amyE, a divIC deletion at its normal locus, and the spectinomycin resistance gene (spr) in place of divIC. Western blotting and PCR techniques were used to ensure that the amyE locus, containing either BCC or CCC, was not replaced by PL171 DNA. Both of the new BCC hybrid and CCC control strains containing the spr gene (SU386 and SU387, respectively; see Fig. 2) grew normally at 30°C, but growth at 45°C was still very poor (Fig. 3B). While low levels of the B/C/C protein were detected in SU386 (approximately one-fourth of the wild-type level), normal levels of C/C/C were produced in SU387 (Fig. 4B; compare lanes 1, 3, and 4). Note that the B/C/C protein was smaller than C/C/C, as expected (see Fig. 1). It is concluded that the inability of SU387 to grow well at high temperatures is not due to low cellular DivIC levels. One possibility is that ts355 is more temperature sensitive than 168 (SU5), even with div-355 corrected to wild type, possibly due to extra mutations in the mutagenized strain.
As with DivIB, a more drastic change to DivIC was made by replacing both the cytoplasmic and transmembrane domains with those from TolR. The RRC hybrid was constructed in a similar fashion to RRB and cloned into the amyE integrative vector pPY18, which was recently constructed in this laboratory (11). This placed RRC at amyE under Pspac control. In order to circumvent potential problems associated with use of ts355, the hybrid was inserted into an SU5 isogenic div-355 strain (SU347). As a control, wild-type divIC (CCC) was also cloned into pPY18 and inserted into amyE as with RRC. Transformants were obtained at 30°C. However, the RRC transformants failed to grow at 45°C in the presence of IPTG (not shown). The div-355 gene was inactivated in the RRC hybrid and CCC control strains (by using PL171 DNA) to give strains SU391 and SU392, respectively (see Fig. 2). Both SU391 and SU392 grew at 30°C in the presence of IPTG. Thus, like DivIB, the entire cytoplasmic and membrane portion of DivIC is not essential for division. SU391, however, failed to grow at 45°C (Fig. 3B). This suggests that the N-terminal and transmembrane domains may play some other role that is important only at high temperatures, although other scenarios are possible (see below). Note that both R/R/C and C/C/C proteins were produced at close to wild-type levels in SU391 and SU392 (Fig. 4B; compare lanes 1, 5, and 6). The smaller R/R/C hybrid protein migrated further than the wild-type C/C/C protein, as expected. Also, a faint extra band, corresponding to a smaller protein, was observed. This band is most probably a proteolytic fragment of R/R/C because, when the strain was grown in the absence of IPTG, it was not present (see below).
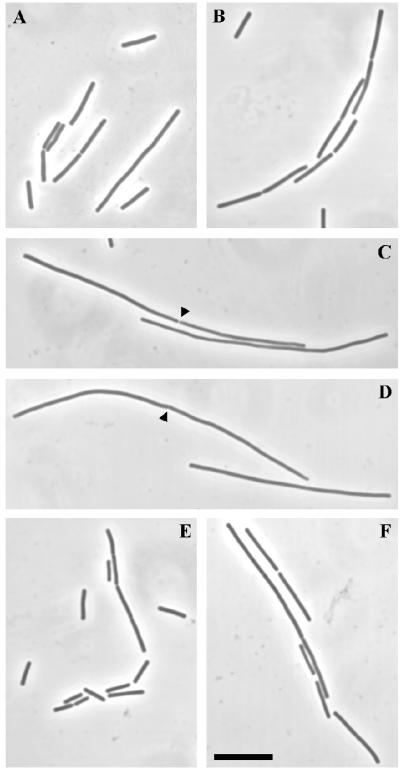
The temperature sensitivity of the RRC strain (SU391) could be due to instability of the hybrid protein at high temperatures. SU391 and SU392 were grown at 30°C in L broth with IPTG to mid-exponential phase and then diluted 1 in 3 into fresh L broth (with and without IPTG) and grown for a further 45 min at 45°C. Samples were taken just prior to the temperature shift and after 45 min at 45°C for cell lysis and Western blotting. In the presence of IPTG, very similar levels of both the R/C/C and C/C/C proteins were produced at the two temperatures, with no evidence of increased proteolytic degradation (Fig. 5; compare lane 1 with lane 2 and lane 4 with lane 5). Fig. 5 also shows that in the absence of IPTG very little C/C/C protein was produced in SU392 (lane 3) and the hybrid R/R/C protein was barely detectable (lane 6) (similar results were obtained at 30°C; not shown). Samples of the same cultures were taken for fixation and cell length measurements both before and 45 min after the temperature shift. Fig. 6 shows microscopic images of the cells. At 30°C, SU391 cells (Fig. 6A) were 7.75 ± 0.25 (mean ± standard error of the mean) μm in length and slightly longer than those of SU392 (5.67 ± 0.20 μm; Fig. 6B). At 45°C, however, the SU391 rapidly filamented in either the presence or absence of IPTG (34 ± 1.48 and 42 ± 2.15 μm, respectively; Fig. 6C and D), indicative of a rapid block in cell division. SU392 cells remained short at 45°C in the presence of IPTG (4.55 ± 0.14 μm; Fig. 6E) and were slightly filamentous in the absence of IPTG (7.80 ± 0.27 μm; Fig. 6F). Thus, very little wild-type DivIC is needed for cell division, even at high temperature. In fact, colonies of SU392 were produced when plated out in the absence of IPTG at 45°C (not shown). It appears that the cytoplasmic and transmembrane domains of DivIC may play an important role for division at high temperatures.
FIG. 5.
Effect of a growth temperature shift on the stabilities and levels of the C/C/C and R/R/C proteins in SU392 and SU391. After growth at 30°C in L broth containing 1 mM IPTG to mid-exponential phase, the cultures were diluted (1 in 3) into fresh L broth (with and without IPTG) at 45°C. Samples were taken for Western blotting (as well as for fixation and microscopy) both immediately prior to and 45 min after the temperature shift. Equal amounts of protein were loaded into each well, as revealed by Coomassie blue staining of a gel containing the same samples (not shown). Lane 1, SU392 (CCC), 30°C, with IPTG; lane 2, SU392 (CCC), 45°C, with IPTG; lane 3, SU392 (CCC), 45°C, without IPTG; lane 4, SU391 (RRC), 30°C, with IPTG; lane 5, SU391 (RRC), 45°C, with IPTG; lane 6, SU391 (RRC), 45°C, without IPTG. Bracket denotes the position of migration of DivIC and hybrid proteins. Standards shown are expressed in kilodaltons.
FIG. 6.
Phase-contrast images of SU391 (RRC) and SU392 (CCC), grown at 30°C or 45°C. See the legend to Fig. 5 for the experimental approach. A, C, and D are images of SU391 grown at either 30°C (panel A) or 45°C with and without IPTG (panels C and D, respectively). B, E, and F are images of SU392 grown at either 30°C (panel B) or 45°C with and without IPTG (panels E and F, respectively). Arrowheads in panels C and D indicate septa. Scale bar = 10 μm.
DivIC localizes to the septal site via its external domain.
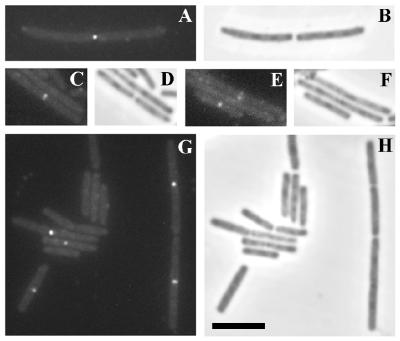
The abilities of the R/R/C and C/C/C proteins in SU391 and SU392 to localize to the division site were compared by IFM, by using affinity-purified polyclonal anti-DivIC antibodies raised against the external domain of DivIC (26). The strains were grown at 30°C in L broth with IPTG and harvested at mid-exponential growth. Localization of the R/R/C protein in SU391 was observed in both preseptal and postseptal cells. Examples are shown in Fig. 7A, C, and E, which show respectively one, one, and two cells with localizations. Examples of localizations in SU392 are shown in Fig. 7G. The patterns of the localizations in both cases were identical to those observed previously in the wild-type 168 strain (26). While the R/R/C hybrid protein could clearly localize to the division site in SU391, the intensity of the localizations appeared to be significantly lower than for SU392, and the frequency was reduced by ∼50% (∼200 cells were scored for each strain). This raises the possibility that less of the hybrid R/R/C protein is targeted to the septum. Note that the R/R/C protein in SU391 is as abundant as the C/C/C protein in SU392 (Fig. 4B and 5). A possible explanation for the decreased level of localization was that a smaller fraction of the R/R/C protein was present in the membrane, due to a difficulty of the TolR sequence in allowing translocation of the protein and maintaining it in the membrane. Subcellular fractionation of SU391, grown at 30°C in L broth with IPTG, was performed. Cytoplasm, membrane, and cell wall (external) material were Western blotted and examined for the presence of the hybrid protein (data not shown). The majority of the protein was present in the membrane fraction, with a small proportion present in the cell wall fraction. Thus, there was certainly enough R/R/C hybrid protein in the membrane for localization to occur at wild-type levels. Accessibility of the external domain of the R/R/C hybrid protein to proteinase K (data not shown) was consistent with the protein being correctly oriented in the membrane.
FIG. 7.
DivIC immunofluorescence in SU391 (RRC) and SU392 (CCC). Both strains were grown at 30°C in L broth plus 1 mM IPTG to mid-exponential phase before fixing and processing for FITC immunostaining. A through F are images of SU391. G and H are images of SU392. Both FITC fluorescence images (panels A, C, E, and G) and phase-contrast images (panels B, D, F, and H) are shown. Scale bar = 5 μm.
The cytoplasmic and membrane domains of DivIB and DivIC are not involved specifically in sporulation.
It has been established here that the cytoplasmic and membrane domains of DivIB and DivIC are not needed for division during vegetative growth (at ≤30°C for DivIC) other than to anchor the proteins correctly in the membrane. However, it was considered possible that these domains are needed for asymmetrical division during sporulation. The hybrid RRB and RRC strains (SU388 and SU391) and the respective BBB and CCC controls (SU320 and SU392), as well as the wild-type 168 strain, were grown and sporulated in Schaeffer’s sporulation medium with IPTG at 30°C. At 22 h after the end of exponential growth, the wild type produced ∼4.5 × 108 spores/ml. The four hybrid and control strains sporulated to 58 to 85% of this level. It is concluded that the cytoplasmic and membrane domains of DivIB and DivIC are not involved specifically in asymmetrical division during sporulation.
DISCUSSION
The simple bitopic nature of the membrane-bound division proteins DivIB and DivIC enables alteration of the sequence of each domain (cytoplasmic, transmembrane, or external) without disrupting the structure or conformation of the other domains. This conveniently allows identification of functional parts of the protein. Hybrid divIB and divIC genes were constructed, such that either just their cytoplasmic-encoding domain or their cytoplasmic-plus-transmembrane-encoding domains were changed completely but still allowed proper membrane insertion of the protein products. All hybrids were functional, at least at 30°C, in the absence of the wild-type gene, allowing hybrid-expressing strains to grow and sporulate normally. DivIC is essential for division of the wild type at 30°C. The DivIB hybrids were functional at 47°C, where DivIB is also essential for wild-type division. Thus, neither the cytoplasmic nor transmembrane domain of DivIB or DivIC needs to be conserved in order for these proteins to participate successfully in either vegetative or sporulation division. By elimination, the external C-terminal domain must carry out the essential function of each protein.
These observations rule out a role for both DivIB and DivIC in signal transduction, a possibility which was raised for the DivIB homolog, FtsQ of E. coli (6). Also, neither of these proteins is likely to interact directly or indirectly with FtsZ, FtsA, or some other cytoplasmic division protein. Since the essential functions of DivIB and DivIC reside solely within their external domain, these proteins are probably involved in the regulation of septal (cross wall) peptidoglycan synthesis. For DivIB such a role is possibly indirect, in view of its dispensability at low temperatures. In support of a role in peptidoglycan synthesis, neither DivIB nor DivIC has a homolog in wall-less bacteria. Also, the great abundance of DivIB and DivIC (14,000 and 50,000 molecules per cell, respectively [26, 31]) compared to membrane-bound division proteins in E. coli (50 to 200 molecules per cell) may reflect the need for high levels of peptidoglycan synthesis to form the thick septal wall.
Domain replacement of E. coli bitopic membrane-bound division proteins has been performed previously (9, 14). The cytoplasmic domain of FtsQ (DivIB homolog) could not be replaced by a foreign sequence and retain function, although the transmembrane segment could be changed (14). The importance of the FtsQ cytoplasmic domain, in contrast to that of DivIB, may reflect differences between the functions of the two proteins. FtsQ is an essential protein, while DivIB is needed only at high temperatures. Also, DivIB is approximately 100-fold greater in abundance than FtsQ. The N-terminal domain of FtsQ may play a very specific role in division, consistent with the essential nature and small abundance of the protein. The high degree of homology between the N-terminal DivIB sequences of B. subtilis and B. licheniformis is surprising, in light of the lack of importance of this domain. Recently, a homolog of divIC has been identified in Listeria monocytogenes, whose protein product has 36% identity with B. subtilis DivIC (25). The C-terminal domain of DivIC is more highly conserved (45% identity) than the N-terminal and transmembrane segments (27 and 25% identities, respectively). This is consistent with the sole importance of the external domain for DivIC function.
While neither DivIB nor DivIC appears to have a role in signal transduction between the cytoplasm and the exterior of the cell during the division process, it is possible that one or more of the other membrane division proteins fulfills such a role. The most likely candidates in B. subtilis are the transmembrane proteins FtsL and PBP2B. It is noteworthy that the PBP2B homolog in E. coli, FtsI, has a requirement for conservation of its N-terminal and transmembrane domains (14), consistent with such a role. It is also possible that the candidate integral membrane division protein (designated YlaO), which is homologous to E. coli FtsW (21), could also fulfill such a signalling role.
Both DivIB and DivIC most probably not only act but also localize to the division site through their external domains. This has been shown directly for DivIC, where obvious localizations were observed with the R/R/C hybrid protein. Due to the presence of a leucine zipper motif in the external domains of DivIC (27) and FtsL (10) and the fact that the stability of DivIC is dependent on FtsL (10), it is tempting to suggest that these two proteins interact directly via these motifs. It would be interesting to know if the R/R/C protein is likewise unstable in the absence of FtsL. The R/R/C hybrid-producing strain was not viable at high temperatures, and formed long filaments, even though an adequate level of hybrid protein was present. This raises the possibility that the cytoplasmic and/or transmembrane segment may be important for DivIC function at high temperatures. Perhaps these regions, although nonessential, may interact with other proteins and enhance the ability of DivIC to localize at temperatures where more localized protein is necessary. Consistent with such a possibility, a much lower intensity of DivIC localization was observed with the R/R/C hybrid. However, the extra four residues of TolR protein predicted to be external to the membrane in the hybrid may displace the external DivIC domain so as to affect its function and localization at high temperatures.
A question not addressed in the present study is whether the external domains of DivIB and DivIC need to be anchored at all to the membrane in order to function. This question was raised and addressed for E. coli FtsN, and it was concluded that such anchoring was unnecessary (9). The membrane-spanning segments of DivIB and DivIC may be needed to ensure that the proteins are oriented correctly in order to facilitate interactions with other division proteins, which may be weak. Future work could be designed to test this as well as to identify what regions of the external domains are necessary for function and/or localization.
ACKNOWLEDGMENTS
We thank Petra Levin and Richard Losick for providing strain PL171 and Anthony Weiss for useful advice.
This work was supported by the Australian Research Council.
REFERENCES
- 1.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beall B, Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation and temperature sensitivity. J Bacteriol. 1989;171:6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Bouché J-P, Pichoff S. On the birth and fate of bacterial division sites. Mol Microbiol. 1998;29:19–26. doi: 10.1046/j.1365-2958.1998.00874.x. [DOI] [PubMed] [Google Scholar]
- 6.Carson M J, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie G E, Farnham P J, Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci USA. 1981;78:4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 27–74. [Google Scholar]
- 9.Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel R A, Harry E J, Katis V L, Wake R G, Errington J. Characterization of the essential cell division gene ftsL (yllD) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- 11.Duggin I G, Andersen P A, Smith M T, Wilce J A, King G F, Wake R G. Site-directed mutants of RTP of Bacillus subtilis and the mechanism of replication fork arrest. J Mol Biol. 1999;286:1325–1335. doi: 10.1006/jmbi.1999.2553. [DOI] [PubMed] [Google Scholar]
- 12.Errington J. Efficient Bacillus subtilis cloning system using bacteriophage vector φ105J9. J Gen Microbiol. 1984;130:2615–2628. doi: 10.1099/00221287-130-10-2615. [DOI] [PubMed] [Google Scholar]
- 13.Gonzy-Tréboul G, Karmazyn-Campelli C, Stragier P. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J Mol Biol. 1992;224:967–979. doi: 10.1016/0022-2836(92)90463-t. [DOI] [PubMed] [Google Scholar]
- 14.Guzman L M, Weiss D S, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harry E J. Ph.D. thesis. Sydney, Australia: University of Sydney; 1992. [Google Scholar]
- 16.Harry E J, Partridge S R, Weiss A S, Wake R G. Conservation of the 168 divIB gene in Bacillus subtilis W23 and B. licheniformis, and evidence for homology to ftsQ of Escherichia coli. Gene. 1994;147:85–89. doi: 10.1016/0378-1119(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 17.Harry E J, Stewart B J, Wake R G. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol Microbiol. 1993;7:611–621. doi: 10.1111/j.1365-2958.1993.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 18.Harry E J, Wake R G. Cloning and expression of a Bacillus subtilis division initiation gene for which a homolog has not been identified in another organism. J Bacteriol. 1989;171:6835–6839. doi: 10.1128/jb.171.12.6835-6839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harry E J, Wake R G. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- 20.Henner D J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- 21.Henriques A O, Glaser P, Piggot P J, Moran C P., Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 1998;28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 22.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 23.Ishino F, Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum forming reaction sequence. Biochem Biophys Res Commun. 1981;101:905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 24.Kampfenkel K, Braun V. Membrane topologies of the TolQ and TolR proteins of Escherichia coli: inactivation of TolQ by a missense mutation in the proposed first transmembrane segment. J Bacteriol. 1993;175:4485–4491. doi: 10.1128/jb.175.14.4485-4491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kathariou, S. 1998. Personal communication.
- 26.Katis V L, Harry E J, Wake R G. The Bacillus subtilis division protein DivIC is a highly abundant membrane-bound protein that localizes to the division site. Mol Microbiol. 1997;26:1047–1055. doi: 10.1046/j.1365-2958.1997.6422012.x. [DOI] [PubMed] [Google Scholar]
- 27.Levin P A, Losick R. Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol. 1994;176:1451–1459. doi: 10.1128/jb.176.5.1451-1459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 29.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Morrison D A. Transformation and preservation of competent bacterial cells by freezing. In: Wu R, editor. Methods in enzymology. New York, N.Y: Academic Press; 1979. pp. 326–331. [DOI] [PubMed] [Google Scholar]
- 30.Muller M M, Vianney A, Lazzaroni J-C, Webster R E, Portalier R. Membrane topology of the Escherichia coli TolR protein required for cell envelope integrity. J Bacteriol. 1993;175:6059–6061. doi: 10.1128/jb.175.18.6059-6061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowland S L, Katis V L, Partridge S R, Wake R G. DivIB, FtsZ and cell division in Bacillus subtilis. Mol Microbiol. 1997;23:295–302. doi: 10.1046/j.1365-2958.1997.2141580.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sharp P M, Higgins D G, Shields D C, Devine K M. Protein-coding genes: DNA sequence database and codon usage. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 557–569. [Google Scholar]
- 34.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanouri A, Daniel R A, Errington J, Buchanan C E. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]