Abstract
Tail-associated lysozyme of bacteriophage T4 (tail lysozyme), the product of gene 5 (gp 5), is an essential structural component of the hub of the phage baseplate. It is synthesized as a 63-kDa precursor, which later cleaves to form mature gp 5 with a molecular weight of 43,000. To elucidate the role of the C-terminal region of the precursor protein, gene 5 was cloned and overexpressed and the product was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting, analytical ultracentrifugation, and circular dichroism. It was shown that the precursor protein tends to be cleaved into two fragments during expression and that the cleavage site is close to or perhaps identical to the cleavage site in the infected cell. The two fragments, however, remained associated. The lysozyme activity of the precursor or the nicked protein is about 10% of that of mature gp 5. Both the N-terminal mature tail lysozyme and the C-terminal fragment were then isolated and characterized by far-UV circular dichroism and analytical ultracentrifugation. The latter remained trimeric after dissociation from the N-terminal fragment and is rich in β-structure as predicted by an empirical method. To trace the fate of the C-terminal fragment, antiserum was raised against a synthesized peptide of the last 12 C-terminal residues. Surprisingly, the C-terminal fragment was found in the tail and the phage particle by immunoblotting. The significance of this finding is discussed in relation to the molecular assembly and infection process.
The product of gene 5 (gp 5) or tail lysozyme of phage T4 constitutes a structurally essential part of the hub of the baseplate and also functions as lysozyme upon infection of the host bacterium. It was first identified as a band by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by Vanderslice and Yegian (18), and its position in the hub assembly pathway was later worked out by Kikuchi and King (7, 8). Another assembly scheme for the hub was also proposed by Kozloff and Zorzopulos (9) based on a different method, but this scheme is not compatible with that of Kikuchi and King and the assembly pathway remained to be further investigated (3). Kao and McClain (6) in 1980 first showed that the lysozyme activity associated with the phage particle was due to gp 5 by isolating a temperature-sensitive pseudorevertant of the gene e amber mutant, T4D.5ts1. Subsequently, Nakagawa et al. (12) isolated gp 5 from the phage tail and characterized the lysozyme activity. As is the case for T4 lysozyme (gp e), gp 5 has N-acetylmuramidase activity, but the optimal pH for maximum activity of the tail lysozyme is 5.8, 1 pH unit lower than that for gp e.
The nucleotide sequence of gene 5 was determined by Mosig et al. (11). They found that the open reading frame of gene 5 encoded 575 amino acid residues and that the central segment of gp 5 has a striking similarity to T4 lysozyme. However, the overexpressed protein from the cloned gene 5, which had the expected molecular weight of 63,000, did not possess lysozyme activity. Since this protein is larger by approximately 20 kDa than the gp 5 isolated from the phage tail, the authors surmised that gp 5 is expressed as a precursor and that the 20-kDa peptide is cleaved off later to allow activity.
There are a number of mutants with distinct phenotypes which have a mutation in gene 5 (4, 6, 20). All the mutation sites were recently determined by sequencing the genomic DNA from each mutant (15). Among them, the 5ts1 mutant described above was identified as Gly322Asp, which is located in the lysozyme domain.
In this paper, we present evidence, obtained by immunoblotting, that the 20-kDa peptide remains associated with the tail of the phage particle. Our experimental results also indicated that the cleavage does not involve any T4 phage protein product but gp 5.
MATERIALS AND METHODS
Bacteria and bacteriophages.
All the bacteria and bacteriophages used in this work are listed in Table 1. Amber mutant 5amB256 was obtained from W. B. Wood, University of Colorado.
TABLE 1.
Bacterial strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or phage | Relevant properties and use | Source or reference |
|---|---|---|
| E. coli strains | ||
| BE | Su−, nonpermissive for T4 amber; selective plating and growth of am+ phage | Our collection |
| B40(Su1) | Su+; growth of T4 amber mutant | Our collection |
| BL21(DE3) | Carries T7 RNA polymerase in λDE3 integrated into the chromosome of BL21 | Novagen (14) |
| XL1-Blue | Transform and keep plasmids in this work | Our collection (2) |
| JM109 | Cell wall for turbidity assay | Our collection (21) |
| Plasmids | ||
| pET29a | Expresses genes under T7 promoter, Kmr | Novagen |
| pSZ146 | Contains gene 5 in pET29a | This work |
| pSZ202-2 | Contains gene 5 in pET29a, expresses gene 5 with histidine tag | This work |
| T4 phages | ||
| T4D | Wild type | Our collection |
| 5amB256 | Gene 5 amber mutant | W. B. Wood |
| 23amH11 | Gene 23 amber mutant | Our collection |
Plasmid construction.
Gene 5 of phage T4 was amplified by PCR from the T4 genome with primers that were designed to add a BamHI and a SalI site (plus a 4-base clamp) to both ends of the PCR products. Thus, the first primer began with 5′-CGGGATCCGGAGAAATCTTATGAAATG-3′ (BamHI site in bold) and the second one began with 5′-TTGTCGACTTAGCCAATGTCAATCCTCG-3′ (SalI site in bold). PCR was performed with Pfu DNA polymerase (Stratagene) for 30 cycles with T4 dc DNA (Takara). The PCR products were purified with a Qia-Tip (Qiagen), digested with BamHI and SalI, ligated to the expression vector pET29a (Novagen), and transformed into Escherichia coli XL1-Blue. The gene 5 insert was verified to have the correct sequence by DNA sequencing. This expression vector, named pSZ146, was introduced into E. coli BL21(DE3). For the C-terminal histidine tag, site-directed mutagenesis was carried out by PCR mutagenesis from pSZ146 by using the QuikChange site-directed mutagenesis kit (Stratagene). The PCR primers were 5′-GACATTGGCTCAGTCGACAAGCTTGC-3′ and 5′-GCAAGCTTGTCGACTGAGCCAATGTC-3′ (bold letters indicate the mutation site and the serine codon alternate for the stop codon). The resulting vector was digested with SalI and XhoI and ligated to delete extra peptide between the end of whole gene 5 and the histidine tag. This expression vector, named pSZ202-2, was introduced into E. coli BL21(DE3).
DNA sequencing.
A Perkin-Elmer–Applied Biosystems 373A DNA sequencer was used for DNA sequencing in combination with the Taq DyeDeoxy Terminator cycle-sequencing kit. The plasmid DNA was prepared with a Qia-Tip. In the cycle-sequencing reactions, the manufacturer’s instructions were modified to use 0.15 to 0.3 pmol of PCR product and 10 to 20 pmol of sequencing primer.
Expression and isolation of fusion proteins.
Six-histidine-tagged precursor gp 5 (pre-gp5-his) fusion proteins in E. coli was produced as follows. E. coli BL21(DE3) harboring pSZ202-2 was grown in Luria-Bertani broth at 37°C overnight. The culture was diluted 20-fold with the same medium and incubated at 37°C for 4 h, and expression of pre-gp5-his was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. After further incubation for 3 h at 37°C, the cells were harvested by centrifugation.
The cell pellets were resuspended in 40 ml of buffer (50 mM HEPES [pH 7.9], 10% glycerol, 0.1% Triton X-100, 0.5 M KCl, 5 mM imidazole) on ice. The cells were then lysed by sonication (Branson Sonifier 250). The solution was centrifuged at 28,000 × g for 30 min (Tomy). The supernatant was loaded onto a preequilibrated 5-ml Ni-nitrilotriacetate (NTA)-agarose (Qiagen) column at 4°C. pre-gp5-his was eluted from the column with a 5 to 250 mM imidazole gradient. EDTA was added to each collected fraction to give a final concentration of 5 mM.
Isolation of mature gp 5 from histidine-tagged proteins.
The pre-gp5-his as isolated above turned out to contain 70% of nicked product; the cleavage site was between Ser351 and Ala352 (see Results). The N-terminal fragment turned out to be the mature gp 5 (gp 5*). To isolate gp 5*, the pre-gp5-his as prepared above was mixed with buffer D (2 M guanidine-HCl, 50 mM Tris-HCl [pH 8.0]) in a 50-ml plastic tube and 5 ml of Ni-NTA-agarose was added. The tube was rotated overnight at 4°C, and then the sample mixture with the resin was loaded on a column. gp 5* was found in the flowthrough fractions. The column was washed with buffer D, and the histidine-tagged protein was eluted from the column at an imidazole concentration of 200 mM. The eluted solution was collected into fractions. After elution, 0.5 M EDTA (pH 8.0) was added to each fraction to give a final concentration of 5 mM.
SDS-PAGE.
SDS-PAGE was carried out as specified by Laemmli (10) in a vertical minislab gel (9 by 7.5 cm) and stained with Coomassie brilliant blue R-250 by the method of Wyckoff et al. (19).
Amino-terminal sequence analysis.
Automatic Edman degradation was carried out with a gas-phase sequencer (Shimadzu PPSQ-21 protein sequencer). Phenylthiohydantoin derivatives of amino acids were analyzed with an on-line high-performance liquid chromatography system.
Immunoblotting.
Immunoblotting was carried out by the method of Harlow and Lane (5). Proteins were run on an SDS-PAGE gel and electroblotted onto an Immobilon membrane (Millipore Corp.). Anti-gp 5* antiserum or anti-C-terminal peptide (QYTIDGSRIDIG) antiserum was used as the primary antibody; alkaline phosphatase-labeled F(ab′)2 goat anti-rabit immunoglobulin G heavy plus light chains (Zymed) was used as the secondary antibody. Nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate-p-toluidine salt were used for staining.
Preparation of tails.
Tails were obtained from lysates of 23amH11-infected E. coli BE cells by the method of Tschopp et al. (16), except that the lysate was precipitated by ammonium sulfate at 40% saturation (0.224 g/ml) to remove ribosomes and membrane fragments. The ammonium sulfate precipitation was repeated three times, and the resultant suspension was dialyzed against 0.1 M phosphate buffer (pH 7.2), before being subjected to ultracentrifugation. The dialyzed solution was centrifuged at 140,000 × g for 90 min, and the pellet was resuspended in a small volume of 0.1 M phosphate buffer (pH 7.2). The concentrated tail solution was then layered on a 10 to 30% linear sucrose gradient containing 0.1 M phosphate buffer (pH 7.2) and centrifuged for 2.5 h at 200,000 × g. The tail peak fractions were collected and dialyzed against 0.1 M phosphate buffer (pH 7.2) to remove sucrose.
Preparation of cell walls for turbidity assay of lysozyme.
The preparation of cell walls was based on the method of Tsugita et al. method (17). E. coli JM109 was cultivated in 300 ml of Luria-Bertani broth with 40 ml of UV buffer (0.021 M Na2HPO4, 0.011 M KH2PO4, 0.068 M NaCl, 0.029 M K2SO4, 1 mM MgSO4, 0.1 mM CaCl2, 0.01% gelatin) and 10 ml of 10% glucose at 37°C. At an absorbance at 600 nm of 0.5, the cells were centrifuged at 2,000 × g for 10 min. Cell pellets were resuspended and washed with ice-cold UV buffer and then centrifuged at 2,000 × g for 10 min. Recovered cell pellets were resuspended and washed with ice-cold water and then centrifuged at 2,000 × g for 10 min. Finally, the cell pellets were frozen in liquid nitrogen and lyophilized.
Turbidity assay.
The turbidity assay was based on the method of Tsugita et al. (17). The cell wall preparation was dissolved in 50 mM Tris (pH 7.5), and the optical density at 450 nm was adjusted to about 0.7. A 1-ml volume of the cell wall solution was poured into a cuvette and quickly mixed with 20 μl of various concentrations of sample solution. The cuvette was immediately placed in a spectrophotometer (Beckman DU640), and the time course of solution clearing was recorded at 450 nm for 1 min.
CD and secondary-structure estimation.
The far-UV circular dichroism (CD) spectrum was measured at 25°C on a J-720 spectropolarimeter (Jasco) in a 1-mm-path quartz cell. The spectrum was analyzed for estimation of the secondary structure by CONTIN, a computer program developed by Steven W. Provencher (13).
Analytical ultracentrifugation.
Sedimentation equilibrium analysis was carried out with an Optima XL-I analytical ultracentrifuge (Beckman). The sedimentation equilibrium profile was monitored at 280 nm, and the data were analyzed with the software provided by the manufacturer.
RESULTS
Overexpression and isolation of pre-gp5-his.
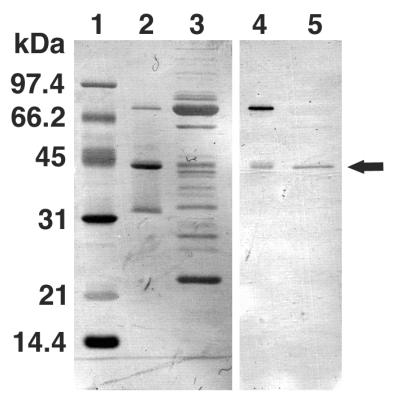
To isolate and characterize the precursor tail lysozyme protein gp 5, gene 5 was cloned into an expression vector with a histidine tag at the 3′ end of the gene and the gene was overexpressed and purified on a nickel column (see Materials and Methods). SDS-gel electrophoresis of the purified protein revealed three bands with molecular weights of 75,000, 43,000, and 32,000; the largest bands corresponded to the expected pre-gp5-his (Fig. 1). The N-terminal sequences of these three bands as determined by Edman degradation were MEMISNN-, MEMISNN-, and AMAATVA-, respectively, indicating that the last two bands were the cleavage products of the first and that the two fragments were held together after cleavage. The 43-kDa fragment was observed to migrate in SDS-gel electrophoresis at a rate identical to that of the mature tail lysozyme (Fig. 1). This indicated that the cleavage upon maturation probably occurred at the identical site, namely, between Ser351 and Ala352, and that the cleavage does not require the expression of any other phage gene.
FIG. 1.
Overexpression and isolation of the tail lysozyme, gp 5. After SDS-PAGE, overexpressed proteins were transferred onto a polyvinylidene difluoride membrane and stained with Coomassie brilliant blue R250 (lanes 1 to 3) or subjected to immunoblotting (lane 4 and 5). Lanes: 1, molecular mass standard; 2 and 4, overexpressed proteins after Ni-NTA column elution; 3 and 5, tail proteins. Arrow, gp 5* (mature gp 5). The proportional staining density of each band reflects the efficiency of transfer to PVDF membrane and does not necessarily represent the stoichemistry.
Isolation and characterization of the N-terminal 43-kDa and C-terminal 32-kDa fragments.
Since it was difficult to separate the intact precursor protein from the cleaved protein, we proceeded to purify the N-terminal fragment as follows. The overexpressed protein which contained both intact and nicked precursor protein was retained on the nickel column. The immobilized nicked protein was denatured with a wash of 2 M Gdn-HCl, which caused the N-terminal protein to dissociate and flow through the column. Fractions of the N-terminal protein were collected, and the Gdn-HCl was removed by dialysis. After elution with 2 M Gdn-HCl, the C-terminal 32-kDa fragment which remained bound to the column was eluted by 200 mM imidazole.
Maturation of gp 5.
It was shown by SDS-PAGE that some of the pre-gp5-his was cleaved into 43- and 32-kDa proteins immediately after expression (Fig. 1). To prove that the 43-kDa protein is identical to the mature gp 5 in the tail, the 43-kDa protein and the tail that contained the mature gp 5 were analyzed simultaneously by SDS-gel electrophoresis in which the tail lysozyme was detected by immunoblotting. Figure 1 shows that the 43-kDa protein and gp 5 are indistinguishable in size and also that the 43-kDa protein is stained by anti-gp 5 antibody. Since the C-terminal sequence of the mature gp 5 has not been determined, this is not a strict proof, but it is very likely that the two proteins are identical and that the 32-kDa protein is the C-terminal domain with six histidine residues at the C terminus (C-term-his). Maturation of gp 5 occurs immediately after expression by cleavage between Ser351 and Ala352. The fact that the 43-kDa protein was eluted together with the 32-kDa C-term-his protein from an Ni-NTA column indicated that the two fragments remained together as a complex after cleavage.
Presence of the C-terminal fragment in the tail and phage particle.
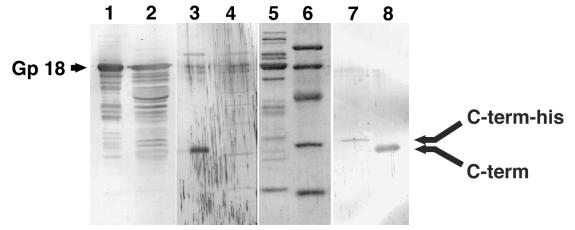
To elucidate the fate of the C-terminal polypeptide after cleavage, antiserum was raised against the C-terminal peptide of precursor gp 5 (see Materials and Methods). By using this antiserum, tail proteins and the lysate of 5am phage-infected cells as well as overexpressed pre-gp5 were examined by immunoblotting. As expected, the third band, which was not stained by anti-mature gp 5 or gp 5* antiserum, was stained with this antiserum and no band was detected in the lysate of 5am phage-infected cells (Fig. 2, lanes 3 and 4). It was unexpected that the tail revealed one stained band (Fig. 2, lane 8). This indicated that the C-terminal polypeptide stayed as a structural component after cleavage. It turned out that the C-terminal polypeptide stays in the phage particle, as shown below.
FIG. 2.
Analysis of the C-terminal fragment of gp 5. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane and stained with Coomassie brilliant blue R250 (lanes 5 and 6) or subjected to immunoblotting (lanes 1 to 4, 7, and 8). Anti-gp 18 antiserum was used in lanes 1 and 2, anti C-terminal peptide (QYTIDGSRIDIG) antiserum was used in lanes 3, 4, 7, and 8. Lanes: 1, 3, 5, and 8, tail proteins; 2 and 4, 5am phage particle; 6, molecular mass standard; 7, pre-gp5-his complemented 5am phage.
This prompted us to check if the C-terminal fragment is also present in the phage particle. For that purpose, E. coli BL21(DE3), which harbored a plasmid with the pre-gp5-his gene, was infected with T4D.5amB256, a gene 5 amber mutant that has an amber mutation at codon 10. Almost the same number of phages was produced as in the case where E. coli B40(Su1) without plasmid was infected by the same amber phage (data not shown). The complemented 5am phage was then purified by differential centrifugation from the lysate and analyzed by immunoblotting. Figure 2, lane 7, shows that the complemented phage particle has histidine-tagged C-terminal fragment. This observation supported our notion that the cleavage of pre-gp5-his after expression reflects the process of in vivo maturation and that the C-terminal polypeptide stays in the virion.
Lysozyme activity of gp 5 and suppression by the C-terminal fragment.
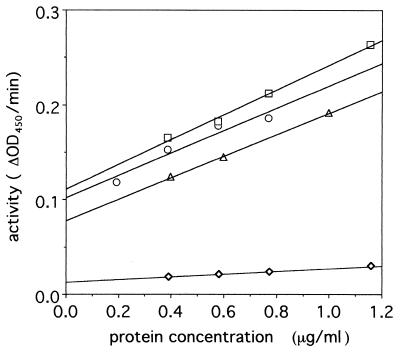
The lysozyme activity of the precursor protein and that of the 43-kDa fragment were compared. The lysozyme activity of the 43-kDa fragment was 10 times higher than that of the precursor proteins (Fig. 3). This indicated that the C-terminal fragment inhibits the lysozyme activity. At present it is not clear whether precursor gp 5 activity was due to the presence of some nicked protein or to the inherent activity of the intact precursor protein.
FIG. 3.
Turbidity assay of lysozyme activity. The lysozyme activity of gp 5* (□), gp 5* mixed with isolated C-term-his (○), hen egg lysozyme standard (▵), and nicked pre-gp5-his (◊) are shown. The activities of 1 μg of protein for the lysozyme proteins were 0.13, 0.12, 0.11, and 0.014, respectively. The assay was carried out as described in Materials and Methods. OD450, optical density at 450 nm.
CD spectra of the gene products.
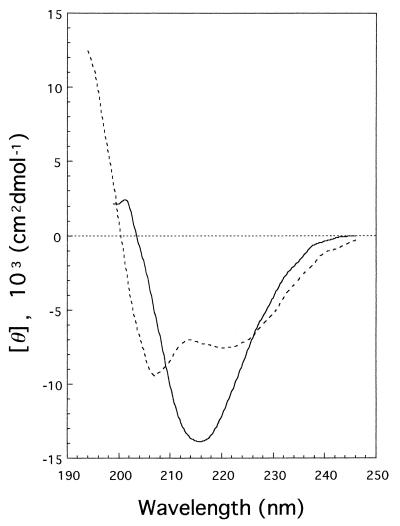
CD spectra of mature gp 5 and the C-terminal fragment were measured in the far-UV region (Fig. 4) and were analyzed by CONTIN (13) to estimate the secondary structure. The gp 5* was estimated to have 27% α-helix, 31% β-sheet, and 24% β-turn, whereas the C-term-his is rich in β-structure, with 11% α-helix, 58% β-sheet, and 0% β-turn.
FIG. 4.
Far-UV CD spectrum of gp 5* (---) and C-term-his (—). The secondary structures of gp 5* and C-term-his, as estimated by CONTIN, were 27% α-helix, 31% β-sheet, and 24% β-turn and 11% α-helix, 58% β-sheet, and 0% β-turn, respectively.
Sedimentation equilibrium analysis.
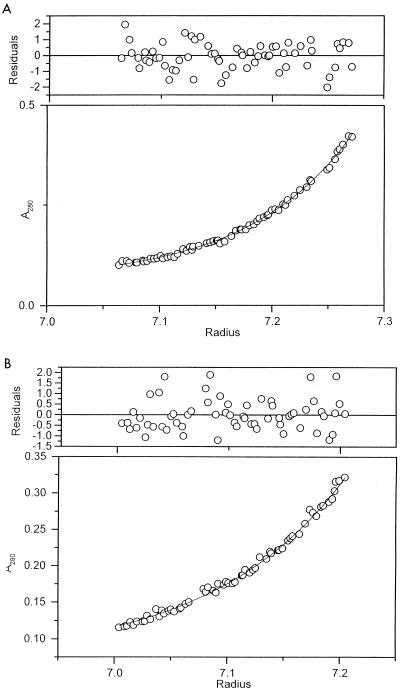
To establish the subunit structure of the tail lysozyme, the molecular weights and sedimentation coefficients of gp 5* and nicked pre-gp5-his were measured by sedimentation analysis. Sedimentation equilibrium analysis revealed that their molecular weights were 194,400 ± 10,410 and 40,700 ± 4,500 (mean ± standard deviation), respectively (Fig. 5). The former value is close to 189,300, which is about three times the molecular weight of monomer pre-gp5-his, 63,100, estimated from the amino acid sequence. On the other hand, the molecular weight for monomer gp 5* is close to that estimated from the amino acid sequence, namely, 38,810. These results indicate that pre-gp5-his is a trimer whereas gp 5* is a monomer.
FIG. 5.
Sedimentation equilibrium of pre-gp5-his (A) and gp 5* (B). Molecular weights of 194,400 ± 10,410 and 40,700 ± 4,500 for pre-gp5-his and gp 5*, respectively, were obtained, which indicated that the former is a trimer, and the latter is a monomer. Residuals are the difference between the experimental data and the fitted curve and have to be evenly scattered throughout the cell.
DISCUSSION
Many proteins are known to be expressed as precursor proteins from which an extra peptide is cleaved off at some stage after biosynthesis. Before cleavage, the extra peptide may function as a signal for protein transport, a suppressor of enzymatic activity, or a helper of protein folding, as in the case of intramolecular chaperones. Major head proteins, including the scaffolding protein gp 22, are known to be cleaved by the phage-encoded prohead protease, gp21. Prohead protease itself is self-cleaved. Cleavage of the head proteins is a prerequisite for head expansion and DNA packaging. In contrast, gp 5 is the only gene product among tail proteins for which processing is suggested. Mosig et al. (11) showed that the overexpressed pre-gp5 has no lysozyme activity and surmised that if the precursor domain were cleaved off it would inhibit lysozyme activity during morphogenesis of the phage. The data in the present paper basically supported their notion. In fact, the enzyme activity of pre-gp5-his or its nicked products is only 1/10 that of gp 5*, the mature gp 5. In addition, cleavage occurs immediately after expression. Mosig et al. (11) speculated that processing of pre-gp5 requires a phage-induced protein(s). Our observation, however, indicates that the processing occurs either via an E. coli enzyme or autolytically.
In this study we found that the C-terminal fragment stayed in the baseplate and also in the final virion after cleavage. However, a sedimentation equilibrium study has revealed that the pre-gp5-his or its nicked products are trimeric whereas gp 5* is monomeric. The C-terminal fragment is thus necessary not only for the suppression of the lysozyme activity but also for trimer formation. The functional difference between gp 5* and the C-terminal fragment is also reflected in the difference in the CD spectra in that the C-terminal fragment has much more β-structure (58%) than does gp 5* (31%). It is noted that the C-terminal region contains 12 repeated VXGXXXXX sequences, which are predicted to form β-structure. The question then arises whether the C-terminal fragment also functions in the penetration process of the infection process, but this remains to be elucidated.
Although a strict proof awaits determination of the C-terminal sequence of the mature gp 5 in the baseplate, our present results strongly suggest that the cleavage site we observed for the overexpressed pre-gp5-his protein, between Ser351 and Ala352, is identical to the site used for in vivo maturation of the gene product. The observed molecular weight of gp 5* by SDS-PAGE, i.e., 43,000, may appear much higher than the calculated value of 38,800, but many other tail proteins also give higher molecular weights by SDS-PAGE than the calculated values. For example, the calculated molecular weights of gp 48 and gp 28 are 39,700 and 17,300, and observed values are 44,000 & 24,000, respectively. The immunoblotting analysis reveals that gp 5* is indistinguishable in size from authentic gp 5 in the tail (Fig. 1). The other evidence that gp 5* is the mature form of gp 5 is based on isoelectric focusing gel electrophoresis (data not shown). The isoelectric point (pI) of gp 5* is around 8.0, which is identical to that of the mature gp 5 in the tail (12). We propose that gp 5 forms a trimer immediately after synthesis and that the trimerized pre-gp5 is then cleaved between Ser351 and Ala352, with the C-terminal fragment remaining in the complex. Whether cleavage occurs through a self-cleavage or via E. coli protease activity remains to be elucidated. The C-terminal domain thus stays as a part of the baseplate structure until infection, when it is supposedly released together with gp 5*. We surmise that in some way the C-terminal domain helps the tail lysozyme diffuse into the periplasm.
5am phage which had been complemented with pre-gp5-his retained the C-terminal fragment with the histidine tag (Fig. 2), indicating that pre-gp5-his, which has six C-terminal histidine residues, was functional. This supports the idea that the C-terminal fragment is located beneath the baseplate and that the C-terminal domain does not interact with other baseplate proteins.
Upon infection, the C-terminal fragment is supposedly released from the baseplate and gp 5 is thus activated. Once gp 5 is converted to the active form, gp 5*, the lysozyme activity is not suppressed by addition of the C-terminal fragment (Fig. 3), indicating that gp 5* no longer interacts with the C-terminal fragment. In this regard, it is interesting that the spackle (Sp) protein, now identified as gp 61.3 (22), is suggested to bind to gp 5 during superinfection exclusion, when the cell is resistant to lysis from without (1). Sp protein may act as lysozyme inhibitor in place of the C-terminal fragment during assembly.
ACKNOWLEDGMENTS
This work was supported in part by a grant to F.A. from the Ministry of Education, Science, and Culture of Japan. N.C.G. was supported in part by NIH grant GM21967 to B. W. Matthews.
REFERENCES
- 1.Abedon S T. Lysis and the interaction between free phages and injected cells. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 397–405. [Google Scholar]
- 2.Bullock W O, Fernandez J M, Short J M. XL-1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 3.Coombs D H, Arisaka F. T4 tail structure and function. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 259–291. [Google Scholar]
- 4.Hall D H, Sargent R G, Trofatter K F, Russell D L. Suppressors of mutations in the bacteriophage T4 gene coding for both RNA ligase and tail fiber attachment activities. J Virol. 1980;36:103–108. doi: 10.1128/jvi.36.1.103-108.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 471–510. [Google Scholar]
- 6.Kao S H, McClain W H. Baseplate protein of bacteriophage T4 with both structural and lytic functions. J Virol. 1980;34:95–103. doi: 10.1128/jvi.34.1.95-103.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975;99:695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi Y, King J. Genetic control of bacteriophage T4 baseplate morphogenesis. II. Mutants unable to form the central part of the baseplate. J Mol Biol. 1975;99:673–694. doi: 10.1016/s0022-2836(75)80179-3. [DOI] [PubMed] [Google Scholar]
- 9.Kozloff L M, Zorzopulos J. Dual functions of bacteriophage T4D gene 28 product: structural component of the viral tail baseplate central plug and cleavage enzyme for folyl polyglutamates. I. Identification of T4D gene 28 product in the tail plug. J Virol. 1981;40:635–644. doi: 10.1128/jvi.40.3.635-644.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Mosig G, Lin W, Franklin J, Fan W H. Functional relationships and structural determinants of two bacteriophage T4 lysozymes: a soluble (gene e) and a baseplate-associated (gene 5) protein. New Biol. 1989;1:171–179. [PubMed] [Google Scholar]
- 12.Nakagawa H, Arisaka F, Ishii S. Isolation and characterization of the bacteriophage T4 tail-associated lysozyme. J Virol. 1985;54:460–466. doi: 10.1128/jvi.54.2.460-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provencher S W, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 14.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S, Hoshida K, Arisaka F. Mapping of functional sites on the primary structure of the tail lysozyme of bacteriophage T4 by mutational analysis. Biochim Biophys Acta. 1998;1384:243–252. doi: 10.1016/s0167-4838(98)00016-8. [DOI] [PubMed] [Google Scholar]
- 16.Tschopp J, Arisaka F, van Driel R, Engel J. Purification, characterization and reassembly of the bacteriophage T4D tail sheath protein P18. J Mol Biol. 1979;128:247–258. doi: 10.1016/0022-2836(79)90128-1. [DOI] [PubMed] [Google Scholar]
- 17.Tsugita A, Inouye M, Terzaghi E, Streisinger G. Purification of bacteriophage T4 lysozyme. J Biol Chem. 1968;243:391–397. [PubMed] [Google Scholar]
- 18.Vanderslice R W, Yegian C D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974;60:265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- 19.Wyckoff M, Rodbard D, Chrambach A. Polyacrylamide gel electrophoresis in sodium dodecyl sulfate-containing buffers using multiphasic buffer systems: properties of the stack, valid Rf- measurement, and optimized procedure. Anal Biochem. 1977;78:459–482. doi: 10.1016/0003-2697(77)90107-5. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, Uchida H. Organization and function of bacteriophage T4 tail. I. Isolation of heat-sensitive T4 tail mutants. Virology. 1973;52:234–245. doi: 10.1016/0042-6822(73)90412-1. [DOI] [PubMed] [Google Scholar]
- 21.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 22.Yonesaki, T. Personal communication.