Abstract
Klebsiella pneumoniae porin genes were analyzed to detect mutations accounting for the porin deficiency observed in many β-lactam-resistant strains. PCR and Southern blot analysis revealed the existence of a third porin gene in addition to the OmpK36 and OmpK35 porin genes previously described. This new porin gene was designated ompK37 and is present in all of the clinical isolates tested. The OmpK37 porin gene was cloned, sequenced, and overexpressed in Escherichia coli. In contrast to that of the major porins, OmpK37 porin expression was only detectable by Western blot analysis in porin-deficient β-lactam-resistant strains, suggesting strong down regulation under standard laboratory conditions. Functional characterization suggested a narrower pore for the OmpK37 porin than for K. pneumoniae porins OmpK36 and OmpK35. This correlated with the susceptibility to certain β-lactam antibiotics, since a K. pneumoniae strain expressing porin OmpK37, but not porin OmpK36 or OmpK35, was less susceptible to β-lactam antibiotics than the same strain expressing either porin OmpK36 or OmpK35.
The outer membrane of gram-negative bacteria plays a significant role in a variety of functions; it serves as a diffusion barrier to extracellular solutes and interacts with the bacterial environment. This membrane is composed of a bilayer containing phospholipids, lipopolysaccharide, and outer membrane proteins (OMPs). One family of OMPs, the porins, are present in large amounts in the outer membrane and form water-filled channels that permit the diffusion of small hydrophilic solutes across the outer membrane. Porins are generally divided into two classes: nonspecific porins (e.g., OmpC and OmpF), which permit the general diffusion of small polar molecules (<600 Da), and specific porins (e.g., LamB), which facilitate the diffusion of specific substrates.
We have defined two major porins in Klebsiella pneumoniae designated OmpK36 (3) and OmpK35 (14). Their characterization revealed that porins OmpK36 and OmpK35 are the homologues of porins OmpC and OmpF from Escherichia coli, respectively. Furthermore, porins OmpK36 and OmpK35 allow the diffusion of a wide variety of molecules, including bacterial nutrients and antimicrobials.
β-Lactam antibiotics are currently used in the treatment of infections with K. pneumoniae (30). This microorganism is a major nosocomial pathogen causing pneumonia, urinary tract infections, and bacteremia, particularly in immunocompromised patients. Antimicrobial treatment is critical for these patients; however, multiantibiotic-resistant strains emerge frequently in the hospital (20, 25, 27). Resistance to β-lactams is associated mainly with the expression of plasmid- or chromosome-encoded β-lactamases able to inactivate β-lactams. However, since β-lactam antibiotics penetrate the outer membrane of many gram-negative bacteria through porins, antibiotic resistance can also be caused by porin loss or deficiency (23). Porin loss as a mechanisms of antimicrobial resistance has been described in many species (1, 19). Recently, we demonstrated that this antimicrobial resistance strategy also operates in K. pneumoniae (15, 18). In these studies, we isolated clinical strains resistant to β-lactam antibiotics with a characteristic in common: they simultaneously lacked expression of the OmpK35 and OmpK36 porins.
To study the molecular mechanisms causing porin loss in K. pneumoniae, we PCR amplified the porin genes from porin-deficient K. pneumoniae clinical isolates. Interestingly, in addition to the OmpK36 and OmpK35 porin genes, we amplified a third DNA fragment. Sequence analysis of this unexpected DNA fragment revealed the existence of a new porin gene in K. pneumoniae. Functional characterization of this new porin revealed a narrower pore than those of porins OmpK35 and OmpK36, which did not allow penetration by certain β-lactams, causing resistance to these antimicrobial agents.
MATERIALS AND METHODS
Strains, plasmids, and media.
The K. pneumoniae strains and plasmids used in this study are indicated in Table 1. Escherichia coli DH5α was used for cloning experiments. Strains were grown in Luria-Bertani medium supplemented with 50-mg/liter kanamycin or ampicillin when required.
TABLE 1.
Strains and plasmids included in this study
| K. pneumoniae strain or plasmid | Relevant phenotype | Source or reference |
|---|---|---|
| Porin-sufficient strainsa | ||
| SD8 | OmpK36+ OmpK35+ OmpK37− | 13 |
| CSUB10S | OmpK36+ OmpK35− OmpK37− | 4 |
| Porin-deficient strainsb | ||
| CSUB10R | OmpK36− OmpK35− OmpK37− | 4 |
| LB4 | OmpK36− OmpK35− OmpK37+ | 18 |
| LB66 | OmpK36− OmpK35− OmpK37+ | 15 |
| Plasmids | ||
| pBluescript KS(−) | High-copy-number cloning vector, Apr | Stratagene |
| pGEM-T | PCR fragment cloning vector, Apr | Invitrogen |
| pWSK29 | Low-copy-number cloning vector, Apr | 29 |
| pCSI2 | Kmr cassette-containing vector | 9 |
| pSUV7 | pACYC184 with cloned ompK36 | 3 |
| pSHA25K | pWSK29 with cloned ompK36 gene and Kmr cassette from pCSI2 | This work |
| pSHA16 | pWSK29 with cloned ompK35 | Our laboratory |
| pSHA16K | pWSK29 with cloned ompK35 gene and Kmr cassette from pCSI2 | This work |
| pQE1 | pWSK29 carrying ompK37-containing EcoRI fragment | This work |
| pQE1K | pQE1 with cloned Kmr cassette from pCSI2 | This work |
| pQE3 | pBluescript KS(−) carrying ompK37-containing BamHI fragment from pQE1 | This work |
| pQE3K | pQE3 with cloned Kmr cassette from pCSI2 | This work |
| pQE7 | pWSK29 carrying ompK37 transcribed by lac promoter | This work |
| pQE7K | pQE7 with cloned Kmr cassette from pCSI2 | This work |
| pQE31 | pBluescript KS(−) containing ompK37 and 700 bp of ompK37 upstream flanking sequence | This work |
| pQE31K | pQE31 with cloned Kmr cassette from pCSI2 | |
| pQE33 | pBluescript KS(−) containing ompK37 and 200 bp of ompK37 upstream flanking sequence | This work |
| pQE33K | pQE33 with cloned Kmr cassette from pCSI2 | This work |
Porin-sufficient strains are strains expressing either porin OmpK36 or OmpK35 or both.
Porin-deficient strain are strains lacking expression of porins OmpK36 and OmpK35.
DNA procedures.
Plasmid DNA was isolated by using the Wizard Miniprep Kit (Promega) in accordance with the manufacturer’s instructions. Isolation of genomic DNA, transformation, and electroporation were carried out by standard techniques (5). T4 DNA ligase and restriction endonucleases were used by following the manufacturer’s recommendations (Pharmacia). DNA fragments prepared by restriction enzyme digestion were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. DNA fragments were recovered from agarose gels by using the Qiaquick gel extraction kit (Qiagen). Southern blot analysis and probe labeling and detection were carried out by using the ECL kit (Amersham). OmpK36, OmpK35, and OmpK37 porin gene probes were obtained from strain SD8 by PCR with primers U681 and L1316. DNA sequencing was performed by using an automated sequencing apparatus (Applied Biosystems).
PCR.
PCR amplifications were performed in a Thermoline Amplitron 1 thermal cycler by using Taq polymerase (Pharmacia) with 30 cycles of amplification (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C).
The primers used to amplify porin genes were U681 (5′-CGGTTACGGCCAGTGGGAATA-3′) and L1316 (5′-GACGCAGACCGAAATCGAACT-3′). They anneal to sequences conserved in both ompK36 and ompK35 located 215 and 850 bp downstream of the ompK36 start codon (3), respectively. Three amplicons of about 600, 650, and 700 bp were obtained from strain SD8 with the above primers and cloned in pGEM-T. Primer INTF (5′-GCAGTATCAGGGCAAAAAC-3′), which anneals 506 bp downstream of the ompK36 start codon, and primer L1316 were used for PCR amplification of plasmids pSUV7 and pSHA16, containing the cloned ompK36 and ompK35 genes, respectively (Table 1) and the pGEM-T clone containing the 700-bp amplicon described above. The amplicons obtained with primers INTF and L1316, which contain sequences specific for the three porin genes studied, were used as specific probes for Southern blotting.
The ompK37 gene was amplified without its native promoter using primers complementary to regions situated in the 5′ region (5′-CCGGATCCTAAAGCATGAGTTC-3′) and in the 3′ region (5′-GGGGATCCGCATCAGAACTGG-3′) of the gene. Restriction sites (BamHI, underlined) were incorporated for cloning. The primers used to amplify the ompK37 promoter region were L879 (5′-CCTGAACGTTGTATTCCCACTG-3′) and UP37 (5′-CCAAGCTTGACGGAAAACGTCAA-3′). They anneal to sequences located 225 bp downstream and 933 bp upstream of the ompN start codon (accession no. AF035618), respectively.
Isolation of OMPs and porins.
Bacterial cell envelopes containing cytoplasmic and outer membranes were obtained by French press cell lysis and centrifugation. OMPs were isolated as sodium lauryl sarcosinate-insoluble material (11). Porins were isolated as described before (2) by a combination of methods (22, 24). Electrophoretic analysis of OMPs was performed in 11.5% acrylamide–0.5% bisacrylamide–0.1% sodium dodecyl sulfate (SDS) gels. Samples were boiled for 5 min in Laemmli’s sample buffer before electrophoresis. Coomassie blue-stained OMP gels were analyzed by densitometry using the Whole Band Analyzer Program (Bioimage). Purity of the isolated porins was confirmed by SDS-polyacrylamide gel electrophoresis (PAGE) analysis and N-terminal sequencing (Applied Biosystems).
OmpK37 porin expression in K. pneumoniae clinical isolates was also analyzed by Western blotting. For this purpose, SDS-PAGE gels were transferred to Immobilon-P filters (Millipore) by using the buffers and conditions described by Towbin et al. (28), except that 1 A was applied for 1 h. Filters were blocked in 1% bovine serum albumin in phosphate-buffered saline (PBS). After washing, the filters were sequentially incubated with anti-OmpK37 serum (see below) diluted 1:100 and with alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G (1:5,000; Sigma). The filters were developed as previously described (6). All of the incubations were carried out at room temperature for 1 h in 1% bovine serum albumin–0.05% Tween 20–PBS, and after incubations with the antiserum, washing steps with 0.05% Tween 20–PBS were performed.
Antiserum.
New Zealand rabbits were subcutaneously injected three times every 2 weeks with 80 μg of purified OmpK37 and bled 2 weeks after the last injection. To avoid cross-reaction with porins OmpK36 and OmpK35, antiserum was rendered monospecific (anti-OmpK37) by affinity chromatography on octadecyl silica-immobilized OmpK37 (7). Briefly, antiserum was incubated with octadecyl silica-immobilized OmpK37 overnight at 4°C and poured into a 5-ml tuberculin syringe plugged with glass wool. To remove nonspecifically bound antibodies, the column was washed successively with PBS, PBS–5% dioxan, and PBS. Specific antibodies against OmpK37 were eluted with 0.1 M glycine–0.15 M NaCl, pH 2.6.
Liposome swelling assay.
Liposomes were reconstituted with purified porins as described previously (17), except that 9.3 μmol of acetone-extracted egg phosphatidylcholine, 0.3 μmol of dicetylphosphate, and 3 μg of purified proteins were used. The rates of sugar diffusion were tested in 5 mM Tris-HCl (pH 7.5) and 15% (wt/vol) dextran T-40.
Susceptibility testing.
MICs of antimicrobial agents were determined by microdilution in accordance with the National Committee for Clinical Laboratory Standards recommendations (21) with cation-adjusted Mueller-Hinton broth (Difco). MICs were also determined by using E-test strips in accordance with the manufacturer’s (AB Biodisk) recommendations.
Nucleotide sequence accession number and homology searches.
The sequence of the K. pneumoniae SD8 ompK37 gene has been deposited in EMBL under accession no. AJ011502. DNA and protein sequence analyses were performed by using BLASTN and BLASTP. Pairwise comparisons of mature porin sequences to obtain their degrees of identity and similarity were performed with the blast 2 sequences tool from the National Center for Biotechnology Information using the default parameters. These sequences were aligned with ClustalW, analyzed with Protdist (BIONJ algorithm) (12), and represented with Treeview of the Phylip package.
RESULTS
Identification, cloning, and sequencing of the ompK37 gene from K. pneumoniae.
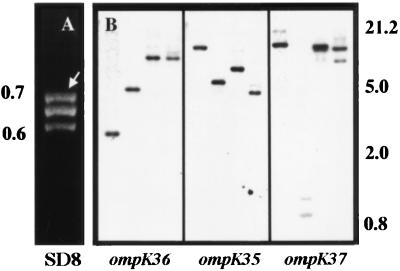
In the course of an investigation to detect and characterize mutations accounting for the loss of porin expression seen in some antimicrobial-resistant strains, we PCR amplified internal porin sequences from K. pneumoniae clinical isolates. Electrophoretic analysis of the PCR amplicons (Fig. 1A) detected the existence of three DNA fragments in strain SD8, suggesting the presence of a new porin gene, in addition to the classical porins OmpK36 and OmpK35 described in K. pneumoniae. These results were confirmed by Southern blot analysis of chromosomal DNA from strain SD8 with specific probes for ompK36, ompK35, and the putative new porin gene (Fig. 1B). As expected, the ompK36 and ompK35 probes detected DNA fragments with different sizes, indicating the existence of two separated porin genes. The probe for the putative new porin gene detected DNA sequences located in chromosomal regions different from those observed for the ompK36 and ompK35 genes. Together, these results indicated the existence of a new porin gene in strain SD8 that we designated ompK37. This new porin gene was present in all of the K. pneumoniae clinical isolates analyzed, as we confirmed by Southern blot analysis using the ompK37 probe described above (data not shown). The DNA fragment corresponding to the ompK37 gene from strain SD8 was cloned and sequenced. Since the deduced amino acid sequence predicts the OmpK37 protein to be synthesized with a signal peptide, the processing site was confirmed by N-terminal protein sequencing which yielded the sequence AEIYNKDGNKLDLYGKVD. The mature OmpK37 porin consists of 353 amino acid residues which yield a protein of 39,491 Da. The comparison between the amino acid sequences of known enterobacterial porins and the deduced protein sequence of OmpK37 clearly supported our previous deduction that OmpK37 belongs to the porin family. A standard BLASTP database search revealed that OmpK37 does not belong to the previously described porin family OmpC, OmpF, PhoE, or Lc/NmpC (16). Rather, we observed a distinct group of porins consisting of K. pneumoniae OmpK37 and the recently described porins OmpS2 (10) from Salmonella typhi and OmpN (26) from E. coli. The percentages of identity and similarity of OmpK37 with the OmpS2 and OmpN porins are 80 and 88% and 77 and 85%, respectively. In contrast, other closely related porins from K. pneumoniae presented lower scores of 70 and 78% and 58 and 68% for OmpK36 and OmpK35, respectively.
FIG. 1.
PCR and Southern blot analysis of the K. pneumoniae SD8 chromosome. (A) Resolution by agarose gel electrophoresis of PCR products obtained by amplification of internal sequences of porin genes from K. pneumoniae SD8 with primers U681 and L1316. The arrow indicates the PCR fragment corresponding to the OmpK37 porin gene. (B) Southern blot analysis of porin genes ompK36, ompK35, and ompK37 of strain SD8. Chromosomal DNA was digested with EcoRI, EcoRV, HindIII, and KpnI (from left to right in each panel) and hybridized with a probe specific for each gene. Molecular masses are indicated in kilobases on the left for panel A and on the right for panel B.
Based on sequence alignment (Fig. 2) and on the tridimensional structure of the E. coli OmpF porin (8), we predicted the secondary structure of the OmpK37 porin. It consists of 16 β-strands highly conserved in all porins, eight short periplasmic turns, and eight highly variable extracellular loops. L3 and L4 are the most- and least-conserved loops in different porins, respectively; with L4 having an insertion of 16 amino acids in OmpK37 with respect to OmpF. Both the highly conserved motif PEFGGD and charged residues R37, R75, R124, and D106 and E110, which are opposed across the pore in OmpF, are also present in OmpK37.
FIG. 2.
Alignment of the OmpK37 sequence with selected sequences from other enterobacterial porins from E. coli (porins OmpC, OmpF, OmpN, PhoE, and NmpC) and S. typhi (OmpS1 and OmpS2). Porins were selected on the basis of their high degree of amino acid identity with OmpK37. Protein sequences were derived from nucleotide sequences. Secondary-structural motifs are those of the OmpF structure (8).
Expression of the OmpK37-encoding gene.
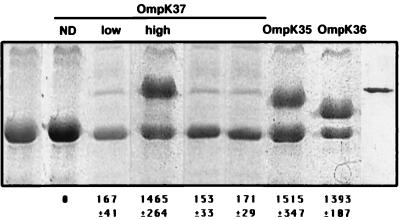
For the construction of an OmpK37 expression plasmid, we cloned the corresponding ompK37 DNA fragment from strain SD8 in plasmid pWSK29, giving plasmid pQE1. Plasmids encoding porins were tagged with a kanamycin resistance cassette to allow their cloning and selection in the multiresistant background of porin-deficient K. pneumoniae CSUB10R. Transformation of this strain with pQE1K and SDS-PAGE analysis of the OMPs did not detect any extra protein compared to the OMPs from strain CSUB10R (Fig. 3). To express and isolate the OmpK37 porin, we overexpressed the ompK37 gene by using two different strategies. First, we subcloned from pQE1 a 5-kb BamHI fragment containing ompK37 in the high-copy-number plasmid pBluescript, giving plasmid pQE3 (pQE3K). Second, we cloned ompK37 without its own promoter behind the lac promoter of pWSK29, giving plasmid pQE7 (pQE7K). In CSUB10R transformed with pQE3K or pQE7K, we detected a new OMP of about 39 kDa by SDS-PAGE (Fig. 3).
FIG. 3.
SDS-PAGE analysis of OMPs isolated from strain CSUB10R and strain CSUB10R expressing different K. pneumoniae porins. Purified OMPs (5 μg) from strain CSUB10R and its derived clones carrying plasmids pQE1K, pQE3K, pQE7K, pQE31, pQE33, pSHA16K, and pSHA25K (form left to right, lanes 1, 2, 3, 4, 5, 6, 7, and 8, respectively) were resolved by SDS-PAGE, Coomassie blue stained, and analyzed by densitometry. The porin expressed by each clone is indicated at the top of each lane. Densitometric analysis results for the corresponding porin are indicated in arbitrary units at the bottom of each lane and are the mean ± the standard deviation of at least three independent experiments. Purified OmpK37 is shown in lane 9. ND, not detected.
To determine more precisely the limit of the upstream region required for maximal expression of the OmpK37 porin, we generated different constructions with DNA fragments including the OmpK37-encoding sequence and adjacent chromosomal DNA sequences extending various distances upstream of the ompK37 start codon. Each construction was cloned into K. pneumoniae CSUB10R, and the OMPs were analyzed by SDS-PAGE and Coomassie blue staining. OmpK37 porin expression was essentially the same for cells harboring pQE31K, a construct that included DNA sequences extending approximately 700 nucleotides upstream of the ompK37 start codon, as for cells with pQE33K, a construct including sequences extending 200 nucleotides upstream (Fig. 3). These results suggest that the entire −35, −10 promoter region required for maximal expression of OmpK37 is included in the upstream 200 bp adjacent to the ompK37 start codon.
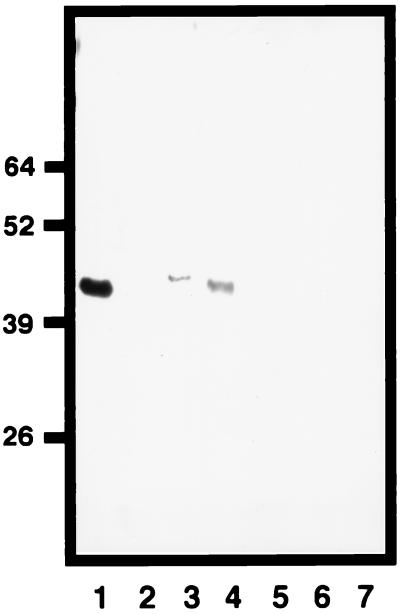
We investigated the natural OmpK37 porin expression among clinical isolates. Although the ompK37 gene is present in all of the K. pneumoniae isolates analyzed, expression of the OmpK37 porin was not detected by SDS-PAGE and Coomassie blue staining in any isolate. However, we were able to detect OmpK37 expression by Western blot analysis with anti-OmpK37 serum in some strains. As shown in Fig. 4, OmpK37 porin expression was detected in porin-deficient (OmpK36− OmpK35−) strains LB4 and LB66. However, not all of the porin-deficient strains studied expressed OmpK37 (e.g., strain CSUB10R). Furthermore, among 10 porin-sufficient strains (OmpK36+, OmpK35+, or both), we did not detect OmpK37 expression. This is the case for strain SD8 (Fig. 4), which was used to clone ompK37.
FIG. 4.
Western blot analysis of OmpK37 expression using monospecific antiserum. K. pneumoniae SD8, LB4, LB66, and CSUB10R were subjected to porin isolation methods, and the resulting materials were analyzed in lanes 2 to 5, respectively. Purified porins OmpK37, OmpK36, and OmpK35 were included as controls in lanes 1, 6, and 7, respectively. Molecular masses in kilodaltons are indicated on the left.
To investigate whether the variation in OmpK37 expression was associated with changes in the promoter region sequence, we analyzed the nucleotide sequence of this region of the K. pneumoniae chromosome from strains LB4, LB66, SD8, and CSUB10R. Comparison of the region extending 200 nucleotides upstream of the ompK37 start codon required for maximal expression of the porin revealed no nucleotide sequence differences.
Functional characterization of OmpK37: its role in resistance to β-lactam antibiotics.
We reconstituted purified porin OmpK37 into liposomes and studied the rate of sugar permeation (Table 2). Sugar uptake by liposomes containing porin OmpK37 resembled that by liposomes containing K. pneumoniae porins OmpK36 and OmpK35: nonspecific diffusion through the pore depended on the molecular mass of the sugar. However, diffusion through OmpK37 was moderately lower than through porins OmpK36 and OmpK35, suggesting the existence of a narrower pore in the OmpK37 porin. Assays with liposomes containing E. coli porins OmpF and OmpC were performed as controls in our experiments with results similar to those previously reported in the literature (26) (data not shown).
TABLE 2.
Rates of sugar permeation through various K. pneumoniae porins
| Sugar | Mol wt | Mean rate ± SDa
|
||
|---|---|---|---|---|
| OmpK37 | OmpK36 | OmpK35 | ||
| Galactose | 180.2 | 72.3 ± 4.2 | 88.0 ± 2.2 | 91.8 ± 2.3 |
| Mannose | 180.2 | 49.3 ± 3.2 | 84.0 ± 3.2 | 61.9 ± 1.3 |
| GluNAc | 221.1 | 29.6 ± 0.9 | 44.2 ± 6.4 | 42.0 ± 4.2 |
| Lactose | 360.3 | NDb | ND | 5.1 ± 1.3 |
| Trehalose | 378.3 | ND | ND | ND |
Expressed as a percentage of the value for arabinose. Standard deviations correspond to at least five measurements with four different liposome preparations.
ND, not detectable by the method used.
Since β-lactam antibiotics pass the outer membrane through porins, we studied the role of porin OmpK37 in susceptibility to antimicrobials (Table 3) . We compared the MICs for porin-sufficient strain K. pneumoniae CSUB10S and the clonally related porin-deficient variant strain CSUB10R (either alone or transformed with cloned porins). As described before, due to its porin deficiency, strain CSUB10R is more resistant to β-lactams than is parental strain CSUB10S (4). As expected, for K. pneumoniae CSUB10R expressing OmpK36 or OmpK35, the MICs reverted to values similar to those observed for strain CSUB10S. However, CSUB10R expressing porin OmpK37 from plasmid pQE7K was less susceptible to cefotaxime and cefoxitin than was CSUB10R expressing either OmpK36 or OmpK35. These results were not due to differences in porin expression, since similar levels of expression were achieved from plasmids pSH25K, pSH16K, and pQE7K, encoding porins OmpK36, OmpK35, and OmpK37, respectively (Fig. 3). The difference in the MIC for CSUB10R expressing OmpK37 versus OmpK36 or OmpK35 was more pronounced when CSUB10R was transformed with plasmid pQE1K or pQE3K. Expression of OmpK37 from these two plasmids is either very low (pQE3K) or not detectable (pQE1K) by SDS-PAGE analysis and Coomassie staining (Fig. 3). Under those circumstances, MICs are very similar to those for CSUB10R. In contrast, MICs of meropenem and imipenem, which are small zwitterionic compounds, were similar to those for the original sensitive strain, independently of the porin expressed. Only when OmpK37 was expressed in low amounts (pQE1K and pQE3K) were the meropenem and imipenem MICs close to those observed for resistant strain CSUB10R. In summary, CSUB10R encoding the OmpK37 porin was less susceptible to certain β-lactam antibiotics than were clonally related strains encoding OmpK35 or OmpK36, suggesting lower penetration of those antibiotics through OmpK37.
TABLE 3.
MICs of β-lactam antibiotics for clonally related K. pneumoniae strains expressing different porins
| Strain | Plasmid | Porin phenotypea | Porin expressionb | MIC (μg/ml)c of:
|
|||
|---|---|---|---|---|---|---|---|
| CTX | FOX | MP | IP | ||||
| CSUB10S | OmpK36+ OmpK35− OmpK37− | 4 | 2 | 0.06 | 0.125 | ||
| CSUB10R | OmpK36− OmpK35− OmpK37− | 512 | 128 | 2 | 0.5 | ||
| CSUB10R | pSHA25K | OmpK36+ OmpK35− OmpK37− | High | 4 | 2 | 0.06 | 0.06 |
| CSUB10R | pSHA16K | OmpK36−OmpK35+ OmpK37− | High | 4 | 4 | 0.12–0.06 | 0.06 |
| CSUB10R | pQE1K | OmpK36− OmpK35−OmpK37+ | Not detectable | 512 | 64 | 2 | 0.25 |
| CSUB10R | pQE3K | OmpK36− OmpK35−OmpK37+ | Low | 256 | 64–32 | 2 | 0.25 |
| CSUB10R | pQE7K | OmpK36− OmpK35−OmpK37+ | High | 128 | 32 | 0.06 | 0.125 |
Analyzed by SDS-PAGE and Coomassie blue staining. In strain CSUB10R(pQE1K), OmpK37 expression was detected by Western blot analysis with anti-OmpK37 serum. Plasmid-encoded porins are in boldface.
The level of each expressed porin from the corresponding plasmid was determined by SDS-PAGE analysis and Coomassie blue staining (Fig. 3).
CTX, cefotaxime; FOX, cefoxitin; M, meropenem; IP, imipenem.
DISCUSSION
Porins play a crucial role in the interactions between the environment and bacteria. In addition, or probably as a consequence, they are present in large amounts in the outer membrane of gram-negative bacteria. Since E. coli major porins OmpC and OmpF were defined, a large number of OmpC- or OmpF-type porins have been described in other enterobacterial species. We reported the existence of two major porins, OmpK36 and OmpK35, in K. pneumoniae. They are homologous to OmpC and OmpF, and they are expressed in large amounts in most K. pneumoniae clinical isolates independently of the isolation source (13).
We focused our attention on the role of these porins in penetration by antibiotics. As has been reported for other species, porin loss is an important cause of resistance to some antimicrobials, particularly β-lactam antibiotics (23). This phenomenon was clearly shown by us in K. pneumoniae both in vitro and in vivo (15, 18). As a result of these investigations, we isolated clinical strains resistant to most of the β-lactam antibiotics currently used to treat K. pneumoniae infections. These isolates had a characteristic in common: they simultaneously lacked expression of porins OmpK35 and OmpK36. To characterize the mutations causing loss of porin expression and also to identify putative new porins that may replace the functions of the lost porins, we scrutinized the K. pneumoniae genome.
We identified, cloned, and sequenced a new porin gene that was designated ompK37. The amino acid sequence revealed that this new porin of K. pneumoniae is highly homologous to porins OmpS2 from S. typhi and OmpN from E. coli. Both OmpS2 and OmpN were previously described as quiescent porins (10, 26). They were not detected in the outer membrane of strains grown under standard laboratory conditions. Furthermore, OmpN was overexpressed in order to isolate it and characterize its pore properties (26). In our case, OmpK37 expression was detected by a more sensitive analysis. However, OmpK37 was not detected in all of the strains tested. Among all of the clinical isolates analyzed, OmpK37 expression was detected preferentially in porin-deficient strains. OmpK37 porin overexpression was also achieved through the lac promoter. Together, the results obtained with porins OmpK37, OmpS2, and OmpN indicate that in the enterobacterial species in which these porin genes have been detected, they are subjected to strong down regulation. Regulation most probably occurs at the transcriptional level and by trans-acting mechanisms, since promoter substitution causes overexpression of ompK37. In addition, we have shown that the 200-bp nucleotide sequence immediately upstream of the start codon is sufficient for maximal expression of the OmpK37 porin. This region was identical among strains expressing or not expressing OmpK37, suggesting the possibility that the trans-acting factor(s) may influence ompK37 transcription by interacting with sequences further upstream.
Porins OmpK37, OmpS2, and OmpN are not expressed or are expressed at very low levels under standard laboratory conditions. However, these porins may be expressed under other conditions, and under these circumstances, we still do not know about their possible roles. In previous reports, no functions were attributed to the quiescent porins mentioned. Since OmpK37 expression was detected mainly in porin-deficient β-lactam-resistant strains, we decided to characterize its possible role in permeation by β-lactams.
For this purpose, we expressed similar amounts of each K. pneumoniae porin (OmpK37, OmpK36, or OmpK35) on the outer membrane of a porin-deficient clinical isolate expressing β-lactamases. MICs of β-lactam antibiotics were higher for the strain that expressed OmpK37 than for those that expressed OmpK36 or OmpK35. This phenomenon was particularly relevant with cefotaxime and cefoxitin, the MICs of which were similar to those for the resistant strain. The molecular masses of cefotaxime and cefoxitin and their charges allow them to penetrate the outer membrane more efficiently through the OmpK36 and OmpK35 porins than through OmpK37, suggesting narrower pore for this new porin. Similar conclusions may be deduced from the sugar penetration experiments, in contrast to the results obtained by Prilipov et al. (26), who reported that sugar diffusion through OmpN was similar to that through OmpF and OmpC. However, we believe that porin expression in the natural host (K. pneumoniae) and MIC determination represent a biological phenomenon more representative than the liposome swelling assays, where porin properties may be altered (30). A detailed comparison between the OmpK36 three-dimensional structure and the predicted OmpK37 secondary structure revealed an insertion of one bulky residue (Tyr-118) located in loop 3 of OmpK37. Since loop 3 is involved in the formation of the pore, this insertion may contribute to the narrower pore of OmpK37.
Meropenem and imipenem belong to the carbapenem antibiotic class. They are zwitterionic compounds, and their molecular masses are lower than those of the β-lactams cefoxitin and cefotaxime. These antibiotics are more active than the β-lactams described above against porin-deficient strains. In our experiments, MICs of carbapenems did not vary between strains expressing different porins. Our data supports the idea that treatment with β-lactam antibiotics can select strains deficient in high-expression porins OmpC and OmpF (or OmpK36 and OmpK35 in K. pneumoniae). In these strains, expression of quiescent porins or alternative porins like OmpK37, which essentially allows penetration by carbapenems but not other β-lactams, may explain why many strains resistant to β-lactams can still be treated with carbapenems. In concordance with these findings, we have detected expression of OmpK37 in strains deficient in porins OmpK36 and OmpK35, although we could not detect its expression in all of the porin-deficient strains studied. It is also possible that, in some strains, downregulation or even loss of OmpK37 may lead to an increase in the levels of resistance to carbapenems. The MICs obtained with strains expressing low amounts of OmpK37 support this hypothesis.
In summary, we have identified and characterized a new porin of K. pneumoniae. Its expression is very low under standard laboratory conditions. However, OmpK37 expression may be stimulated under conditions (e.g., antibiotic pressure) in which its functional characteristics (a narrower pore) may be advantageous over those of the “classical” OmpC-OmpF-type porins. Under these circumstances, OmpK37 may function as a porin for the uptake of substrates in strains that lack OmpK35 and OmpK36 as a result of selection for β-lactam resistance. A better characterization, currently in progress in our laboratory, of the OmpK37 expression regulatory mechanisms, will improve our knowledge of its role in resistance or sensitivity to antibiotics.
ACKNOWLEDGMENTS
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT). S.H.A. and A.D. were supported by predoctoral fellowships from the CICYT and CSIC-CAROB, respectively. S.A. was supported by a postdoctoral contract from the CICYT.
We thank Tilman Schirmer for his comments on the structure of OmpK37, Darryl A. León for his help in sequence analysis, and the Centro de Investigaciones Biológicas for DNA and protein sequencing. Members of the UIB also thank J. Lalucat for continuous support.
REFERENCES
- 1.Aggeler R, Then R L, Ghosh R. Reduced expression of outer-membrane proteins in β-lactam-resistant mutants of Enterobacter cloacae. J Gen Microbiol. 1987;133:3383–3392. doi: 10.1099/00221287-133-12-3383. [DOI] [PubMed] [Google Scholar]
- 2.Albertí S, Marqués G, Camprubí S, Merino S, Tomás J M, Vivanco F, Benedí V J. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun. 1993;61:852–860. doi: 10.1128/iai.61.3.852-860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albertí S, Rodríguez-Zuiñones F, Schirmer T, Rummel G, Tomás J M, Rosenbusch J P, Benedí V J. A porin from Klebsiella pneumoniae: sequence homology, three dimensional structure, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardanuy C, Liñares J, Domínguez M A, Hernández-Allés S, Benedí V J, Martínez-Martínez L. Outer membrane profiles of clonally related Klebsiella pneumoniae isolated from clinical samples and activity of cephalosporins and carbapenems. Antimicrob Agents Chemother. 1998;42:1636–1640. doi: 10.1128/aac.42.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1997. [Google Scholar]
- 6.Blake M S, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 7.Chiong M, Lavandero S, Ramos R, Aguillón J C, Ferreira A. Octadecyl silica: a solid phase for protein purification by immunoadsorption. Anal Biochem. 1991;197:47–51. doi: 10.1016/0003-2697(91)90353-u. [DOI] [PubMed] [Google Scholar]
- 8.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Mora M, Oropeza R, Puente J L, Calva E. Isolation and characterization of ompS1, a novel Salmonella typhi outer membrane protein-encoding gene. Gene. 1995;158:67–72. doi: 10.1016/0378-1119(95)00171-2. [DOI] [PubMed] [Google Scholar]
- 11.Filip C, Fletcher G, Wulf J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 13.Hernández-Allés S, Albertí S, Alvarez D, Martínez-Martínez L, Gil J, Tomás J M, Benedí V J. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology. 1999;145:673–679. doi: 10.1099/13500872-145-3-673. [DOI] [PubMed] [Google Scholar]
- 14.Hernández-Allés S, Albertí S, Rubirés X, Merino S, Tomás J M, Benedí V J. Isolation of FC3-11, a bacteriophage specific for the Klebsiella pneumoniae porin OmpK36, and its use for the isolation of porin-deficient mutants. Can J Microbiol. 1996;41:399–406. [Google Scholar]
- 15.Hernández-Allés, S., V. J. Benedí, L. Martínez-Martínez, A. Pascual, A. Aguilar, J. T. Tomás, and S. Albertí. Development of resistance in Klebsiella pneumoniae during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob. Agents. Chemother., In press. [DOI] [PMC free article] [PubMed]
- 16.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 17.Lee E-H, Collatz E, Trias J, Gutmann L. Diffusion of β-lactam antibiotics into proteoliposomes reconstituted with outer membranes of isogenic imipenem-susceptible and -resistant strains of Enterobacter cloacae. J Gen Microbiol. 1992;138:2347–2351. doi: 10.1099/00221287-138-11-2347. [DOI] [PubMed] [Google Scholar]
- 18.Martínez-Martínez L, Hernández-Allés S, Albertí S, Tomás J M, Benedí V J, Jacoby G A. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:342–348. doi: 10.1128/aac.40.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medeiros A A, O’Brien T F, Rosenberg E Y, Nikaido H. Loss of OmpC porin in a strain of Salmonella typhimurium causes increased resistance to chephalosporins during therapy. J Infect Dis. 1987;156:751–757. doi: 10.1093/infdis/156.5.751. [DOI] [PubMed] [Google Scholar]
- 20.Meyer K S, Urban C, Eagan J A, Berger B J, Rahal J J. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med. 1993;119:353–358. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 22.Nikaido H. Proteins forming large channels from bacterial and mitochondrial outer membranes: porins and phage lambda receptor proteins. Methods Enzymol. 1983;97:85–100. doi: 10.1016/0076-6879(83)97122-7. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nurminen M. A mild procedure to isolate 34K, 35K, and 36K porins of the outer membrane of Salmonella typhimurium. FEMS Microbiol Lett. 1978;3:331–334. [Google Scholar]
- 25.Philippon A, Labia R, Jacoby G A. Extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prilipov A, Phale P S, Koebnik R, Widmer C, Rosenbusch J P. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J Bacteriol. 1998;180:3388–3392. doi: 10.1128/jb.180.13.3388-3392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice L B, Willey S H, Papanicolau A, Medeiros A A, Eliopoulos G M, Moellering R C, Jacoby G A. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemother. 1990;34:2193–2199. doi: 10.1128/aac.34.11.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R W, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 30.Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]