Abstract
We found in the Escherichia coli genome sequence a homologue of RER2, a Saccharomyces cerevisiae gene required for proper localization of an endoplasmic reticulum protein, and designated it rth (RER2 homologue). The disruption of this gene was lethal for E. coli. To reveal its biological function, we isolated temperature-sensitive mutants of the rth gene. The mutant cells became swollen and burst at the nonpermissive temperature, indicating that their cell wall integrity was defective. Further analysis showed that the mutant cells were deficient in the activity of cis-prenyltransferase, namely, undecaprenyl diphosphate synthase, a key enzyme of the carrier lipid formation of peptidoglycan synthesis. The cellular level of undecaprenyl phosphate was in fact markedly decreased in the mutants. These results are consistent with the fact that the Rer2 homologue of Micrococcus luteus shows undecaprenyl diphosphate synthase activity (N. Shimizu, T. Koyama, and K. Ogura, J. Biol. Chem. 273:19476–19481, 1998) and demonstrate that E. coli Rth is indeed responsible for the maintenance of cell wall rigidity. Our work on the yeast rer2 mutants shows that they are defective in the activity of cis-prenyltransferase, namely, dehydrodolichyl diphosphate synthase, a key enzyme of dolichol synthesis. Taking these data together, we conclude that the RER2 gene family encodes cis-prenyltransferase, which plays an essential role in cell wall biosynthesis in bacteria and in dolichol synthesis in eukaryotic cells and has been well conserved during evolution.
In eukaryotic cells, organelles of the secretory pathway are interconnected by dynamic vesicular traffic. From the endoplasmic reticulum (ER), a variety of proteins are transported to the Golgi apparatus by carrier vesicles and then to their final destinations. On the other hand, in order to maintain the ER-specific functions, many resident ER proteins have to be correctly retained in the ER. Two mechanisms of ER localization have been proposed. One is static retention, which prevents ER proteins from being exported from the ER, and the other is dynamic retrieval, which returns ER proteins from the Golgi apparatus to the ER (12, 14, 15).
The Saccharomyces cerevisiae RER2 gene was originally identified as being involved in this ER protein localization (12). We isolated mutants of two complementation groups that mislocalize an ER membrane protein, Sec12p, and designated them rer1 and rer2 (for retention or retrieval of ER proteins). The rer2 mutants were temperature sensitive for growth and showed pleiotropic phenotypes: in addition to ER protein mislocalization, they showed slow growth, defects in N- and O-glycosylation, sensitivity to hygromycin B, and abnormal accumulation of membranes, including the ER and Golgi apparatus (16). The rer2 disruptant could grow very slowly, but the double disruption of RER2 and SRT1, a homologue of RER2, was a lethal event (16).
In the course of our study of the RER2 gene, we found that RER2 homologues exist not only in eukaryotes but also in prokaryotes. In Escherichia coli, the RER2 homologue had been registered in the genome sequence as a hypothetical open reading frame, o253. We designated it rth (for RER2 homologue) and decided to investigate its biological function to obtain a clue to the roles of the RER2 gene family. In this paper, we will present evidence that the E. coli rth gene encodes undecaprenyl diphosphate synthase (UDS), which is responsible for the synthesis of undecaprenyl phosphate, the carrier lipid required for the biosynthesis of peptidoglycan in the cell wall. In parallel, we showed that the yeast Rer2 protein is required for the synthesis of dolichol, the polyprenyl compound involved in protein glycosylation (16). The conserved essential function of the Rer2 family as cis-prenyltransferase will be discussed.
MATERIALS AND METHODS
Materials.
[1-14C]isopentenyl diphosphate (IPP) (specific activity, 2.04 TBq/mol) was purchased from Amersham Co. Nonlabeled IPP and all-E-farnesyl diphosphate (FPP) were synthesized by phosphorylation of the corresponding prenols (9). Solanesol (all-E-nonaprenol) and dodecaprenyl phosphate were purchased from Sigma Chemical Co. Ficaprenols were provided by Nisshin Flour Milling Co. Thin-layer chromatography (TLC) plates (silica gel 60 F254 and LKC-18) were purchased from Merck Co. and Whatman Co., respectively.
Bacterial strains, plasmids, and culture media.
The E. coli strains used were MG1655 (rph ilvG rfb) (2), DH1 (recA1 endA1 gyrA96 thi-1 hsdR17 supE44) (13), and N1126 (thi leu pro malA his thy cysC lacZ ara mtl xyl str spc polA12) (19). The plasmids used were pUC19 (20), pUC4K (20), pBR322 (13), pJK274 (11), pJK282 (11), and pJK286 (10).
Bacteria were grown routinely in Luria Bertani (LB) broth and antibiotic medium 3 (Difco). Because the rth(Ts) mutants isolated in this work showed clearer temperature sensitivity on the antibiotic medium 3 plate at 42°C, this medium was used for the temperature-sensitive growth test, for observation of cell shape, and for quantitation of cellular isoprenoids. Where relevant, antibiotics were added to 20 μg/ml for ampicillin (Ap), 17 μg/ml for chloramphenicol (Cm), and 50 μg/ml for kanamycin (Km).
Plasmid construction.
For cloning of the rth cdsA region, the HindIII DNA fragments containing the rth+ and cdsA+ genes were prepared by PCR with the oligonucleotides 127-1 (5′-CCAAGCTTCTGCGGACGTCTGTTTATGG-3′) and 127-2 (5′-CCAAGCTTAAACGCTCAACGCGAACACC-3′) as primers and the rth+ cdsA+ BamHI fragments were prepared with the oligonucleotides 127-3 (5′-CCGGATCCCTGCGGACGTCTGTTTATGG-3′) and 127-4 (5′-CCGGATCCAAACGCTCAACGCGAACACC-3′) as primers. MG1655 cells were used directly as templates, and Takara Ex Taq polymerase (Takara Shuzo Co., Kyoto, Japan) was used as the enzyme. The rth+ cdsA+ HindIII fragment was inserted into the HindIII site of pUC19, and the SmaI Cmr cassette of pJK211 was inserted into the resultant plasmid at the HpaI site in the rth gene to obtain pUC19-rth::Cmr cdsA+. pJK211 was constructed by inserting the BamHI Cmr cassette of pJL3-4779 (11), after treatment with Klenow fragments, into the HincII site of pUC8 and by converting the HindIII site of pUC8 into a SmaI site by using Klenow fragments and a SmaI linker. The rth+ cdsA+ mini-F plasmids were constructed by inserting the rth+ cdsA+ HindIII PCR fragment into the HindIII site of pJK286 (mini-F Apr) and inserting the rth+ cdsA+ BamHI PCR fragment into the BamHI site of pJK282 (mini-F Kmr). The rth+ cdsA+ BamHI fragment was also inserted into the BamHI site of pJK2039 to obtain pJK2039-rth+ cdsA+. A multicopy vector, pJK2039, was constructed by ligating the PvuII Kmr fragment of pJK274 (11) with the NruI-ScaI fragment of pBR322. The SmaI Cmr cassette of pJK211 was inserted into the HpaI site in the rth gene of pJK2039-rth+ cdsA+ to obtain pJK2039-rth::Cmr cdsA+. The PstI fragment containing both the Kmr marker and a part of the cdsA gene of pJK2039-rth+ cdsA+ was replaced with the Kmr PstI fragment of pUC4K to construct pJK2039-rth+ cdsA−.
Disruption of the rth gene.
For disruption of the rth gene, the E. coli polA12 strain, N1126, was first transformed with pJK282 (mini-F Kmr)-rth+ cdsA+. The cells were further transformed with pUC19-rth::Cmr cdsA+ and then incubated at 42°C. Among the Cmr transformants, Aps colonies that were devoid of the pUC plasmid were selected. By eliminating the strains with the Cmr marker inserted into mini-F plasmids, we obtained the desired rth disruptant strain, N1126 rth::Cmr/pJK282-rth+ cdsA+. To construct MG1655rr (rth::Cmr recA::Tn10)/pJK282-rth+ cdsA+, MG1655 was transformed with pJK282-rth+ cdsA+ and the rth gene of the resultant strain was disrupted by P1 transduction with N1126 rth::Cmr/pJK282-rth+ cdsA+ as a donor. The recA gene was inactivated by P1 transduction with GL787L as a donor (4).
Isolation of temperature-sensitive mutants.
The rth(Ts) mutants were isolated by plasmid shuffling with a mini-F plasmid as a vector (10). The rth cdsA region was mutagenized by error-prone PCR in the presence of 0.2 or 0.4 mM MnCl2 (21) with the primers 127-1 and 127-2 and the plasmid pJK282-rth+ cdsA+ as the template. The amplified HindIII fragments were cloned into a mini-F vector, pJK286 (Apr), and the resultant plasmids were introduced by electroporation into the rth disruptant, MG1655rr/pJK282 (Kmr)-rth+ cdsA+. Among the Apr transformants, temperature-sensitive mutants were selected on antibiotic medium 3 plates containing ampicillin at 42°C and then confirmed for kanamycin sensitivity at 30°C to make sure that the original Kmr mini-F plasmid was absent.
Preparation of enzyme fractions.
Cells from a 100-ml culture were disrupted in 2 ml of 100 mM potassium phosphate (pH 7.4) and 10 mM 2-mercaptoethanol with a Tomy UD-200 ultrasonic disintegrator eight times for 15 s at 30-s intervals. After centrifugation, a supernatant fraction was obtained and used as a cell homogenate.
For fractionation, protamine sulfate (final concentration, 0.4% [wt/vol]) was added to the homogenate, and the mixture was centrifuged. The supernatant was dialyzed against buffer A (10 mM potassium phosphate [pH 7.5], 1 mM 2-mercaptoethanol) for 12 h. The resulting solution containing 16 mg of protein was applied to 2 ml of DEAE-Toyopearl 650M column equilibrated with buffer A. Elution was carried out with 4 ml of buffer A containing 60 mM NaCl, 6 ml of buffer A containing 120 mM NaCl, and then 4 ml of buffer A containing 300 mM NaCl. Fractions (2 ml) were collected and assayed for enzyme activities.
Prenyltransferase reaction and product analysis.
The reaction mixture contained, in the final volume of 0.2 ml, 2.0 nmol of [1-14C]IPP (1.1 × 105 dpm; specific activity, 0.92 TBq/mol), 1.0 nmol of FPP, 0.2 μmol of MgCl2, 0.2 mg of Triton X-100, 10 μmol of potassium phosphate (pH 7.5), and the enzyme fraction. After incubation at 30 or 40°C for 30 min, the reaction was stopped by heating it at 95°C for 3 min.
The products were extracted with 1-butanol, and the radioactivity was measured. Prenyl diphosphates in the extract were hydrolyzed with phosphatase by the method of Fujii et al. (6). The products of hydrolysis were extracted with hexane and analyzed by TLC as described in the legend to Fig. 3.
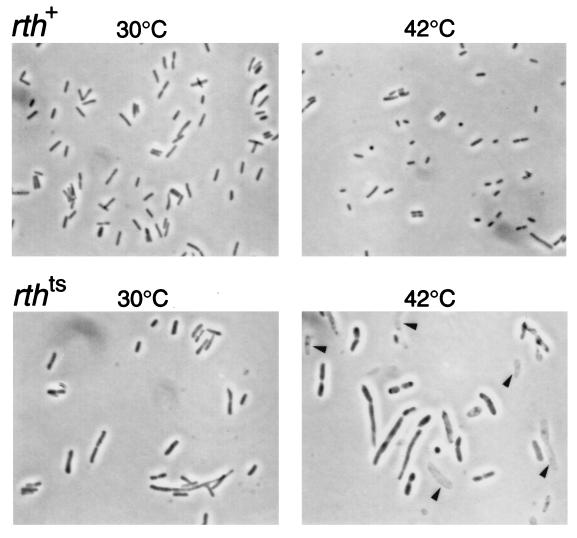
FIG. 3.
Cell shapes of the rth-6 mutant. The upper panels show the rth+ strain, MG1655rr/pJK286-rth+ cdsA+; the lower panels show the rth-6 mutant, MG1655rr/pJK286-rth-6 cdsA+. The cells were incubated at the indicated temperatures for 2 h and observed without fixation under a microscope equipped with phase-contrast optics. The magnifications are the same for all of the panels (a 100× objective was used). The arrowheads in the panel with the rth mutant incubated at 42°C indicate ghosts.
Extraction of isoprenoids.
Isoprenoids were extracted from cells as described previously (8), with a slight modification. About 10 mg (dry weight) of cells was harvested by centrifugation and washed once with 0.85% NaCl. After the NaCl solution was removed as completely as possible, the cells were suspended in 2 ml of methanol. Solutions of ubiquinone-10, phylloquinone, and dodecaprenyl phosphate were added to the suspension as internal standards for isoprenoid quantitation. Isoprenoid quinones were extracted from this mixture with hexane. One milliliter of 60% KOH was added to the residual methanol-water layer, and the mixture was heated in a boiling-water bath for 60 min. The hydrolysis products, polyprenyl monophosphates, were extracted with diethyl ether. The diethyl ether extract was washed with 5% acetic acid and dried under N2.
Analysis of isoprenoids.
The hexane extract containing isoprenoid quinones was loaded on a 0.4-g column of neutral alumina (grade III). Menaquinone and demethylmenaquinone were eluted with 2.5% diethyl ether in hexane, and ubiquinone was eluted with 7.5% diethyl ether in hexane. These compounds were analyzed by high-performance liquid chromatography (HPLC) on a reversed-phase octyldecyl silane (ODS) column (Hitachi 3056; 4.0 by 150 mm; eluent, 2-propanol-methanol, 1:1 [vol/vol]; flow rate, 1 ml/min). Menaquinone and demethylmenaquinone were monitored at 248 nm with a spectrophotometric detector, and their amounts were calculated from the ratio of the peak area to the internal standard, phylloquinone. Ubiquinones were monitored at 275 nm, and the ratio of the peak area to the internal standard, ubiquinone-10, was used for the quantitation.
The diethyl ether extract containing polyprenyl phosphate was suspended in chloroform and loaded on a 0.3-g column of silicic acid containing 6% water. The column was washed with chloroform, and prenyl phosphates were eluted with chloroform-methanol (1:4 [vol/vol]). The eluate was dried under N2 and taken up in 2-propanol-methanol (1:1 [vol/vol], containing 10 mM phosphoric acid) and analyzed by HPLC on the same column as described above. The eluent was 2-propanol-methanol (1:1 [vol/vol], containing 10 mM phosphoric acid), and the flow rate was 1 ml/min. A wavelength of 210 nm was used for monitoring prenyl phosphates.
RESULTS AND DISCUSSION
rth is essential for cell growth.
The E. coli gene rth (open reading frame o253 [3]), which is located at 4.2 min, encodes a protein of 253 amino acid residues which exhibits extensive homology to the yeast Rer2 protein (Fig. 1A). The yeast rer2 mutant showed interesting pleiotropic phenotypes, including ER protein mislocalization, glycosylation deficiency, and accumulation of abnormal membranes. Although these phenotypes are unique to eukaryotes, the family of genes is evolutionarily conserved from prokaryotes to higher eukaryotes. Therefore, we were puzzled by but interested in the functions of this gene family. To obtain a clue to their biological roles, we decided to examine the function of the E. coli counterpart, rth.
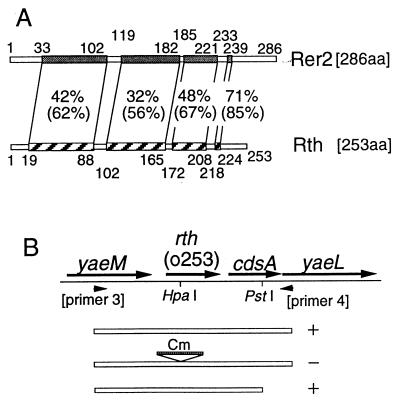
FIG. 1.
E. coli rth gene. (A) Homology between the S. cerevisiae Rer2 protein and the E. coli Rth protein. The polypeptides of Rer2 and Rth are shown as boxes. The shaded and striped boxes represent homologous domains, and the percent values indicate identity and similarity (in parentheses) between the two domains. The numbers along the polypeptides indicate the amino acid (aa) residue numbers. (B) Chromosomal inserts in plasmids used for complementation test. The arrows indicate open reading frames in the rth region. The arrowheads represent the PCR primers used for cloning. The chromosomal regions cloned into the vector pJK2039 are shown as three open boxes: top, the PCR fragment amplified with primers 3 and 4; middle, the same PCR fragment with a Cmr cassette inserted into the HpaI site within the rth gene; bottom, the PCR fragment lacking the 3′ region of the cdsA gene. Plus and minus signs denote complementation, positive and negative, respectively, for both rth-2 and rth-6 mutations.
First, we disrupted the chromosomal rth gene by inserting a Cmr cassette in the presence of a complementing rth+ cdsA+ mini-F plasmid, pJK282 (mini-F Kmr)-rth+ cdsA+. Downstream of the rth gene, the cdsA gene, which codes for CDP-diglyceride synthase, is transcribed in the same orientation as rth (Fig. 1B). At that time, it was not clear whether the cdsA gene was essential for cell growth or whether these rth+ and cdsA+ genes were organized into one transcriptional unit. If the cdsA gene was an essential gene and was transcribed from the promoter of the rth gene, an insertion mutation into rth would also disrupt the function of the cdsA gene, and this insertion mutation should be complemented by an rth+ cdsA+ plasmid but not by an rth+ cdsA plasmid. We considered this possibility and used pJK282-rth+ cdsA+ as the complementing plasmid.
We performed P1 transduction to transfer the disrupted rth gene (rth::Cm) from this disruptant to other strains. As shown in Table 1, we obtained transductants at high frequencies when the recipient strain carried the rth+ cdsA+ plasmid but not in control strains without this plasmid. There was about a 300-fold difference between the transduction frequencies of the strains with and without the rth+ cdsA+ plasmid, suggesting that rth and/or cdsA are essential for cell growth. Even in the absence of the rth+ cdsA+ plasmid, Cmr colonies appeared at low frequencies. Among these transductants, 87 of 100 Cmr colonies were Kmr, suggesting that the Kmr complementing plasmid was simultaneously transferred with the chromosomal rth::Cm gene. The rth genes of the residual 13 colonies were examined by PCR with the primers 127-3 and 127-4. DNA bands of the same sizes as those of the rth+ parental strain were amplified, suggesting that these Cmr strains were pseudodisruptants and remained rth+.
TABLE 1.
Transduction of the Cmr marker inserted into the rth gene
| Bacterium/plasmid | Frequency of transductiona |
|---|---|
| MG1655/pJK286 | 3.4 × 10−7 |
| MG1655/pJK286-rth+ cdsA+ | 1.0 × 10−4 |
| MG1655/pJK2039 | 9.1 × 10−7 |
| MG1655/pJK2039-rth+ cdsA+ | 1.2 × 10−4 |
| MG1655/pJK2039-rth+ cdsA− | 7.6 × 10−7 |
Number of Cmr transductants divided by number of plaque-forming P1 particles.
To determine which gene is essential for cell growth, we constructed an rth+ cdsA plasmid, pJK2039-rth+ cdsA−. P1 transduction was performed again as described above with a strain carrying this plasmid as a recipient. As shown in Table 1, transductants were obtained at a much lower frequency than with recipient cells containing pJK2039-rth+ cdsA+. In other words, the plasmid had to carry both rth+ and cdsA+ genes to achieve complete complementation. This result suggests that the insertion mutation in the rth gene affected the expression of the downstream cdsA gene, which is essential for cell growth, at least under these experimental conditions.
We went on to isolate temperature-sensitive mutants by PCR mutagenesis of the rth+ cdsA+ region and by plasmid shuffling (10). Three temperature-sensitive mutants were obtained. Complementation analysis showed that the temperature-sensitive growth of two of the three mutants was complemented by the rth+ cdsA plasmid, indicating that the lethality was due to the rth mutations (Fig. 1B). These two mutants were designated rth-2 and rth-6. The residual mutant was complemented by the rth+ cdsA+ plasmid but not by the rth+ cdsA plasmid. Because all of these mutants contained single missense mutations in the rth gene and were proved to be defective in cis-prenyltransferase activity (see below), the third mutant (designated rth-4) probably had another mutation in the cdsA gene in addition to the rth mutation. Taking these data together, we concluded that rth is an essential gene.
rth mutants show a defect in cell wall rigidity.
We examined the growth of the rth mutant strains at permissive and nonpermissive temperatures. Typical growth curves of rth+ and rth-6 cells at 30 and 42°C are shown in Fig. 2. As shown in Fig. 2A, the growth of the rth-6 mutant was normal at 30°C but significantly retarded at 42°C in LB broth containing 0.5% NaCl. The temperature-sensitive growth was much more evident when the rth-6 mutant was cultured in LB broth containing no NaCl. The mutant cells grew almost normally at 30°C but did not grow at all at 42°C (Fig. 2B). The rth-2 mutant showed growth profiles very similar to those of the rth-6 mutant at both 30 and 42°C. The rth-4 mutant grew more slowly than the other mutants even at 30°C, perhaps due to the additional cdsA mutation (data not shown). These results indicate that the rth(Ts) mutants are sensitive to low osmolarity at high temperature.
FIG. 2.
Growth of rth+ and rth-6 cells. Strains MG1655rr/pJK286-rth+ cdsA+ and MG1655rr/pJK286-rth-6 cdsA+ were grown in LB broth containing 0.5% NaCl at 30°C for 12 h. Then, 0.05 ml of these cultures was inoculated into LB broth containing 0.5% NaCl (A) or without NaCl (B). The cells were grown at 30 or 42°C with shaking. Optical densities at 660 nm (OD660) were measured at 1-h intervals. Open squares, rth+ at 30°C; solid squares, rth+ at 42°C; open circles, rth-6 at 30°C; solid circles, rth-6 at 42°C.
Next we examined the rth(Ts) mutant cells under a microscope after incubation at the nonpermissive temperature. The cells of the three mutants appeared to become swollen and often burst into ghosts. Figure 3 shows an example of the rth-6 mutant. Note that the cells were larger than those of the control strain. From this observation and the sensitivity of rth mutants to low osmolarity, we suspected that the rigidity of the cell wall was impaired in the mutants and that it might be because of a defect in peptidoglycan synthesis. This idea was consistent with the fact that a bacterium with no cell wall, Mycoplasma genitalium, has no RER2 homologue (5).
rth mutants are defective in UDS activity.
In the course of this study, we realized that the E. coli Rth is also homologous with a Micrococcus luteus protein which was very recently reported to have the activity of UDS in vitro (17). This enzyme is a cis-prenyltransferase that adds isoprene units onto FPP to produce undecaprenyl (C55) diphosphate, the precursor of the carrier lipid for peptidoglycan synthesis (Fig. 4).
FIG. 4.
Biosynthesis of isoprenoids.
Our results with the defective cell wall of rth mutants and the report of the Micrococcus homologue led us to examine the properties of UDS in the mutants. Crude cell homogenates were prepared and used as enzyme sources, and the reactions were carried out with [1-14C]IPP and FPP as substrates. The reaction products were extracted and treated with phosphatase, and the resulting prenols were analyzed by normal-phase TLC (Fig. 5A). When the homogenate of the rth+ strain was used, two spots were detected at the same positions as authentic solanesol and ficaprenol, respectively. The former product is all-E-octaprenol, and the latter is the mixture of Z,E-polyprenols described previously (7). When the reactions were carried out with the mutant lysates, in contrast, the spot corresponding to the mixture of Z,E-polyprenols was scarcely detected at either 30 or 40°C. These results indicate that the mutants are defective in UDS activity but retain the activity of another prenyltransferase, ODS.
FIG. 5.
Prenyltransferase activity. (A) Prenol products synthesized in an in vitro assay. Cell homogenates were prepared from the strains MG1655rr/pJK286 (mini-F Apr)-rth+ cdsA+ (lanes 1 and 2), MG1655rr/pJK286-rth-2 cdsA+ (lanes 3 and 4), MG1655rr/pJK286-rth-4 cdsA(Ts) (lanes 5 and 6), and MG1655rr/pJK286-rth-6 cdsA+ (lanes 7 and 8). The homogenates (0.1 mg of protein) were incubated with 10 μM [1-14C]IPP and 5 μM FPP in a final volume of 0.2 ml at 30°C (lanes 1, 3, 5, and 7) or at 40°C (lanes 2, 4, 6, and 8) for 30 min. The resulting prenols were extracted and analyzed by TLC on a silica gel 60F254 plate with a benzene-ethyl acetate (4:1 [vol/vol]) solvent system and by autoradiography. For more details, see Materials and Methods. The arrowheads indicate the positions of authentic prenols: Fi, ficaprenols; So, solanesol (all-E-nonaprenol); Ori, origin; SF, solvent front. (B) Fractionation of cell homogenates. The homogenates were prepared from the same strains shown in panel A, rth+ (open circles), rth-2 (solid triangles), rth-4 (solid squares), and rth-6 (solid circles), and fractionated by DEAE-Toyopearl column chromatography. The prenyltransferase assay was carried out at 30°C with 50 μl of each fraction as the enzyme source in a final volume of 0.1 ml. The dotted line indicates the NaCl concentration of the elution buffer. (C) Prenol products of fraction 3. The cell homogenates prepared from the strains shown in panels A and B, rth+ (lanes 1 and 2), rth-2 (lanes 3 and 4), rth-4 (lanes 5 and 6), and rth-6 (lanes 7 and 8), were fractionated as described for panel B, and fraction 3 of each sample was incubated with [1-14C]IPP and FPP in the presence of 0.04 mg of Triton X-100 in the 0.2-ml mixture. The reaction was carried out at either 30 (lanes 1, 3, 5, and 7) or 40°C (lanes 2, 4, 6, and 8) for 30 min (lanes 1 and 2) and 3 h (lanes 3 to 8). The resulting prenols were analyzed by TLC on a reversed-phase LKC-18 plate with an acetone-water (19:1 [vol/vol]) solvent system and autoradiography. The arrowheads indicate the positions of authentic prenols: So, solanesol (all-E-nonaprenol); C50, decaprenol (ficaprenol-50); C55, undecaprenol (ficaprenol-55); C60, dodecaprenol (ficaprenol-60); Ori, origin; SF, solvent front.
To confirm the lesion of UDS in the mutants, we fractionated the homogenates with a DEAE-Toyopearl column and separated the activity of UDS from that of ODS. UDS and ODS are known to be eluted with 120 and 300 mM NaCl, respectively (7). As shown in Fig. 5B, the UDS activities (fractions 3 and 4) of the three mutants were remarkably reduced; they were as low as 4% or less of that of the rth+ strain. The ODS activities (fractions 6 and 7) of the mutants were not significantly affected. The activities of UDS from the rth mutants were very low at either 30 or 40°C in vitro. This may be because the mutant enzymes were unstable even at 30°C in vitro.
Next, we analyzed the reaction products synthesized by UDS. The assay reaction was carried out for 30 min with fraction 3 from the wild-type strain and for 3 h with the fractions 3 from the mutant cells to obtain enough radioactive products for the TLC analysis. After treatment with phosphatase, the prenols were subjected to reversed-phase TLC (Fig. 5C). The products from the wild-type fraction 3 were mainly C50 (decaprenol), C55 (undecaprenol), and C60 (dodecaprenol). Their diphosphate esters are known to be the bona fide products in the reaction with purified UDS (1). In the case of rth mutant fractions 3, the products were mainly C45 (nonaprenol), C50, and C55, with carbon chains shorter by one isoprene unit than those of the wild-type enzyme. This may be due to the reduced chain elongation activity barely remaining in the mutant UDS.
Undecaprenyl phosphate is decreased in rth mutant cells.
In order to discover the effects of the rth mutations on isoprenoid synthesis in vivo, we further measured the cellular levels of isoprenoids in the mutants. Isoprenoids were extracted from the cells cultured at 30°C and were analyzed by HPLC. As shown in Table 2, the levels of polyprenyl phosphates, particularly undecaprenyl phosphate, were appreciably lower in the mutants than in the wild-type cells, even though the cells were cultured at the permissive temperature. On the other hand, the levels of isoprenoid quinones were not significantly changed or were even higher in the mutants. These results indicate that Rth is indeed responsible for the formation of undecaprenyl phosphate in vivo. It should be noted here that the remaining activities to synthesize polyprenyl phosphates at 30°C are considerable in the mutants in contrast to the in vitro results of the UDS activities (Fig. 5). The major products of the enzyme were C50, C55, and C60 phosphates in the mutant cells, unlike the in vitro results. These observations suggest that the UDS enzymes from the mutants were less stable in vitro, perhaps due to the lack of other cellular components, and show more profound defects in vitro than in vivo. At 42°C, the cellular level of undecaprenyl phosphate was further decreased in the mutants. However, when the cells were incubated at 42°C, the HPLC profiles showed an unidentified peak near dodecaprenyl phosphate, which made the correct quantitation of polyprenyl phosphates difficult (data not shown).
TABLE 2.
Isoprenoid levels in rth mutantsa
| Strainc | Level (μg/g [dry wt] of cells)b
|
||||||
|---|---|---|---|---|---|---|---|
| UQ-8 | DMK-8 | MK-8 | C50-P | C55-P | C60-P | Total Pol-P | |
| rth+ | 870 (14) | 157 (3) | 86 (4) | 28 (± 0.2) | 515 (± 18) | 23 (± 0.8) | 566 (± 18) |
| rth-2 | 1,155 (21) | 240 (6) | 99 (4) | 62 (± 5) | 114 (± 38) | 36 (± 12) | 210 (± 55) |
| rth-4 | 1,074 (40) | 236 (4) | 69 (10) | 59 (± 6) | 132 (± 21) | 88 (± 14) | 279 (± 41) |
| rth-6 | 1,131 (19) | 224 (9) | 68 (0.1) | 45 (± 3) | 60 (± 3) | 29 (± 2) | 134 (± 8) |
Values shown represent averages of results obtained from two (for polyprenyl phosphates) or more (for isoprenoid quinones) assays. The numbers in parentheses indicate standard deviations (for isoprenoid quinones) or ranges (for polyprenyl phosphates).
UQ-8, ubiquinone-8; DMK-8, demethylmenaquinone-8; MK-8, menaquinone-8; C50-P, decaprenyl phosphate; C55-P, undecaprenyl phosphate; C60-P, dodecaprenyl phosphate; Pol-P, polyprenyl phosphate.
rth+ indicates the strain MG1655rr/pJK286-rth+ cdsA+. rth-2, rth-4, and rth-6 are its derivatives harboring the respective mutant alleles.
Conclusions.
The results in this work have clearly shown that Rth is responsible for prenyl chain elongation in carrier lipid synthesis in vivo. From this, together with the fact that Rth is structurally homologous to the Micrococcus UDS, we conclude that Rth functions as the cis-prenyltransferase, UDS, for peptidoglycan synthesis. The development of this work greatly stimulated our study of the yeast Rer2 protein, and in fact, we have now demonstrated that the rer2 mutant is deficient in the yeast cis-prenyltransferase activity required for dolichol synthesis and concluded that the RER2 gene encodes this key enzyme (16). Dolichol is essential for N- and O-linked protein glycosylation in yeast, but interestingly, not all the phenotypes of the rer2 mutant can be explained by the glycosylation deficiency, suggesting a novel physiological role of dolichol.
Our discoveries indicate the amazing evolutionary conservation of the RER2 gene family members for the synthesis of carrier lipids, undecaprenyl phosphate for peptidoglycan formation in the bacterial cell wall and dolichyl phosphate in eukaryotic protein glycosylation, and perhaps more. Recently it was reported that the chloroplast monogalactosyldiacylglycerol synthase is homologous to the E. coli and Bacillus subtilis glycosyltransferase, MurG, which catalyzes the last step of peptidoglycan synthesis in bacteria (18). Although the peptidoglycan is a structure unique to bacteria, some homologues of the enzymes involved in peptidoglycan synthesis appear to function in similar reactions of different biological processes.
ACKNOWLEDGMENTS
We are grateful to T. Koyama of Tohoku University for providing information prior to publication. We also thank M. Wachi of the Tokyo Institute of Technology and H. Hara of Saitama University for helpful discussions and T. Mizuno for her excellent technical assistance.
This work was supported by grants to J.K. from the Ministry of Education, Science, Sports and Culture of Japan.
ADDENDUM IN PROOF
During the review process, a paper describing the bacterial UDS (UppS protein) was published (C. M. Apfel, B. Takács, M. Fountoulakis, M. Stieger, and W. Keck, J. Bacteriol. 181:483–492, 1999).
REFERENCES
- 1.Baba T, Allen C M., Jr Substrate specificity of undecaprenyl pyrophosphate synthase from Lactobacillus plantarum. Biochemistry. 1978;17:5598–5604. doi: 10.1021/bi00619a003. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2460–2488. [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Man B, Shao Y. The complete genome sequence of E. coli K12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Peterson S N, Smith H O, Hutchison III C A, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 6.Fujii H, Koyama T, Ogura K. Efficient enzymatic hydrolysis of polyprenyl pyrophosphate. Biochim Biophys Acta. 1982;712:716–718. [PubMed] [Google Scholar]
- 7.Fujisaki S, Nishino T, Katsuki H. Isoprenoid synthesis in Escherichia coli. Separation and partial purification of four enzymes involved in the synthesis. J Biochem. 1986;99:1327–1337. doi: 10.1093/oxfordjournals.jbchem.a135600. [DOI] [PubMed] [Google Scholar]
- 8.Fujisaki S, Nishino T, Katsuki H, Hara H, Nishimura Y, Hirota Y. Isolation and characterization of an Escherichia coli mutant having temperature-sensitive farnesyl diphosphate synthase. J Bacteriol. 1989;171:5654–5658. doi: 10.1128/jb.171.10.5654-5658.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandutsch A A, Paulus H, Levin E, Bloch K. Purification of geranylgeranyl pyrophosphate synthetase from Micrococcus lysodeikticus. J Biol Chem. 1964;239:2507–2515. [PubMed] [Google Scholar]
- 10.Kato J, Ikeda H. Construction of mini-F plasmid vectors for plasmid shuffling in Escherichia coli. Gene. 1996;170:141–142. doi: 10.1016/0378-1119(95)00865-9. [DOI] [PubMed] [Google Scholar]
- 11.Kato J, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishikawa S, Nakano A. Identification of a gene required for membrane protein retention in the early secretory pathway. Proc Natl Acad Sci USA. 1993;90:8179–8183. doi: 10.1073/pnas.90.17.8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 14.Sato K, Nishikawa S, Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene products as a component involved in ER localization of Sec12p. Mol Biol Cell. 1995;6:1459–1477. doi: 10.1091/mbc.6.11.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato M, Sato K, Nakano A. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J Cell Biol. 1996;134:279–293. doi: 10.1083/jcb.134.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato M, Sato K, Nishikawa S, Hirata A, Kato J, Nakano A. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol Cell Biol. 1999;19:471–483. doi: 10.1128/mcb.19.1.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu N, Koyama T, Ogura K. Molecular cloning, expression and purification of undecaprenyl diphosphate synthase—no sequence similarity between E- and Z-prenyl diphosphate synthases. J Biol Chem. 1998;273:19476–19481. doi: 10.1074/jbc.273.31.19476. [DOI] [PubMed] [Google Scholar]
- 18.Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya K. Cloning of the gene for monogalactosyldiacyl-glycerol synthase and its evolutionary origin. Proc Natl Acad Sci USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanimoto K, Iino T. An essential gene for replication of the mini-F plasmid from origin I. Mol Gen Genet. 1984;196:59–63. doi: 10.1007/BF00334092. [DOI] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 21.Zou C, Fujita N, Igarashi K, Ishihama A. Mapping the cAMP receptor protein contact site on the α subunit of Escherichia coli RNA polymerase. Mol Microbiol. 1991;6:2599–2605. doi: 10.1111/j.1365-2958.1992.tb01437.x. [DOI] [PubMed] [Google Scholar]