SUMMARY
Reprogramming glial cells to convert them into neurons represents a potential therapeutic strategy that could repair damaged neural circuits and restore function. Recent studies show that downregulation of the RNA-binding protein PTBP1 leads to one-step conversion of Müller glia (MG) into retinal ganglion cells (RGCs) with a high efficiency. However, the original study did not perform fate-mapping experiments to confirm MG-to-RGC conversion after Ptbp1 downregulation. To address the fundamental question of whether Ptbp1 downregulation can convert MG into RGCs in the mouse retina, we perform fate-mapping experiments to lineage trace MG independent of the adeno-associated virus (AAV)-mediated labeling system. Here, we report that Ptbp1 downregulation by CRISPR-CasRx or small hairpin RNA is insufficient to convert MG to RGCs. The original conclusion of MG-to-RGC conversion is due to leaky labeling of endogenous RGCs. Our results emphasize the importance of using stringent fate mapping to determine glia-to-neuron conversion in cell reprogramming research.
Graphical Abstract
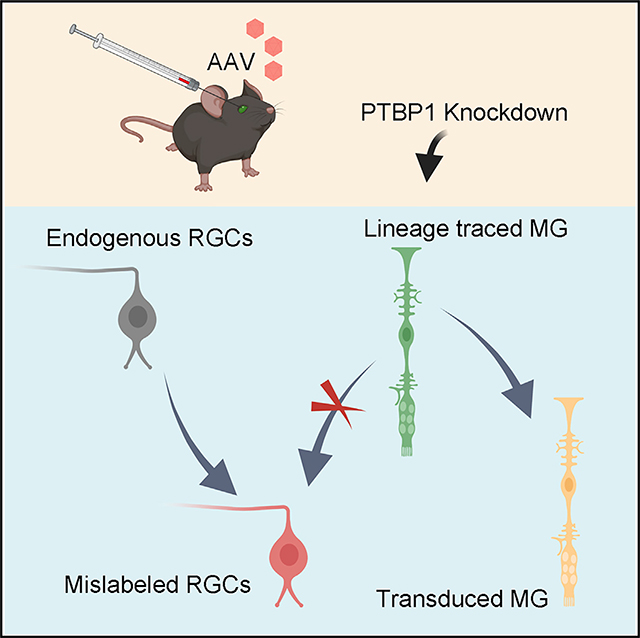
In brief
Leaky labeling of endogenous retinal ganglion cells (RGCs) leads to misinterpretation of glia-to-neuron conversion in the mouse retina. Using stringent fate-mapping experiments, Xie et al. show that lineage-traced Müller glia (MG) are not converted into RGCs after Ptbp1 downregulation by CRISPR-CasRx or small hairpin RNA.
INTRODUCTION
In zebrafish, Müller glia (MG) are a source of retinal stem cells that can replenish neuronal loss and restore vision, establishing a powerful self-repair mechanism (Goldman, 2014). In mammals, however, MG lack regenerative capability, as they do not spontaneously generate new retinal neurons in response to cell loss after injury or in a variety of retinal degenerative diseases (Hoang et al., 2020; Karl and Reh, 2010). Exciting progress made in recent years demonstrates that MG can be reprogrammed in vivo to generate new rod photoreceptors, as well as bipolar and amacrine-like cells, by manipulating the expression levels of transcription factors that control cell proliferation, cell fate specification, and determination (Hoang et al., 2020; Jorstad et al., 2017; Ueki et al., 2015; Yao et al., 2018).
Retinal ganglion cells (RGCs) are the sole output neurons in the retina that transmit visual information from the eye to the brain via their long-distance axon projections bundled together in the optic nerve. Degeneration of RGC somas and axons is a leading cause of vision impairment and blindness in a variety of pathological states, such as retinal ischemia, optic nerve transection, intracranial hypertension, and glaucoma (Levin and Gordon, 2002). Regenerative therapy via in vivo reprogramming of MG to RGCs holds great potential for vision restoration in the treatment of an array of retinal degenerative diseases affecting RGCs (Blackshaw and Sanes, 2021).
PTBP1 (polypyrimidine tract-binding protein 1), an RNA-binding protein, has emerged as a prominent reprogramming factor for neuronal regeneration, as downregulation of its expression confers one-step direct conversion of mouse fibroblasts into functional neurons in vitro (Xue et al., 2013), as well as conversion of astroglial cells into functional brain neurons in vivo (Qian et al., 2020). Recently, Zhou et al. reported that Ptbp1 downregulation can also convert MG into functional RGCs in the adult mouse retina (Zhou et al., 2020), representing potential major advances in the fields of MG reprogramming and RGC axon regeneration: (1) de novo regeneration of RGCs from reprogrammed MG in vivo and (2) full-length regeneration of RGC axons from MG-converted RGCs. However, fate-mapping experiments, the gold standard to definitively confirm the origin of newly generated RGCs, were not performed in the studies. Therefore, we carried out fate-mapping experiments to lineage trace MG and their progeny after Ptbp1 downregulation using either the CRISPR-CasRx-mediated knockdown system developed by the original authors (Zhou et al., 2020) or the small hairpin RNA (shRNA)-based depletion system developed by the other group (Qian et al., 2020). Surprisingly, we found no evidence of MG-to-RGC conversion or axon regeneration after Ptbp1 downregulation using either system in the intact or injured adult mouse retina. Furthermore, our results revealed that the original conclusion of MG-to-RGC conversion after Ptbp1 knockdown was due to leaky adeno-associated virus (AAV)-mediated expression that labeled endogenous RGCs. Our results demonstrate that downregulation of Ptbp1 is not sufficient to convert MG into RGCs in the adult mouse retina, emphasizing the importance of performing stringent fate-mapping experiments in glia-to-neuron reprogramming research.
RESULTS
AAV-based Cre recombination is unsuitable for examining MG-to-RGC conversion
Zhou et al. used an AAV-based Cre recombination system under the control of a GFAP mini-promoter to label MG in reporter mice for reprogramming MG to RGCs after Ptbp1 downregulation (Zhou et al., 2020). To critically examine MG-to-RGC conversion after Ptbp1 knockdown, we first repeated the experiments by Zhou et al. using their AAV-based Cre recombination for MG labeling and CRISPR-CasRx for Ptbp1 downregulation. To clearly detect MG migration to the ganglion cell layer (GCL), an essential step for successful MG-to-RGC conversion, we used Sun1-GFP reporter mice (Rosa-CAG-LSL-Sun1-GFP), which express Sun1-GFP fusion protein at the inner nuclear membrane of targeted cell types after Cre-dependent removal of a floxed STOP cassette (Mo et al., 2015). Following the same experimental methodology described by Zhou et al. (2020), we used rep-cap PHP.eB (Chan et al., 2017) for AAV packaging and production and performed subretinal co-injection of the following AAVs (>1 × 1013 vg/mL): AAV-GFAP-Cre, together with AAV-GFAP-CasRx-Ptbp1 (Ptbp1 downregulation) or AAV-GFAP-CasRx (control) in 5-week-old Sun1-GFP reporter mice. We examined MG-to-RGC conversion at 4 weeks after AAV injection (Figures 1A and 1B). Indeed, we observed that an equivalent number of Sun1-GFP+ cells were located in the GCL and they were co-immunostained with RBPMS, a specific marker for RGCs (Figures 1C–1G), indicative of repeatable results of “MG-to-RGC conversion” after Ptbp1 downregulation (Zhou et al., 2020). However, a similar “conversion” efficiency was also observed in the control group (Figures 1C and 1H–1K). This observation raised the possibility that the “MG-to-RGC conversion” could be caused by leaky expression of AAV-GFAP-Cre that labeled the endogenous RGCs. To examine such a possibility, we performed subretinal injection of AAV-GFAP-Cre under the same experimental conditions. Four weeks after AAV injection, RGCs were labeled with a similar efficiency after AAV-mediated Cre recombination in the absence of Ptbp1 downregulation by CRISPR-CasRx (Figures 1C and 1L–1O). These results indicate that the AAV system used by Zhou et al. labels not only MG but also endogenous RGCs and, therefore, is unsuitable for critical examination of MG-to-RGC conversion.
Figure 1. AAV-mediated Cre recombination labels endogenous RGCs and thus is unsuitable for examining MG-to-RGC conversion.
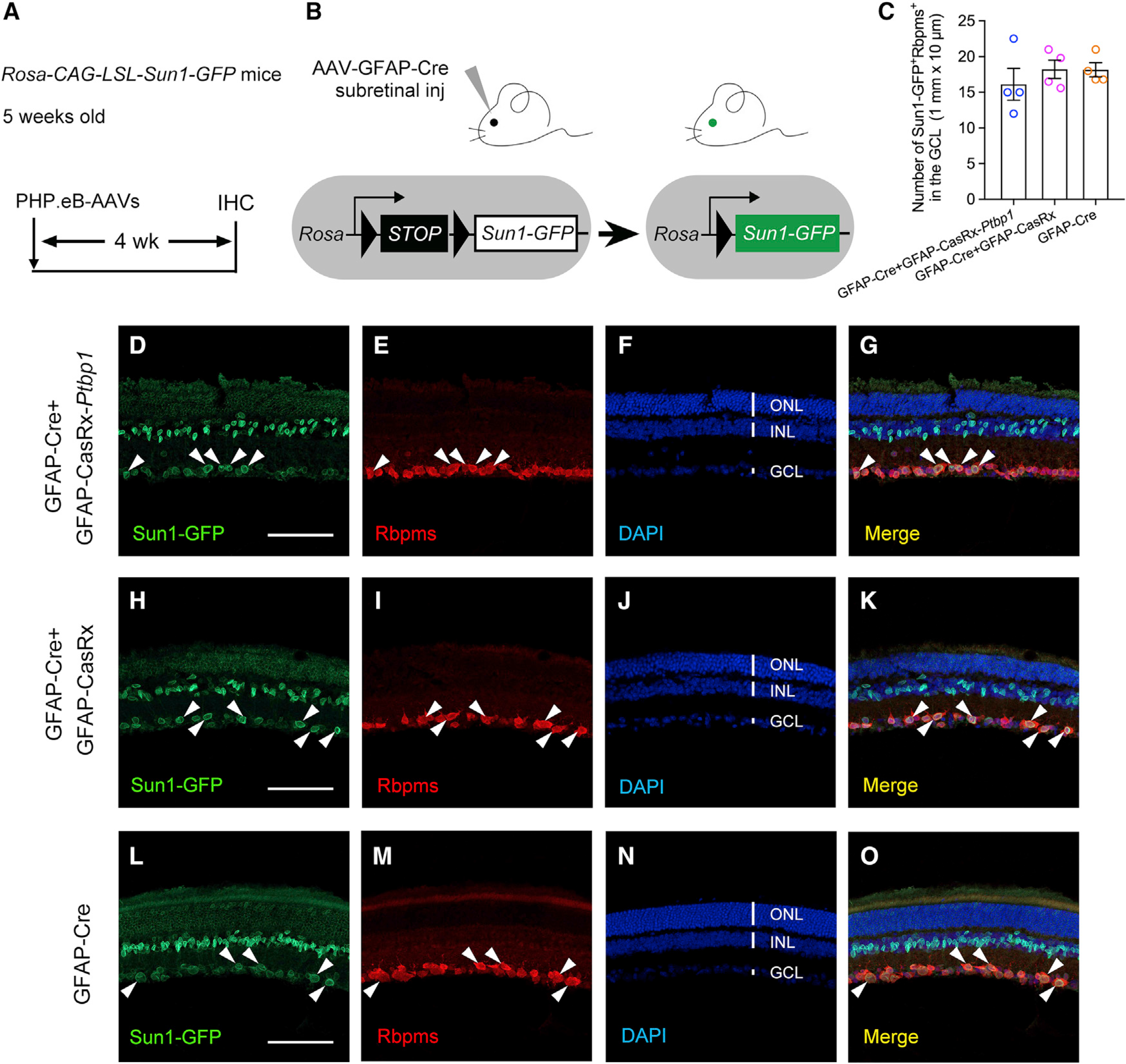
(A) Experimental design for testing MG-to-RGC conversion. Subretinal injection of PHP.eB-AAVs was performed in Rosa-CAG-LSL-Sun1-GFP reporter mice at 5 weeks of age, followed by immunohistochemistry analysis using the pan-RGC marker RBPMS at 4 weeks after AAV injection. IHC, immunohistochemistry.
(B) Schematic illustration showing that AAV-GFAP-Cre subretinal injection results in Sun1-GFP expression after Cre-dependent recombination in Sun1-GFP reporter mice.
(C) Quantification of Sun1-GFP+Rbpms+ cells in the ganglion cell layer (GCL) at 4 weeks after AAV injection. n = 4 retinas per group. Data are presented as the mean ± SEM. No significant difference was found between any two groups, p > 0.5, unpaired t test.
(D–G) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry after Ptbp1 knockdown in retinas receiving GFAP-Cre and GFAP-CasRx-Ptbp1. White arrowheads: Sun1-GFP-labeled cells were RBPMS positive in the GCL.
(H–K) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving GFAP-Cre and GFAP-CasRx. White arrowheads: Sun1-GFP-labeled cells were RBPMS positive in the GCL.
(L–O) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving GFAP-Cre only. White arrowheads: Sun1-GFP-labeled cells were RBPMS positive in the GCL.
(D–O) Scale bars, 100 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. See also Figure S1.
Following the same experimental methodology described by Zhou et al. (Zhou et al., 2020), we next examined whether Ptbp1 knockdown can promote axon regeneration in Rosa-CAG-LSL-tdTomato (Ai9) reporter mice at 4 weeks after AAV injection (Figures S1A and S1B). Indeed, we observed tdTomato+ axons in the optic nerve (Figure S1C), indicative of repeatable results of “axon regeneration” after Ptbp1 downregulation (Zhou et al., 2020). However, we also observed similar levels of tdTomato+ axons in retinas receiving control AAVs (Figure S1D) or AAV-GFAP-Cre only (Figure S1E), but not in retinas without AAV injection (Figure S1F). The tdTomato+ axons in the optic nerve were mostly likely from the mislabeled endogenous tdTomato+ RGCs in the retinas receiving AAV-GFAP-Cre (Figures S1G–S1J). Therefore, the AAV-based Cre recombination system used by Zhou et al. is unsuitable for critical examination of axon regeneration. Taken together, the fundamental question of whether MG can be converted to RGCs after Ptbp1 downregulation remains unanswered.
Ptbp1 downregulation by CRISPR-CasRx fails to convert MG into RGCs in the intact mouse retina
To unambiguously address the question of whether Ptbp1 downregulation can convert MG to RGCs, we resorted to fate-mapping experiments to trace the lineages of MG and their progeny. First, we generated Sun1-GFP fate-mapping mice by crossing the Glast-CreERT transgenic line (Wang et al., 2012) with the Rosa-CAG-LSL-Sun1-GFP reporter line, resulting in specific labeling of MG for lineage tracing following tamoxifen administration. This fate-mapping technique was successfully used to examine bipolar and amacrine-like retinal neurons converted from reprogrammed MG in injured retinas (Hoang et al., 2020; Jorstad et al., 2017). Five-week-old Sun1-GFP fate-mapping mice were given daily intraperitoneal injection of tamoxifen for 6 consecutive days to induce Sun1-GFP expression (Figures S2A and S2B). Two weeks after tamoxifen administration, Sun1-GFP+ cells were exclusively localized in the middle of the inner nuclear layer (INL) (Figure S2C), and they were co-immunostained with the MG nuclear marker Sox2 (Figures S2D–S2F). These results confirmed that Sun1-GFP fate-mapping mice are suitable for lineage tracing experiments targeting mouse MG. Next, we used the Sun1-GFP fate-mapping mice to examine whether Ptbp1 downregulation by CRISPR-CasRx can convert MG into RGCs at 4 weeks after subretinal injection of AAV-GFAP-tdTomato (an infection marker), together with AAV-GFAP-CasRx (control) or AAV-GFAP-CasRx-Ptbp1 (Ptbp1 downregulation) (Figure 2A). As Ptbp1 knockdown efficiency in MG was not quantifiably examined in the original study (Zhou et al., 2020), we decided to first examine the mRNA and protein levels of Ptbp1 after CRISPR-CasRx-mediated knockdown at 2 weeks after AAV injection. MG were purified using fluorescence-activated cell sorting (FACS), and total RNAs were extracted from Sun1-GFP-labeled MG (without AAV) or tdTomato+ AAV-transduced Sun1-GFP-labeled MG (Figures 2B and S3A–S3D). While real-time qPCR results confirmed the expression of either GFAP-CasRx or GFAP-CasRx-Ptbp1 in tdTomato+ AAV-transduced Sun1-GFP-labeled MG (Figure 2C), the knockdown was mild at the mRNA level (Figure 2E). To further examine the knockdown efficiency of Ptbp1 at the protein level, we used Ptbp1 immunohistochemistry to detect and quantify its expression in tdTomato+ AAV-transduced Sun1-GFP-labeled MG. Consistent with a mild knockdown effect at the RNA level, the immunofluorescence intensity of PTBP1 was moderately reduced in Sun1-GFP-labeled MG receiving GFAP-CasRx-Ptbp1 compared with the control (Figures 2D and 2F). Taken together, our results showed a mild knockdown efficiency of Ptbp1 by CRISPR-CasRx in MG. We next examined whether mild downregulation of Ptbp1 can convert MG into RGCs at 4 weeks after AAV injection. After examining a total of seven retinas (13,604 Sun1-GFP+ MG cells) from the control group (Figure 2G), we found that not a single Sun1-GFP-labeled MG cell was RBPMS positive (Figures 2I–2M, quantified in Figure 2H), indicating that the fate-mapping method works stringently with no mislabeling of endogenous RGCs. Surprisingly, after examining a total of nine retinas (18,765 Sun1-GFP+ MG cells) from the Ptbp1 knockdown group (Figure 2G), we also did not observe a single Sun1-GFP-labeled MG that was converted to RBPMS-positive RGCs either (Figures 2N–2R, quantified in Figure 2H). Furthermore, none of the Sun1-GFP-labeled MG migrated to the GCL (Figure 2N), where endogenous RGCs are localized (Figure 2P).
Figure 2. Ptbp1 downregulation by CRISPR-CasRx fails to convert MG into RGCs in Sun1-GFP fate-mapping (Glast-CreERT;Rosa-CAG-LSL-Sun1-GFP) mice.
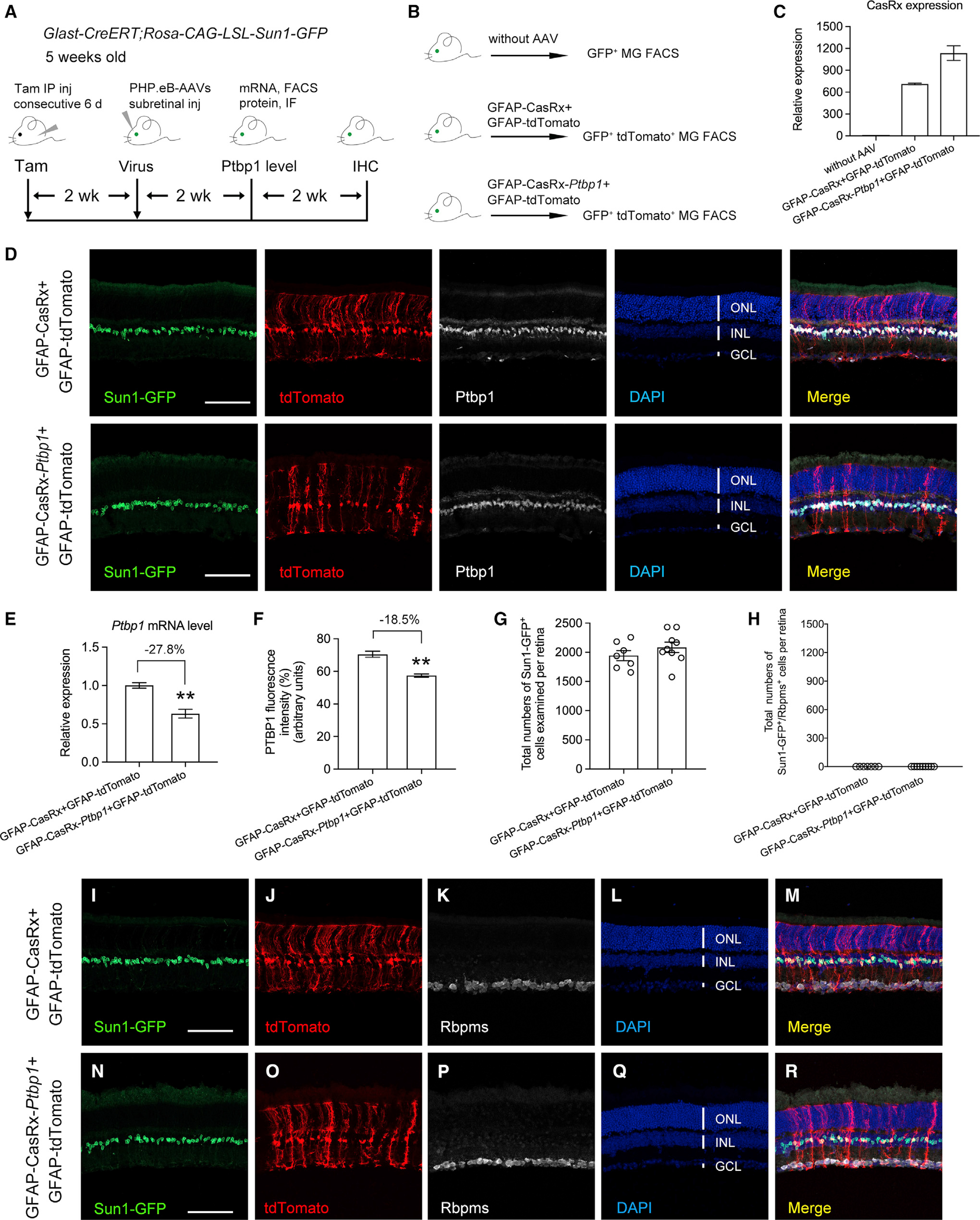
(A) Experimental design for testing MG-to-RGC conversion in Sun1-GFP fate-mapping mice. Subretinal injection of PHP.eB-AAVs was performed in Sun1-GFP fate-mapping mice receiving tamoxifen-induced labeling of MG at 5 weeks of age. Ptbp1 mRNA and protein levels were measured from purified MG using fluorescence-activated cell sorting (FACS) and immunofluorescence (IF), respectively, at 2 weeks after AAV injection. Immunohistochemistry analysis using the pan-RGC marker RBPMS was performed at 4 weeks after AAV injection. IHC, immunohistochemistry.
(B) Experimental design for FACS purification of Sun1-GFP labeled MG with or without AAV infection.
(C) Real-time qPCR analysis of AAV-transduced (tdTomato+) Sun1-GFP labeled MG confirmed the expression of AAV-GFAP-CasRx or AAV-GFAP-CasRx-Ptbp1. The relative expression level of CasRx in Sun1-GFP-labeled MG without AAV infection was set as 1. Data are presented as the mean ± SEM, n = 3.
(D) Confocal images of the indicated markers showing low-efficiency knockdown of Ptbp1 by the CRISPR-CasRx system 2 weeks after AAV infection. Scale bars, 100 μm.
(E) Real-time qPCR analysis of Ptbp1 mRNA levels in AAV-transduced (tdTomato+) Sun1-GFP-labeled MG showing a mild knockdown effect. The relative expression level of Ptbp1 in AAV-GFAP-CasRx (control)- and AAV-GFAP-tdTomato (infection marker)-transduced MG was set as 1. Data are presented as the mean ± SEM, n = 3, **p < 0.01, unpaired t test.
(F) Quantification of PTBP1 immunofluorescence intensity in AAV-transduced (tdTomato+) Sun1-GFP-labeled MG showing low-efficiency knockdown of Ptbp1. n = 4 retinas per group. Data are presented as the mean ± SEM, **p < 0.01, unpaired t test.
(G) Total numbers of Sun1-GFP-labeled MG examined in retinas at 4 weeks after receiving GFAP-tdTomato (an infection marker), together with GFAP-CasRx (control) or GFAP-CasRx-Ptbp1 (Ptbp1 downregulation). Data are presented as mean ± SEM, n = 7–9 retinas per group.
(H) Total numbers of Sun1-GFP-labeled MG that were also immunoreactive for the pan-RGC marker RBPMS per retina after Ptbp1 downregulation.
(I–M) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving GFAP-CasRx and GFAP-tdTomato.
(N–R) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving GFAP-CasRx-Ptbp1 and GFAP-tdTomato.
(I–R) Scale bars, 100 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. See also Figures S2 and S3.
We then examined whether Ptbp1 downregulation by CRI-SPR-CasRx induces MG-to-RGC conversion in tdTomato fate-mapping mice (Figure S4A), generated by crossing the Glast-CreERT transgenic line with the Rosa-CAG-LSL-tdTomato (Ai9) reporter line, resulting in specific labeling of MG for lineage tracing following tamoxifen administration (Rueda et al., 2019). Four weeks after subretinal co-injection of AAV-GFAP-GFP (an infection marker), together with AAV-GFAP-CasRx (control) or AAV-GFAP-CasRx-Ptbp1 (Ptbp1 downregulation), no tdTomato-labeled MG were co-immunostained with RBPMS in the GCL from either the control or the Ptbp1 downregulation group (Figures S4B–S4M), consistent with the results obtained from the Sun1-GFP fate-mapping mice. All these results indicate unsuccessful conversion of MG into RGCs after Ptbp1 downregulation.
To further examine whether mild downregulation of Ptbp1 could lead to robust axon regeneration from MG-converted RGCs as previously reported (Zhou et al., 2020), we generated EYFP fate-mapping mice by crossing the Glast-CreERT transgenic line with the R26R-EYFP reporter line (Figures S5A and S5B), resulting in tamoxifen-induced EYFP expression in the cytoplasm of MG (Figures S5C–S5F), to facilitate the detection of regenerated axons from MG-derived RGCs. We first used EYFP fate-mapping mice to lineage trace MG after Ptbp1 downregulation (Figure S6A). Four weeks after subretinal co-injection of AAV-GFAP-tdTomato (an infection marker), together with AAV-GFAP-CasRx (control) or AAV-GFAP-CasRx-Ptbp1 (Ptbp1 downregulation), no EYFP-labeled MG were co-immunostained with RBPMS in the GCL from either the control or the Ptbp1 downregulation group (Figures S6B–S6M), consistent with the results obtained using both Sun1-GFP and tdTomato fate-mapping mice. Importantly, we did not observe EYFP-labeled RGC axons in the optic nerve after Ptbp1 downregulation (Figures S6N and S6O), indicating that RGC axon regeneration did not occur when MG-to-RGC conversion failed in the first place. Taken together, our fate-mapping experiments employing genetic lineage tracing of MG in all three fate-mapping mice (Sun1-GFP, tdTomato, and EYFP) demonstrate that CRISPR-CasRx-mediated Ptbp1 downregulation is not sufficient to convert MG into RGCs.
Small hairpin RNA-mediated Ptbp1 depletion fails to convert MG into RGCs in the intact mouse retina
Another study recently reported that astroglial cells can be converted to different neuronal subtypes in the brain after Ptbp1 depletion using shRNA (Qian et al., 2020). To examine whether shRNA-mediated Ptbp1 depletion, independent of CRISPR-CasRx-mediated Ptbp1 downregulation, could convert MG to RGCs, we performed fate-mapping experiments to lineage trace MG using the shRNA-based Ptbp1 depletion system obtained from this study (Qian et al., 2020). We used the ShH10 rep-cap to package AAV-CMV-LSL-RFP (control) and AAV-CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion) for efficient transduction of adult mouse MG via intravitreal injection (Elsaeidi et al., 2018; Klimczak et al., 2009; Yao et al., 2016, 2018). The shRNA-based Ptbp1 depletion is a Cre-LoxP system; tamoxifen administration in the Sun1-GFP fate-mapping mice will also activate RFP expression to label transduced MG without the need of an additional infection marker. As shRNA-mediated Ptbp1 depletion has not previously been tested in the mouse retina, we first examined Ptbp1 mRNA and protein levels 2 weeks after tamoxifen treatment in the Sun1-GFP fate-mapping mice receiving AAV-CMV-LSL-RFP (control) or AAV-CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion) injection (Figure 3A). MG were purified using FACS, and total RNAs were extracted from sorted RFP+ AAV-transduced Sun1-GFP-labeled MG (Figures 3B, S3E, and S3F). Real-time qPCR results showed that the shRNA-based system efficiently reduced Ptbp1 mRNA levels (Figure 3C). To further examine the knockdown efficiency of Ptbp1 at the protein level, we quantified PTBP1 immunofluorescence intensity in RFP+ AAV-transduced Sun1-GFP-labeled MG. As expected, the Ptbp1 levels were not altered in Sun1-GFP-labeled MG receiving AAV-CMV-LSL-RFP injection (Figures 3D–3H and 3D′–3H′). By contrast, a significant reduction in PTBP1 was observed in Sun1-GFP-labeled MG receiving AAV-CMV-LSL-RFP-shPtbp1 injection (Figures 3I–3M and 3I′–3M′, quantified in Figure 3N), confirming the efficacy of Ptbp1 depletion in adult mouse MG using the shRNA-based system. We next examined whether MG can be converted to RGCs after Ptbp1 depletion at 4 weeks after tamoxifen treatment of Sun1-GFP fate-mapping mice. As expected, no Sun1-GFP-labeled MG were co-immunostained with RBPMS in a total of six retinas (11,856 Sun1-GFP+ MG cells) receiving AAV-CMV-LSL-RFP injection (Figures 3O–3S, quantified in Figures 3Y and 3Z). However, we also did not detect a single MG that was converted into RGC in a total of eight retinas (16,208 Sun1-GFP+ MG cells) receiving AAV-CMV-LSL-RFP-shPtbp1 injection (Figures 3T–3X, quantified in Figures 3Y and 3Z). Interestingly, Sun1-GFP-labeled MG were all localized in the middle of the INL after Ptbp1 depletion (Figure 3T), indicating that MG-to-RGC conversion may not have been initiated, because migration of MG to the GCL is an essential first step for successful MG-to-RGC conversion.
Figure 3. Ptbp1 downregulation by shRNA-based depletion fails to convert MG into RGCs in Sun1-GFP fate-mapping (Glast-CreERT;Rosa-CAG-LSL-Sun1-GFP) mice.
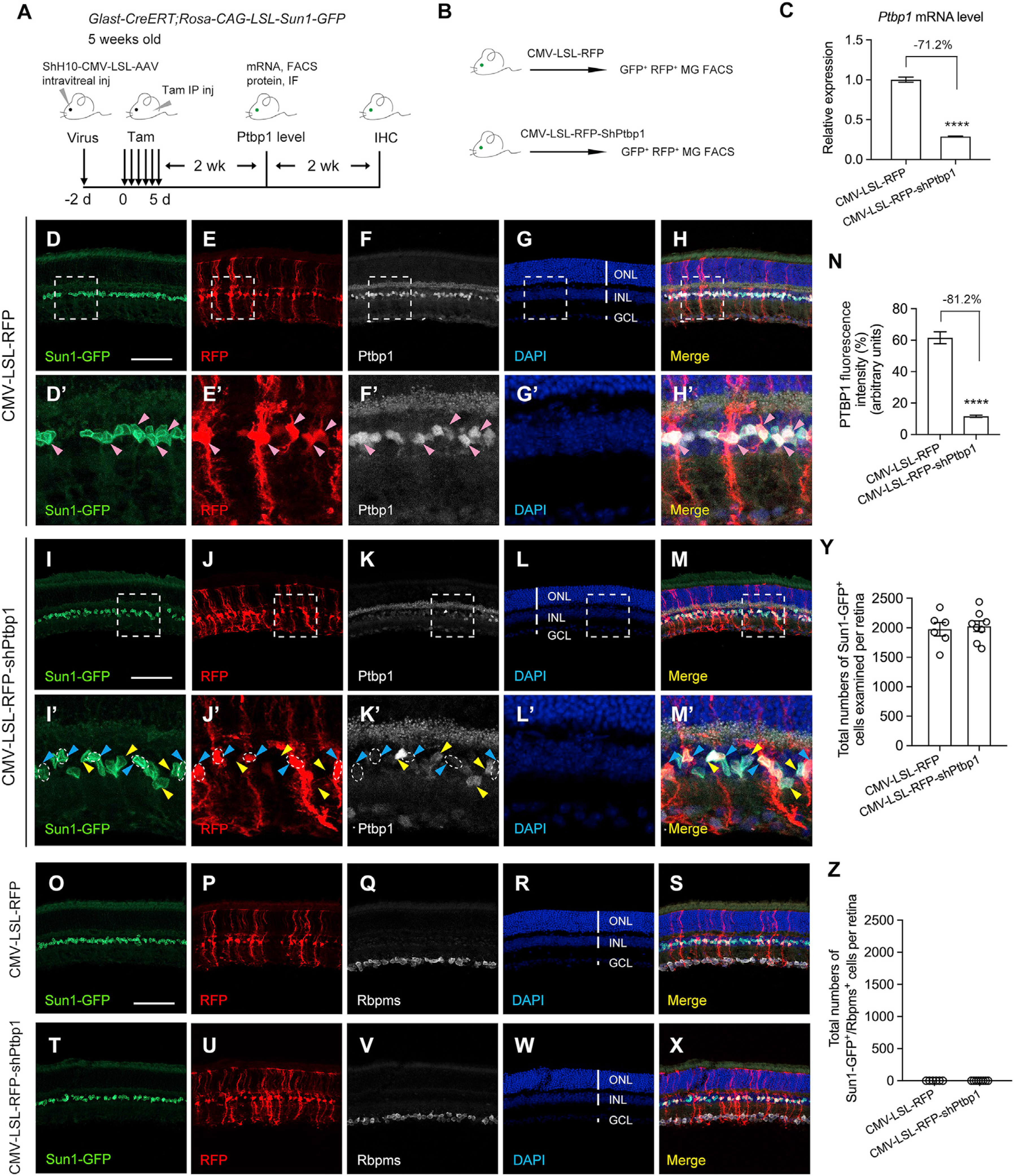
(A) Experimental design for testing MG-to-RGC conversion in Sun1-GFP fate-mapping mice. Intravitreal injection of ShH10-CMV-LSL-AAVs was performed inSun1-GFP fate-mapping mice at 5 weeks of age, followed by tamoxifen administration 2 days later to induce Sun1-GFP expression in MG and Ptbp1 depletion. Fluorescence-activated cell sorting (FACS) and immunofluorescence (IF) were used, respectively, to measure Ptbp1 mRNA and protein levels at 2 weeks after tamoxifen injection. Immunohistochemistry analysis was performed using the pan-RGC marker RBPMS at 4 weeks after tamoxifen injection. IHC, immunohistochemistry.
(B) Experimental design for FACS purification of Sun1-GFP-labeled MG receiving ShH10-CMV-LSL-RFP (control) or CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion).
(C) Real-time qPCR analysis in AAV-transduced (RFP+) Sun1-GFP-labeled MG showing high-efficiency knockdown of Ptbp1. The relative expression level of Ptbp1 in AAV-CMV-LSL-RFP (control) transduced MG was set as 1. Data are presented as the mean ± SEM, n = 3, ****p < 0.0001, unpaired t test.
(D–H) Confocal images showing Ptbp1 expression in Sun1-GFP fate-mapping mice receiving intravitreal injection of ShH10-CMV-LSL-RFP (control). Scale bar, 100 μm. The boxed areas are enlarged in (D′–H′). Pink arrowheads indicate that Ptbp1 levels were maintained in RFP+ Sun1-GFP-labeled MG.
(I–M) Confocal images showing Ptbp1 expression in Sun1-GFP fate-mapping mice receiving intravitreal injection of ShH10-CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion). Scale bar, 100 μm. The boxed areas are enlarged in (I′–M′). Blue arrowheads indicate that Ptbp1 levels were depleted in RFP+ Sun1-GFP-labeled MG. Yellow arrow-heads indicate that Ptbp1 levels were maintained in RFP− Sun1-GFP-labeled MG (non-transduced MG, serving as an internal control for Ptbp1 depletion in transduced MG).
(N) Quantification of PTBP1 immunofluorescence intensity in AAV-transduced (RFP+) Sun1-GFP-labeled MG showing high-efficiency knockdown of Ptbp1. n = 4–5 retinas per group. Data are presented as the mean ± SEM, ****p < 0.0001, unpaired t test.
(O–S) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving CMV-LSL-RFP.
(T–X) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in retinas receiving CMV-LSL-RFP-shPtbp1.
(O–X) Scale bar, 100 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
(Y) Total numbers of Sun1-GFP-labeled MG examined in retinas (4 weeks after tamoxifen injection) receiving CMV-LSL-RFP (control) or CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion). Data are presented as mean ± SEM, n = 6–8 retinas per group.
(Z) Total numbers of Sun1-GFP-labeled MG that were also immunoreactive for the pan-RGC marker RBPMS per retina after Pbtp1 depletion. See also Figures S2 and S3.
Similarly, we used EYFP fate-mapping mice to examine whether shRNA-mediated Ptbp1 depletion could lead to axon regeneration from MG-converted RGCs at 4 weeks after tamoxifen treatment (Figure S7A). Consistent with our results using the Sun1-GFP fate-mapping mice, we did not detect EYFP-labeled MG that were co-immunostained with the RGC marker RBPMS in the GCL from either the AAV-CMV-LSL-RFP (control) or the AAV-CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion) injection (Figures S7B–S7M). Importantly, EYFP-labeled RGC axons were not observed in the optic nerve after Ptbp1 depletion (Figures S7N and S7O), confirming that RGC axon regeneration did not occur when MG-to-RGC conversion failed in the first place. Taken together, using both Sun1-GFP and EYFP fate-mapping mice for genetic lineage tracing, our results demonstrate that shRNA-mediated Ptbp1 depletion is not sufficient to convert MG to RGCs.
Ptbp1 knockdown fails to convert MG into RGCs in a mouse model of NMDA-induced excitotoxic injury
We examined whether Ptbp1 knockdown could convert MG into RGCs after NMDA-induced excitotoxic injury, resulting in a nearly complete loss of RGCs and reduction in the thickness of the inner plexiform layer (Niwa et al., 2016; Zhou et al., 2020). Following the same experimental procedure described by Zhou et al. (2020), we performed intravitreal injection of NMDA (1.5 μL, 200 mM) in tamoxifen-induced Sun1-GFP fate-mapping mice 2 weeks before subretinal injection of AAVs. We next examined whether MG can be converted to RGCs in NMDA-injured retinas at 4 weeks after AAV injection (Figure 4A). After examining a total of three retinas (4,248 Sun1-GFP+ MG cells) from the control group and a total of four retinas (6,194 Sun1-GFP+ MG cells) from the Ptbp1 knockdown group (Figure 4B), even though we observed an overall lack of RBPMS immunoreactivity (Figures 4F and 4K) indicating that NMDA-induced excitotoxicity indeed massively killed endogenous RGCs, not a single Sun1-GFP-labeled MG was RBPMS positive in the absence or presence of Ptbp1 knockdown (Figures 4D–4M, quantified in Figure 4C). Our results demonstrate that NMDA-induced excitotoxic injury did not facilitate MG-to-RGC conversion after Ptbp1 knockdown.
Figure 4. Ptbp1 downregulation fails to convert MG into RGCs in a mouse model of NMDA-induced excitotoxic injury.
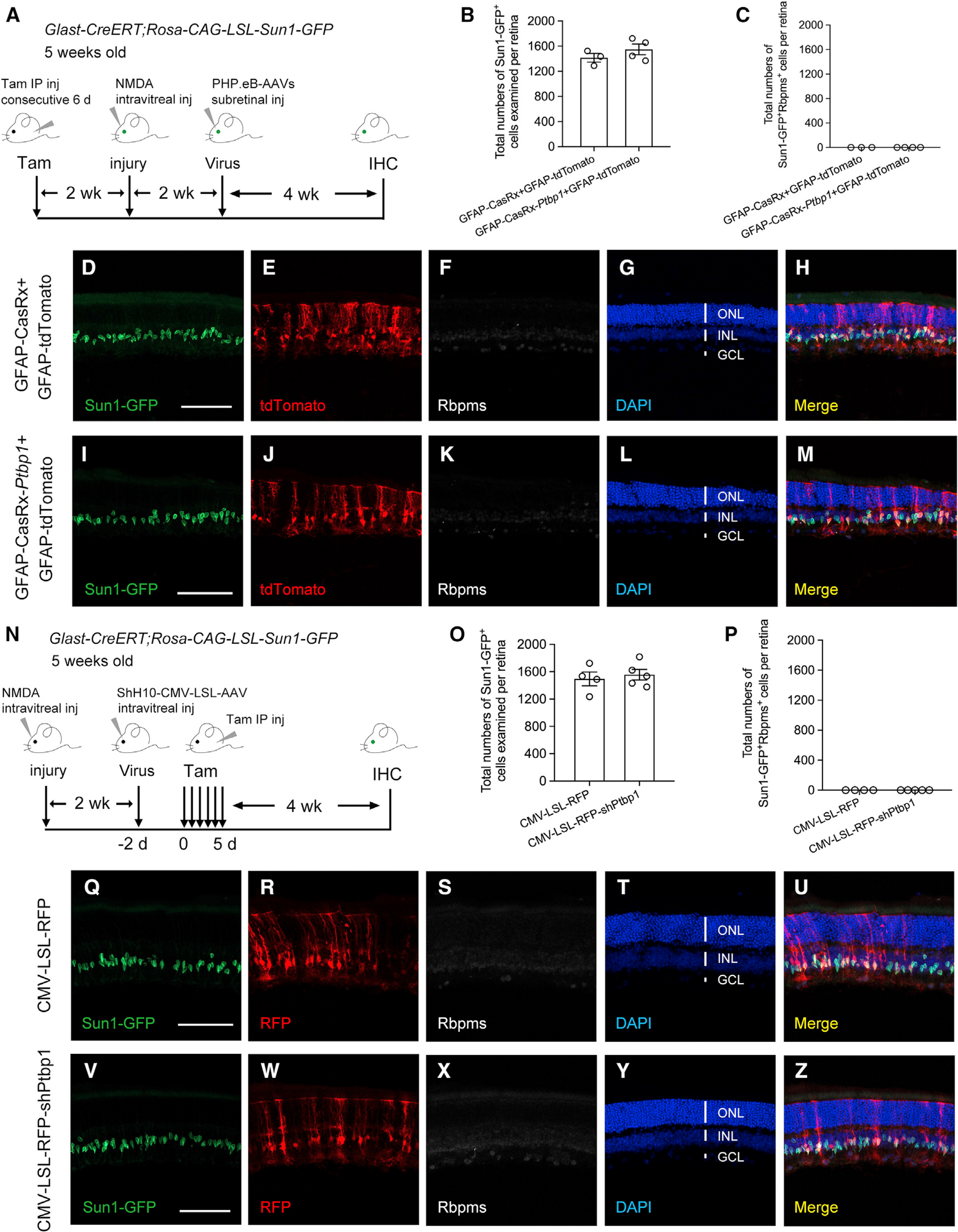
(A) Experimental design for testing MG-to-RGC conversion in NMDA-injured retinas of Sun1-GFP fate-mapping mice. Intravitreal injection of NMDA (1.5 μL, 200 mM) was performed in Sun1-GFP fate-mapping mice receiving tamoxifen-induced labeling of MG at 5 weeks of age, followed by subretinal injection of PHP.eB-AAVs at 2 weeks after injury. Immunohistochemistry analysis using the pan-RGC marker RBPMS was performed at 4 weeks after AAV injection. IHC, immunohistochemistry.
(B) Total numbers of Sun1-GFP-labeled MG examined in NMDA-injured retinas at 4 weeks after receiving GFAP-tdTomato (an infection marker), together with GFAP-CasRx (control) or GFAP-CasRx-Ptbp1 (Ptbp1 downregulation). Data are presented as mean ± SEM, n = 3–4 retinas per group.
(C) Total numbers of Sun1-GFP-labeled MG that were also immunoreactive for the pan-RGC marker RBPMS per retina after Ptbp1 downregulation.
(D–H) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in NMDA-injured retinas receiving GFAP-CasRx and GFAP-tdTomato.
(I–M) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in NMDA-injured retinas receiving GFAP-CasRx-Ptbp1 and GFAP-tdTomato.
(D–M) Scale bars, 100 μm. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
(N) Experimental design for testing MG-to-RGC conversion in NMDA-injured retinas of Sun1-GFP fate-mapping mice. Intravitreal injection of NMDA (1.5 μL, 200 mM) was performed in Sun1-GFP fate-mapping mice at 5 weeks of age, followed by intravitreal injection of ShH10-CMV-LSL-AAVs 2 weeks later. Tamoxifen was administrated 2 days later to induce Sun1-GFP expression in MG and Ptbp1 depletion. Immunohistochemistry analysis was performed using the pan-RGC marker RBPMS at 4 weeks after tamoxifen injection. IHC, immunohistochemistry.
(O) Total numbers of Sun1-GFP-labeled MG examined in NMDA-injured retinas at 4 weeks receiving CMV-LSL-RFP (control) or CMV-LSL-RFP-shPtbp1 (Ptbp1 depletion). Data are presented as mean ± SEM, n = 4–5 retinas per group.
(P) Total numbers of Sun1-GFP-labeled MG that were also immunoreactive for the pan-RGC marker RBPMS per retina after Pbtp1 depletion.
(Q–U) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in NMDA-injured retinas receiving CMV-LSL-RFP.
(V–Z) Confocal images showing expression of Sun1-GFP and RBPMS immunohistochemistry in NMDA-injured retinas receiving CMV-LSL-RFP-shPtbp1.
(Q–Z) Scale bars, 100 μm.
Given the mild downregulation of Ptbp1 mediated by CRISPR-CasRx (Figures 2E and 2F), we further tested whether shRNA-based Ptbp1 depletion, which showed a robust and efficient knockdown of Ptbp1 mRNA and protein levels (Figures 3C and 3N), could convert MG into RGCs after NMDA-induced excitotoxic injury (Figure 4N). As expected, no Sun1-GFP-labeled MG were co-immunostained with RBPMS in a total of four retinas (5,980 Sun1-GFP+ MG cells) in the control group receiving AAV-CMV-LSL-RFP injection (Figures 4Q–4U, quantified in Figures 4O and 4P). Again, we did not detect a single MG that was converted to an RGC in a total of five retinas (7,784 Sun1-GFP+ MG cells) in the Ptbp1 depletion group receiving AAV-CMV-LSL-RFP-shPtbp1 injection (Figures 4V–4Z, quantified in Figures 4O and 4P). Collectively, our results show that Ptbp1 knockdown was not sufficient to convert MG into RGCs in the NMDA-injured mouse retina.
DISCUSSION
Ptbp1 has emerged as a pivotal reprogramming factor capable of converting glial cells to neurons after downregulation of its expression, including conversion of MG to RGCs in the adult mouse retina (Zhou et al., 2020). We first repeated the experiments of Zhou et al. using the same methodology to label MG and found that the PHP.eB-GFAP-Cre was leaky, resulting in mislabeling of endogenous RGCs (Figures 1A–1O). In addition, we also observed other retinal neuronal types, such as photoreceptors localized in the outer nuclear layer (ONL), could be mislabeled, although the mislabeling was weaker in photoreceptors than that in RGCs (Figures 1H and 1L). Consistent with our findings, a recent study reported the ectopic expression of AAV-GFAP-Cre into endogenous neurons in the mouse brain, indicating that the AAV-based Cre recombination system is unsuitable for tracing the lineages of glia cells to determine glia-to-neuron conversion (Wang et al., 2021), emphasizing the importance of performing stringent fate-mapping experiments in glia-to-neuron reprogramming research.
To critically examine Ptbp1-mediated MG-to-RGC conversion in the mouse retina, we generated Sun1-GFP fate-mapping mice that allowed us to lineage trace MG and their progeny after Ptbp1 downregulation using the same CRISPR-CasRx system developed by the original authors (Zhou et al., 2020). Surprisingly, we did not see a single RGC that was converted from MG after a mild downregulation of Ptbp1 by CRISPR-CasRx (Figures 2A–2R). Furthermore, the MG-to-RGC conversion may not have been initiated, as fate mapped MG did not migrate to the GCL, where RGCs reside (Figures 2N and S4I). To unambiguously address the fundamental question of whether sufficient knockdown of Ptbp1 could convert MG to RGCs, we performed additional fate-mapping experiments using the shRNA-based Ptbp1 deletion system developed by an independent group to convert astroglial cells to functional neurons in the brain (Qian et al., 2020). Again, we did not observe a single RGC that was converted from MG after robust and efficient knockdown of Ptbp1 (Figures 3A–3Z), consistent with our fate-mapping results after a mild downregulation of Ptbp1 using the CRISPR-CasRx system. Using two independent systems to reduce Ptbp1 expression in MG, our results provide important evidence that Ptbp1 downregulation is not sufficient to convert MG into RGCs, at least in and of itself.
Zhou et al. also reported long-distance regeneration of RGC axons capable of transmitting visual information from the retina to the primary visual cortex in the adult mouse brain after Ptbp1 downregulation (Zhou et al., 2020). This may represent a major advance in the axon regeneration field, as axons do not regenerate after injury in the adult mammalian central nervous system (He and Jin, 2016). We first repeated their experiments using the same methodology to label MG, and found that the AAV-based Cre recombination system was leaky, resulting in mislabeling of axons in the optic nerve from endogenous RGCs (Figures S1A–S1J). We then generated EYFP fate-mapping mice to monitor RGC axon regrowth after Ptbp1 downregulation. Consistent with our results using the Sun1-GFP or tdTomato fate-mapping mice, we observed no indication of MG-to-RGC conversion, as no EYFP-labeled MG were immunoreactive for the pan-RGC marker RBPMS after Ptbp1 downregulation by CRISPR-CasRx (Figures S6A–S6M) or shRNA-based Ptbp1 depletion (Figures S7A–S7M). Importantly, EYFP-labeled RGC axons were not detected in the optic nerve after mild downregulation of Ptbp1 by CRISPR-CasRx (Figure S6O) or shRNA-based depletion of Ptbp1 (Figures S7O). These results demonstrate that Ptbp1 knockdown cannot lead to RGC axon regeneration to reestablish the retina-brain connection in the absence of successful MG-to-RGC conversion.
Furthermore, Zhou et al. reported that CRISPR-CasRx-mediated Ptbp1 knockdown could replenish RGCs in a mouse model of NMDA-induced excitotoxicity (Zhou et al., 2020). After performing stringent fate-mapping experiments to lineage trace MG, however, we observed no indication of MG-to-RGC conversion in the injured retina, as no Sun1-GFP-labeled MG were immunoreactive for the pan-RGC marker RBPMS after Ptbp1 downregulation by CRISPR-CasRx-mediated knockdown (Figures 4A–4M) or shRNA-based depletion (Figures 4N–4Z). Taken together, our results clearly demonstrate that Ptbp1 knockdown is not sufficient to convert MG into RGCs in normal or NMDA-injured retinas.
Unlike their counterparts in zebrafish, MG are a quiescent pool of retinal stem cells in the mammalian retina. Therefore, finding effective reprogramming strategies to unlock the regenerative capability of MG represents a major challenge in vision neuroscience and regenerative medicine (Goldman, 2014; Karl and Reh, 2010). MG-derived regeneration of RGCs continues to remain as a challenging task in future research endeavors. To avoid incidents of data misinterpretation caused by leaky labeling of endogenous neurons, stringent genetic-based fate-mapping methods should be used across the board in MG reprogramming research. To trace the lineage of MG, fate-mapping mice can be generated by crossing the MG-specific Cre lines, such as the Glast-CreER or the Rlbp1-CreER line, with a Rosa26 reporter line. Using these two MG-specific tamoxifen-inducible Cre mouse lines, Reh and colleagues performed stringent fate-mapping experiments to examine the role of Ascl1 in MG-derived neurogenesis (Jorstad et al., 2017; Ueki et al., 2015). In another study using Glast-CreER;CAG-LSL-Sun1-GFP fate-mapping mice, Hoang et al. profiled the transcriptomic and other changes of MG in NMDA-injured retinas (Hoang et al., 2020). To reemphasize, genetic-based stringent fate-mapping experiments, which do not rely on AAV-mediated expression of a transgene to label targeted glial cells, are required to definitively determine the origin of newly generated neurons regardless of whether the reprogramming is direct conversion of glial cells to neurons or the reprogramming process involves glial cells undergoing initial cell proliferation and subsequent neuronal differentiation (Blackshaw and Sanes, 2021; Boudreau-Pinsonneault and Cayouette, 2018; Martin and Poche, 2019; Xie and Chen, 2022).
Limitations of the study
Given the leaky expression into endogenous RGCs after AAV-GFAP-mediated gene transfer resulting in misinterpretation of MG-to-RGC conversion, future studies are needed to develop new AAV-based tools that are capable of high-efficiency in vivo gene delivery into MG with minimal leakage into endogenous neurons.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bo Chen (bo.chen@mssm.edu).
Materials availability
All materials generated in this study are available upon reasonable request to the lead contact.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
The transgenic mouse line Glast-CreERT (JAX#012586), and reporter mouse lines Rosa-CAG-LSL-Sun1-GFP (JAX#021039), Rosa-CAG-LSL-tdTomato (Ai9, JAX#007909) and R26R-EYFP (JAX#006148) were purchased from The Jackson Laboratory. Hemizygous Glast-CreERT mice and homozygous reporter mice were used for breeding. Glast-CreERT;Rosa-CAG-LSL-Sun1-GFP, Glast-CreERT;Rosa-CAG-LSL-tdTomato and Glast-CreERT;R26R-EYFP fate mapping mice were PCR genotyped and confirmed by using published protocols on Jackson Laboratory. All mice were housed under controlled room temperature and a 12-h light/dark cycle with free access to water and food. Mice at postnatal five weeks of either sex were randomly assigned to different groups for all experiments. All procedures were consistent with animal protocols approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai.
Cell lines
The AAVpro 293 cell line was purchased from Takara Bio USA, Inc., and cultured in DMEM with 10% FBS and 1% penicillin/streptomycin in a 37°C incubator under 5% CO2.
METHOD DETAILS
AAV preparation
AAV-CMV-LSL-RFP and AAV-CMV-LSL-RFP-ShPtbp1 were kindly provided by Dr. Xiangdong Fu (University of California, San Diego) (Qian et al., 2020). AAV-GFAP-CasRx (Addgene #15400) and AAV-GFAP-CasRx-Ptbp1 (Addgene #154001) were deposited from Dr. Hui Yang’s laboratory to Addgene. For AAV-GFAP-Cre construction, the protein-coding region of AAV-GFAP-tdTomato (Yao et al., 2016) was replaced by cDNA for Cre. AAV rep-cap PHP.eB (Addgene #103005) (Chan et al., 2017) or ShH10 (Klimczak et al., 2009), and Helper plasmids were used for co-transfection in AAVpro 293T Cell Line (Takara Bio, 632273). Discontinuous iodixanol gradient ultracentrifugation was used to produce and purify AAV (Grieger et al., 2006). Purified AAVs were concentrated or diluted in PBS. AAV titers, determined by real-time qPCR, were in the range of 1–5 × 1013 genome copies per milliliter.
Intravitreal and subretinal injection
Adult mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) by intraperitoneal injection. ShH10-AAVs and PHP.eB-AAVs were used to delivered into mouse retinas via intravitreal and subretinal injection, respectively. For intravitreal injection, the micropipettes were prepared using a puller and connected with a syringe. The tip was inserted behind the ora serrata. About 1.5 μL NMDA solution (200 mM in PBS), or 1.0 μL of AAV, either ShH10-CMV-LSL-RFP or ShH10-CMV-LSL-RFP-ShPtbp1, was injected into the vitreous body of mouse retinas. For subretinal injection, an incision was made using a 30G needle followed by injection using a Hamilton syringe. PHP.eB-AAVs were injected into the subretinal space. A total volume of 1 μL of AAVs, AAV-GFAP-Cre (0.2 μL) plus AAV-GFAP-CasRx-Ptbp1 (0.8 μL), AAV-GFAP-Cre (0.2 μL) plus AAV-GFAP-CasRx (0.8 μL), or AAV-GFAP-Cre (0.2 μL) plus PBS (0.8 μL), were delivered to the retina via subretinal injection. And a total volume of 1 μL AAVs, AAV-GFAP-tdTomato (or AAV-GFAP-GFP, 0.2 μL) plus AAV-GFAP-CasRx-Ptbp1 (0.8 μL), or AAV-GFAP-tdTomato (or AAV-GFAP-GFP, 0.2 μL) plus AAV-GFAP-CasRx (0.8 μL), were delivered via subretinal injection. After intravitreal or subretinal injection, antibiotics eye ointment was applied to prevent infection.
Tamoxifen intraperitoneal injection
Tamoxifen (Sigma T5648) was dissolved in corn oil at a concentration of 20 mg/mL. To induce Cre recombinase expression, tamoxifen was administered in Glast-CreERT;Rosa-CAG-LSL-Sun1-GFP, Glast-CreERT;Rosa-CAG-LSL-tdTomato or Glast-CreERT; R26R-EYFP mice through intraperitoneal injections at a daily dose of 100mg/kg body weight for six consecutive days.
Retinal cell dissociation and fluorescence-activated cell sorting (FACS)
Retinal dissection was performed in HBSS. Dissected retinas were incubated at 37°C for 20 min in the activated Papain mix composed of 40 μL Papain (Worthington, LS003126), 40 μL Cysteine/EDTA mix (25m M cysteine +5 mM EDTA, pH 6–7) and 320 μL HBSS/HEPES (normal HBSS +10 mM HEPES). Cell pellets were collected after centrifugation for 3 min at 3000 rpm, and treated with 10 μL DNase (Invitrogen, 2 U/μL) in 400 μL HBSS at room temperature by gentle trituration. After additional incubation of 5 min for DNase digestion, dissociated cells were washed with HBSS and collected by centrifugation. Cell pellets were resuspended in an appropriate volume of PBS containing 0.5% BSA for further experiments. To purify Sun1-GFP+, Sun1-GFP+tdTomato+, or Sun1-GFP+RFP+ MG, retinas were isolated from Tamoxifen-induced Glast-CreERT;CAG-LSL-Sun1-GFP (Sun1-GFP MG fate mapping) mice with or without AAV treatment. After retinal dissociation, single-cell resuspension was filtered right before loading on a CSM5L BD cell sorter. DAPI was used as a viability dye to exclude the dead cells. After sorting, cells were subject to RNA isolation and reverse transcription.
RNA isolation, RT-PCR, and real-time qPCR
Total RNA was extracted from sorted retinal cells using TRIzol (Invitrogen) according to the manufacture’s protocol. Extracted RNA was first treated with DNase I (DNA-free™ kit, Invitrogen, AM1906) to remove DNA contamination. Reverse transcription (RT) was then performed by using ProtoScript First Strand cDNA Synthesis Kit (NEB, E6300S). Real-Time quantitative PCR (qPCR) was eventually performed in triplicate with PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, A25742) using an CFX Connect™ real-time PCR detection system (Bio-Rad). A melt curve analysis was performed at the end of reaction to check the reaction specificity. Results were obtained after normalization to the expression level of the housekeeping gene β-actin. Similar results were obtained from two independent biological repeats.
Histology and microscopy
The eyes and optic nerves were removed from perfused mice. Optic nerves were fixed with 4% paraformaldehyde (PFA) in PBS for 2 h at room temperature and then washed 3 × 5 min with PBS before mounted with Fluoromount-G. Retinas were dissected out and fixed with 4% PFA for 1 h at room temperature, followed by overnight incubation at 4°C in PBS containing 30% sucrose. Retinas were frozen in Tissue Plus O.C.T. Compound and sectioned using a Thermo Scientific HM525 NX cryostat at the thickness of 20 μm. Sample slides were washed with PBS before incubation with a blocking buffer containing 5% normal donkey serum, 0.1% Triton X-100 in PBS for 2 h at room temperature. Primary antibodies were added for overnight incubation with antibody dilution buffer containing 1% BSA, 0.1% Triton X-100 in PBS at 4°C. Sections were then washed 3 × 5 min with PBS before 2 h of incubation with secondary antibodies at room temperature in the dark. Retinas were washed 3 × 5 min with PBS and then mounted with Fluoromount-G. Primary antibodies used: goat anti-Sox2 (1:100, Santa Cruz Biotechnology, sc-17320), rabbit anti-Ptbp1 (1:150, Abcam, ab133734), and rabbit anti-Rbpms (1:2000, Millipore, ABN1362). Secondary antibodies used: Alexa Fluor 594 AffiniPure Donkey Anti-Goat IgG (1:500, Jackson ImmunoResearch Labs, 705-585-147), Alexa Fluor 594 AffiniPure donkey anti-rabbit IgG (1:500, Jackson ImmunoResearch Labs, 711-585-152), or Cy5 AffiniPure donkey anti-rabbit IgG (1:500, Jackson Immuno Research Labs, 711-175-152). Cell nuclei were counterstained with DAPI (Thermo Fisher 62248). Confocal images were acquired using a Zeiss LSM 800 microscope. Images were analyzed using ImageJ (Schindelin et al., 2012) and Photoshop.
QUANTIFICATION AND STATISTICAL ANALYSIS
For real-time qPCR experiments measuring the relative expression levels of CasRx or Ptbp1, the indicated “n” represents number of technical repeats; for immunohistochemistry analysis, the indicated “n” represents number of retinas. Quantification data are presented as mean ± SEM. Statistical analysis was performed by unpaired t test using the GraphPad Prism 8. All the statistical details for each experiment were described in the figure legends. A p value ≤ 0.05 was considered significant. Significant differences are indicated by **p < 0.01 and ****p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Goat anti-Sox2 | Santa Cruz Biotechnology | Cat# sc-17320; RRID:AB_2286684 |
| Rabbit anti-PTBP1 | Abcam | Cat# ab133734; RRID:AB_2814646 |
| Rabbit anti-RBPMS | Millipore | Cat# ABN1362; |
| Alexa Fluor 594 AffiniPure Donkey Anti-Mouse IgG (H + L) |
Jackson ImmunoResearch Labs | Cat# 715-585-151; RRID:AB_2340855 |
| Cy5 AffiniPure Donkey Anti-Rabbit IgG (H + L) |
Jackson ImmunoResearch Labs | Cat# 715-585-152; RRID:AB_2340607 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Benzonase Nuclease | Sigma-Aldrich | Cat# E1024-25KU |
| DAPI | Thermo Fisher Scientific | Cat# 62248 |
| HEPES | Fisher BioReagents | Cat# BP310-100 |
| Iodixanol gradient | Alere Technologies | Prod.no.1114542 |
| L-Cysteine | MP Biochemicals | Cat# 101444 |
| N-Methyl-D-aspartic Acid (NMDA) | Millipore Sigma | Cat# 454575 |
| Papain | Worthington-Biochemical | Cat# LS003126 |
| Polyethylenimine (PEI) | Polysciences, Inc. | Cat# 23966-1 |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| TRIzol | Invitrogen | Cat# 15596026 |
| TURBO™ DNase | Invitrogen | Cat# AM2238 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| AAVpro 293T Cell Line | Takara Bio | Cat# 632273 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Glast-CreER: Tg(Slc1a3-cre/ERT)1Nat/J | The Jackson Laboratory | Cat# JAX:012586; RRID: IMSR_JAX:012586 |
| Mouse: R26R-EYFP: B6. 129X1-Gt(ROSA) 26Sortm1(EYFP)Cos/J | The Jackson Laboratory | Cat# JAX:006148; RRID: IMSR_JAX:006148 |
| Mouse: Rosa-CAG-LSL-Sun1-GFP: B6; 129-Gt(ROSA)26Sortm5(CAG–Sun1/sfGFP)Nat/J | The Jackson Laboratory | Cat# JAX:021039; RRID: IMSR_JAX:021039 |
| Mouse: Rosa-CAG-LSL-tdTomato: B6. Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | Cat# JAX:007909; RRID: IMSR_JAX:007909 |
|
| ||
| Oligonucleotides | ||
|
| ||
| β-actin qPCR primer forward: 5′-CTGTCCCTGTATGCCTCTGG-3′ |
Integrated DNA technologies | N/A |
| β-actin qPCR primer reverse: 5′-ATGTCACGCACGATTTCC-3′ |
Integrated DNA technologies | N/A |
| CasRx qPCR primer forward: 5′-AGGCTGGAGAAGATCGTGG-3′ |
Integrated DNA technologies | N/A |
| CasRx qPCR primer reverse: 5′-GGACTCGCCGAAGTACCTCT-3′ |
Integrated DNA technologies | N/A |
| Ptbp1 qPCR primer forward: 5′-AGAGGAGGCTGCCAACACTA-3′ |
Integrated DNA technologies | N/A |
| Ptbp1 qPCR primer reverse: 5′ -ATTGCCATTCCTGCATCCA-3′ |
Integrated DNA technologies | N/A |
| RFP qPCR primer forward: 5′-GGCATCCCCGACTTCTTTA-3′ |
Integrated DNA technologies | N/A |
| RFP qPCR primer reverse: 5′-AGCCGCGTGTTTTCTTCTG-3′ |
Integrated DNA technologies | N/A |
| tdTomato qPCR primer forward: 5′-GAGCAAGGGCGAGGAGGTCA-3′ |
Integrated DNA technologies | N/A |
| tdTomato qPCR primer reverse: 5′-GATGTCGGCGGGGTGCTTC-3′ |
Integrated DNA technologies | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pAAV-CMV-LSL-RFP | Qian et al. (2020) | N/A |
| pAAV-CMV-LSL-RFP-ShPtbp1 | Qian et al. (2020) | N/A |
| pAAV-GFAP-CasRx | Zhou et al. (2020) | Cat# 154000; RRID:Addgene_154000 |
| pAAV-GFAP-CasRx-Pfibp1 | Zhou et al. (2020) | Cat# 154001; RRID:Addgene_154001 |
| pAAV-GFAP-Cre | this study | N/A |
| pAAV-GFAP-tdTomato | this study | N/A |
| pAAV-GFAP-GFP | this study | N/A |
| pUCmini-iCAP-PHP.eB | Chan et al. (2017) | Cat# 103005; RRID:Addgene_103005 |
|
| ||
| Software and algorithms | ||
|
| ||
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism |
| ImageJ | Schindelin et al. (2012) | https://imagej.net/software/fiji |
| Blender 2.9 | Blender | https://www.blender.org |
Highlights.
AAV-based Cre recombination is unsuitable for examining MG-to-RGC conversion
Lineage-traced MG are not converted into RGCs after Ptbp1 downregulation
NMDA-induced injury does not facilitate MG-to-RGC conversion after Ptbp1 downregulation
Stringent fate mapping is required for critical examination of glia-to-neuron conversion
ACKNOWLEDGMENTS
We thank Dr. Xiang-Dong Fu (University of California, San Diego), for providing AAV-CMV-LSL-RFP and AAV-CMV-LSL-RFP-shPtbp1 plasmids and technical advice, and the flow cytometry CoRE (Icahn School of Medicine at Mount Sinai) for technical help with MG purification. This work was supported by National Institutes of Health grants R01 EY024986 and R01 EY028921, an unrestricted challenge grant from Research to Prevent Blindness, the New York Eye and Ear Infirmary Foundation, and The Harold W. McGraw Jr. Family Foundation for Vision Research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110960.
REFERENCES
- Blackshaw S, and Sanes JR (2021). Turning lead into gold: reprogramming retinal cells to cure blindness. J. Clin. Invest. 131. 10.1172/jci146134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau-Pinsonneault C, and Cayouette M (2018). Cell lineage tracing in the retina: could material transfer distort conclusions? Dev. Dynam. 247, 10–17. 10.1002/dvdy.24535. [DOI] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sanchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, and Gradinaru V (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179. 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaeidi F, Macpherson P, Mills EA, Jui J, Flannery JG, and Goldman D (2018). Notch suppression collaborates with Ascl1 and Lin28 to unleash a regenerative response in fish retina, but not in mice. J. Neurosci. 38, 2246–2261. 10.1523/jneurosci.2126-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D (2014). Muller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 15, 431–442. 10.1038/nrn3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, and Samulski RJ (2006). Production and characterization of adeno-associated viral vectors. Nat. Protoc. 1, 1412–1428. 10.1038/nprot.2006.207. [DOI] [PubMed] [Google Scholar]
- He Z, and Jin Y (2016). Intrinsic control of axon regeneration. Neuron 90, 437–451. 10.1016/j.neuron.2016.04.022. [DOI] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, et al. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, and Reh TA (2017). Stimulation of functional neuronal regeneration from Muller glia in adult mice. Nature 548, 103–107. 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, and Reh TA (2010). Regenerative medicine for retinal diseases: activating endogenous repair mechanisms. Trends Mol. Med. 16, 193–202. 10.1016/j.molmed.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak RR, Koerber JT, Dalkara D, Flannery JG, and Schaffer DV (2009). A novel adeno-associated viral variant for efficient and selective intravitreal transduction of rat Muller cells. PLoS One 4, e7467. 10.1371/journal.pone.0007467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LA, and Gordon LK (2002). Retinal ganglion cell disorders: types and treatments. Prog. Retin. Eye Res. 21, 465–484. 10.1016/s1350-9462(02)00012-5. [DOI] [PubMed] [Google Scholar]
- Martin JF, and Poche RA (2019). Awakening the regenerative potential of the mammalian retina. Development 146. 10.1242/dev.182642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384. 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Aoki H, Hirata A, Tomita H, Green PG, and Hara A (2016). Retinal cell degeneration in animal models. Int. J. Mol. Sci. 17, 110. 10.3390/ijms17010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, Zhang X, Xue Y, Maimon R, Dowdy SF, et al. (2020). Reversing a model of Parkinson’s disease with in situ converted nigral neurons. Nature 582, 550–556. 10.1038/s41586-020-2388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda EM, Hall BM, Hill MC, Swinton PG, Tong X, Martin JF, and Poche RA (2019). The hippo pathway blocks mammalian retinal muller glial cell reprogramming. Cell Rep. 27, 1637–1649.e6. 10.1016/j.celrep.2019.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Wilken MS, Cox KE, Chipman L, Jorstad N, Sternhagen K, Simic M, Ullom K, Nakafuku M, and Reh TA (2015). Transgenic expression of the proneural transcription factor Ascl1 in Muller glia stimulates retinal regeneration in young mice. Proc. Natl. Acad. Sci. U. S. A. 112, 13717–13722. 10.1073/pnas.1510595112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Serrano C, Zhong X, Ma S, Zou Y, and Zhang CL (2021). Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 184, 5465–5481.e16. 10.1016/j.cell.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Rattner A, Zhou Y, Williams J, Smallwood PM, and Nathans J (2012). Norrin/Frizzled4 signaling in retinal vascular development and blood brain barrier plasticity. Cell 151, 1332–1344. 10.1016/j.cell.2012.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, and Chen B (2022). Critical examination of muller glia-derived in vivo neurogenesis in the mouse retina. Front. Cell Dev. Biol. 10, 830382. 10.3389/fcell.2022.830382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96. 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Qiu S, Tian L, Snider WD, Flannery JG, Schaffer DV, and Chen B (2016). Wnt regulates proliferation and neurogenic potential of muller glial cells via a lin28/let-7 miRNA-dependent pathway in adult mammalian retinas. Cell Rep. 17, 165–178. 10.1016/j.celrep.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K, Qiu S, Wang YV, Park SJH, Mohns EJ, Mehta B, Liu X, Chang B, Zenisek D, Crair MC, et al. (2018). Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 560, 484–488. 10.1038/s41586-018-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Su J, Hu X, Zhou C, Li H, Chen Z, Xiao Q, Wang B, Wu W, Sun Y, et al. (2020). Glia-to-Neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell 181, 590–603. 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


