Significance
Lymphoid malignancies frequently bear oncogenic chromosomal translocations of the rearranging immune receptor loci; however, the predisposing mechanisms remain unclear. Using a clinically relevant model of T cell leukemia driven by β-catenin activation, our studies show that β-catenin exploits Tcf-1 to promote a novel transcriptional and epigenetic state that down-regulates both homologous recombination (HR) DNA repair and cell cycle regulatory pathways, which promote Tcra/Myc-Pvt1 translocations. Significantly, the HR impairments of these leukemias render them highly sensitive to PARP inhibitors, a finding that could inform future therapeutic options for malignancies with activated β-catenin signaling.
Keywords: genomic instability, T-ALL, β-catenin, Tcf-1
Abstract
Understanding the mechanisms promoting chromosomal translocations of the rearranging receptor loci in leukemia and lymphoma remains incomplete. Here we show that leukemias induced by aberrant activation of β-catenin in thymocytes, which bear recurrent Tcra/Myc-Pvt1 translocations, depend on Tcf-1. The DNA double strand breaks (DSBs) in the Tcra site of the translocation are Rag-generated, whereas the Myc-Pvt1 DSBs are not. Aberrantly activated β-catenin redirects Tcf-1 binding to novel DNA sites to alter chromatin accessibility and down-regulate genome-stability pathways. Impaired homologous recombination (HR) DNA repair and replication checkpoints lead to retention of DSBs that promote translocations and transformation of double-positive (DP) thymocytes. The resulting lymphomas, which resemble human T cell acute lymphoblastic leukemia (T-ALL), are sensitive to PARP inhibitors (PARPis). Our findings indicate that aberrant β-catenin signaling contributes to translocations in thymocytes by guiding Tcf-1 to promote the generation and retention of replication-induced DSBs allowing their coexistence with Rag-generated DSBs. Thus, PARPis could offer therapeutic options in hematologic malignancies with active Wnt/β-catenin signaling.
Genomic instability is a well-characterized hallmark of cancer that drives both malignant transformation and cancer cell plasticity, which underlies treatment escape. It is estimated that a human cell is subjected to ∼70,000 DNA lesions a day and the integrity of its genome depends on a coordinated balance between the generation and faithful repair of these DNA lesions (1). While extensive research into DNA repair pathways has identified many cancer risks and treatment opportunities, the precise molecular mechanism generating large-scale genomic alterations remains incomplete. Chromosomal aberrations and gene fusions are particularly prevalent in hematological malignancies, including up to 80% of T cell acute lymphoblastic leukemia (T-ALL) (2–4). Importantly, T-ALL is an aggressive hematologic malignancy with poor prognosis and limited treatment options that remain largely untargeted (5–7). While dose escalation and combinatorial strategies have improved pediatric cure rates to >90%, these regimens are not well tolerated by adult patients (6). Excitingly, molecular and genomic classifications have led to improved new therapies including antibody and chimeric antigen receptor T cell (CAR-T) regimens that predominately target B cell ALL (6). However, similar success in T-ALL has lagged.
Lymphocytes are unique among somatic cells because their development requires the controlled generation of DNA double strand breaks (DSBs) to recombine the T cell receptor (TCR) and immunoglobulin (Ig) loci. Rag recombinases catalyze these DSBs at recombination signal sequences (RSSs) that flank the V, D, and J segments of these loci. Rag activity occurs during the G0/G1 phases of the cell cycle when the predominant DSB repair pathway is nonhomologous end joining (NHEJ) (8–10). NHEJ is error prone as it directly ligates DNA ends leading to small insertions, deletions, and potentially translocations (11). Homologous recombination (HR), the other major DSB repair pathway, is favored during replication when a sister chromatid is present, providing a template for error-free repair (12). Genome stability during thymocyte development requires a refined coordination between cell cycle and repair mechanisms. This is because T cell development is a highly regulated stepwise progression through precursor stages that require bursts of proliferation followed by G0/G1 arrest to facilitate Rag-mediated receptor rearrangements (13, 14). When DSBs are detected, checkpoint kinases Chek2 or Chek1 signal cell cycle arrest to allow for repair of DNA lesions and maintenance of genome stability (15). In thymocytes, the inability to promptly resolve replication-induced DSBs due to failure of repair pathways and/or checkpoint signaling could allow their persistence into G0/G1. At this phase, which relies on the error-prone NHEJ repair mechanism, these breaks could serve as aberrant substrates during Rag recombination, leading to translocations. Not surprisingly, chromosomal aberrations involving the recombining T cell receptor genes are frequently observed in T-ALL cases (16).
Stabilizing mutations in β-catenin and activation of the Wnt signaling pathway have been described in T cell malignancies (17), including precursor (T-ALL), peripheral (PTCL), cutaneous (CTCL), and adult T cell leukemia (ATL) (18–21). A more recent study showed that β-catenin is required for the initiation of T cell leukemia downstream of Notch signaling (22), which is the most frequently activated pathway due to mutations in T-ALL patients. Additionally, both loss of PTEN and activation of PI3K/Akt signaling, which mark ∼20% of T-ALL (23), stabilize β-catenin by inactivating Gsk3β and preventing the phosphorylation events that initiate its proteasomal degradation (24–26). Mouse models of thymocyte-specific β-catenin activation, via mutation of β-catenin itself, loss of PTEN, or expression of an active AKT, produce genomically unstable T cell leukemias that mirror human disease (24–28). Importantly, β-catenin signaling is required in PTEN-deficient models of T cell transformation (24), highlighting the need to understand the mechanisms by which β-catenin activation drives T cell transformation.
The T cell–specific partners of β-catenin in the context of Wnt signaling are the HMG domain-containing DNA binding proteins Tcf-1 (encoded by Tcf7) and Lef-1. Upon Wnt activation, β-catenin is stabilized and transported to the nucleus where it interacts with DNA-bound Tcf-1 and Lef-1, promoting chromatin accessibility and target gene expression (29). Tcf-1 has essential roles at almost all stages of T cell development and function (30). We and others have shown that Tcf-1 is required for T cell specification (31, 32), progression to the CD4+CD8+ double-positive (DP) stage (33), thymic selection, and the choice of postselection T cell lineages (34). These studies established that Tcf-1 modulates the chromatin landscape and transcription profiles of thymocytes (35). Specifically, we showed that in DP thymocytes Tcf-1 acts both by directly binding its conserved DNA motif or through the indirect coordination with other regulatory proteins (33). Tcf-1 binding can promote either up- or down-regulation of target gene expression depending on sequence context and coordination with binding partners. It remains unclear how oncogenic β-catenin stabilization affects the natural functions of Tcf-1 to promote transformation.
Here we used our previously established mouse model of T-ALL, which relies on conditional stabilization of β-catenin in DP thymocytes, to understand the mechanism by which β-catenin and Tcf-1 cooperate in transforming these cells (24, 25). The leukemias in our model have chromosomal translocations that are identical to aberrations seen in T cell leukemia models resulting from PTEN ablation or constitutive AKT activation (28). We found that conditionally ablating Tcf-1 at the time of β-catenin stabilization abrogates leukemogenesis. This observation allowed us to narrow down on the specific mechanisms of transformation. Our findings indicate that activation of β-catenin redirects Tcf-1 binding and promotes gene expression changes that compromise the replication process and checkpoint responses to replication stress. As a result, DNA damage generated during replication is aberrantly joined with Rag breaks induced during the subsequent G0/G1 phase. The HR impairments of these leukemias render them more vulnerable to Parp inhibitors and provide a therapeutic possibility.
Results
β-Catenin–Induced Translocations Link DSBs from Two Distinct Processes.
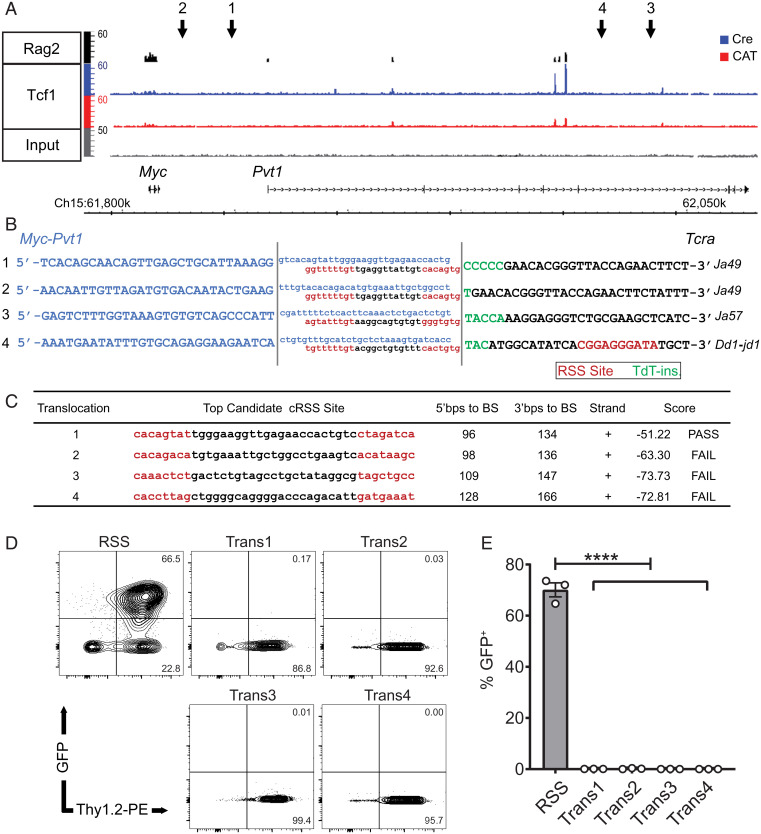
We have previously reported that CD4-Cre–mediated stabilization of β-catenin (36) (CAT mice) produces DP T cell lymphomas (24). These have recurrent translocations in which the Tcra locus on chromosome 14 is illegitimately linked to the Myc-Pvt1 locus on chromosome 15 (25), driving elevated expression of the Myc oncogene and malignant transformation of DP thymocytes. Although lymphomagenesis in CAT mice is dependent on Rag activity, we sought to further understand the mechanisms underpinning the genomic instability that allows for aberrant Tcra/Myc-Pvt1 joins. Given that both Rag2 (37) and Tcf-1 bind the Myc-Pvt1 locus as shown by chromatin immunoprecipitation sequencing (ChIP-seq) profiles from normal DP thymocytes (Fig. 1A), we hypothesized that off-target Rag activity may generate DSBs in the Myc-Pvt1 locus, which are then aberrantly joined with breaks generated during normal Tcra rearrangement. To address this postulate, we made whole genome mate pair libraries from genomic DNA isolated from four CAT lymphomas and next generation sequencing was used to identify discordant reads. The precise translocation breakpoints were then established using Sanger sequencing of regions comprising Tcra/Myc-Pvt1 hybrid reads (Fig. 1B and SI Appendix, Table S1) These analyses revealed that the Tcra breakpoint site involved early rearranging J fragments of the Tcra locus (Ja49, Ja57, and Dd1-jd1). Normal thymocyte development contains inherent danger for genomic stability, as it involves generating DSBs and joining pairs of recombination signal sequence (RSS) sites with either 12- or 23-bp spacers (according to the 12/23 rule), which flank V, D, and J fragments. Additionally, enzymes such as terminal deoxynucleotidyl transferase (TdT) then mediate gap filling at coding joins that is critical for increasing receptor diversity for adaptive immunity. As expected, the Tcra break site of the translocations contained a proximal canonical RSS, suggesting these breaks were Rag generated. Furthermore, small insertions of nontemplated nucleotides (N-nucleotides) next to RSS sites are evidence of TdT activity (Fig. 1B). In contrast, no canonical RSS sequences were identifiable within the Myc-Pvt1 breakpoint sites. As Rag can also generate DSBs at alternative sites known as cryptic RSS (cRSS) sites, we employed the reference database and prediction tool RSSsite (38) to evaluate the 200-bp region flanking the Myc-Pvt1 breakpoints, accounting for potential resection from DSB processing enzymes prior to translocation formation. Although only one low-quality cRSS site was identified in four separate lymphomas (Fig. 1C), we also functionally tested the top scoring sequences from each lymphoma using a Rag-activity reporter assay. We prioritized 23-bp spacer cRSSs that would pair with the 12-bp RSS sites identified in the Tcra site of the translocation (Fig. 1C). The potential cRSS site from each lymphoma was synthesized and cloned into the pMX-INV-GFP Rag retroviral recombination reporter (39). In pMX-INV, Rag activity mediates inversion of an antisense green fluorescent protein (GFP) cDNA flanked with RSS sites and initiates GFP expression. These vectors were used to transduce (v)-Abl kinase-transformed pre-B-cell lines (a gift of B. P. Sleckman, UAB, Birmingham, AL). Treatment of these transduced and highly proliferative pre-B cell lines with the Abl kinase inhibitor, STI571, blocks the G1-to-S transition and rapidly initiates Rag expression and activity (SI Appendix, Fig. S1) (39). In contrast to a control vector containing a bona fide RSS site, these experiments recovered no GFP expression in any of the potential Myc-Pvt1 cRSSs, demonstrating that the cloned sequences were not Rag substrates (Fig. 1 D and E). These findings indicate that the illegitimately repaired DSBs at the Myc-Pvt1 site of the translocation breakpoint result from an alternative, Rag-independent mechanism(s).
Fig. 1.
The Tcra translocation break site is Rag mediated, while the Myc-Pvt1 is Rag independent. (A) ChIP-seq tracks of Rag2 (37) binding in wild-type (WT) DP thymocytes (Top) as well as Tcf-1 binding and enrichment of H3K4m3, and H3K27Ac in Cre and CAT DP thymocytes in the Myc-Pvt1 locus. Arrows 1 through 4 show the location of break sites from sequenced Tcr/Myc-Pvt1 translocations in four CAT lymphomas. (B) Tcra/Myc-Pvt1 breakpoint sequences from whole genome sequencing (uppercase) and flanking endogenous region (lowercase) are shown, highlighting heptamers and nonamers of RSS sites (red) with 12-bp spacers and TdT-inserted N-nucleotides at junctions (green). (C) Highest scoring potential cryptic RSS (cRSS) site from Myc-Pvt1 break points ±200 bp using RSSsite (https://www.itb.cnr.it/rss/index.html) (38) (BS, break site). (D) Representative flow cytometry dot plots of GFP+ cells after a 3-d STI571 (imatinib) treatment of v-Abl-transformed pre-B cells containing pMX-INV-GFP vectors with cloned cRSSs from Myc-Pvt1 sites (Above) or a control, canonical RSS site. Vector also harbors an IRES-Thy1.2, marking successful viral transduction of Rag-reporter cells. (E) Representative flow cytometric histograms of GFP+Thy1.2+ cells (Left) and cumulative analysis from three independent experiments (Right); data are represented as the mean ± SEM, and statistical testing is depicted as two-sided, unpaired t test; ****P ≤ 0.0001.
Tcf-1 Is Essential for β-Catenin–Mediated Transformation of DP Thymocytes.
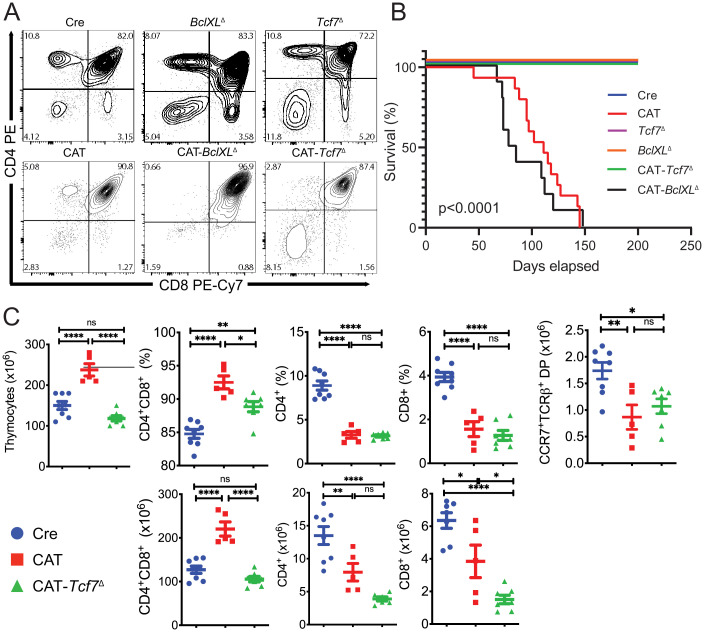
We next investigated processes that are altered by β-catenin activation and may contribute to transformation. Our earlier studies showed that thymocytes with activated β-catenin are less sensitive to irradiation-induced cell death. We further linked this to overexpression of BclXL, the main antiapoptotic mediator in DP thymocytes (40), by showing that BclXL inhibitors abrogated the survival advantage of CAT thymocytes (25). Therefore, to determine whether enhanced DP thymocyte survival contributes to their transformation, we conditionally ablated BclXL in DP thymocytes simultaneously with activation of β-catenin by crossing CAT mice with mice carrying a BclXLfl allele (41). Compound CD4-Cre/Ctnnbex3fl/BclXLfl/fl (CAT-BclXLΔ), mice had the same developmental block in the transition of DP thymocytes to the single-positive (SP) stages described in our previous reports (24) and succumbed to leukemia with the same frequency and latency as CAT mice (Fig. 2 A and B and SI Appendix, Fig. S2). Thus, the BclXL-mediated increase in the survival of CAT DP thymocytes does not contribute to their developmental defect or their transformation in vivo. Consequently, we speculated that the developmental block could be mediated by the DNA binding partner of β-catenin, Tcf-1, and may be contributing to transformation. We therefore produced compound Cd4-Cre/Ctnnb1ex3fl/Tcf7fl/fl (CAT-Tcf7Δ) mutant mice with conditional ablation of Tcf-1 in CAT DP thymocytes. Tcf-1 deletion did not resolve the developmental block, and CAT-Tcf7Δ thymocytes had reduced numbers of CCR7+TCRβ+ postselected DPs as well as fewer CD4+ and CD8+ SP thymocytes, as seen in CAT mice (Fig. 2 A and C). Surprisingly, ablation of Tcf-1 completely abrogated leukemogenesis (Fig. 2B). This finding indicates that Tcf-1 specifically mediates a fraction of the processes that are altered by β-catenin activation in DP thymocytes. Importantly, since these Tcf-1–mediated functions are critical for leukemogenesis, the CAT-Tcf7Δ mouse model offers a valuable tool to specifically narrow down on the mechanisms involved in transformation.
Fig. 2.
Ablation of Tcf7 rescues CAT lymphomas but a developmental block remains. (A) Representative flow cytometry contour plots of live thymocytes from 6- to 8-wk-old mice stained for CD4 and CD8 to show developmental progression. (B) Kaplan–Meier survival curve analysis for Cre (CD4-Cre, n = 10), CAT (CD4-Cre/Ctnnbex3fl/ex3fl, n = 15), Tcf7Δ (CD4-Cre/Tcf7fl/fl, n = 10), BclXLΔ (CD4-Cre/BclXLfl/fl, n = 10), and those with codeletions CAT-Tcf7Δ (CD4-Cre/Ctnnbfl/f/Tcf7fl/fl, n = 5) or CAT-BclXLΔ (CD4-Cre/Ctnnbfl/f/BclXlfl/fl, n = 10). (C) Flow cytometric histograms indicating the percentage (Top) and total number (Bottom) of thymocytes in the indicated late developmental stages in Cre (n = 8), CAT (n = 5), and CAT-Tcf7Δ (n = 7) mice; data are represented as the mean ± SEM, and statistical testing is depicted as two-sided, unpaired t tests; ns > 0.05. *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001.
Aberrant β-Catenin Uses Tcf-1 to Down-Regulate Genome Maintenance Pathways.
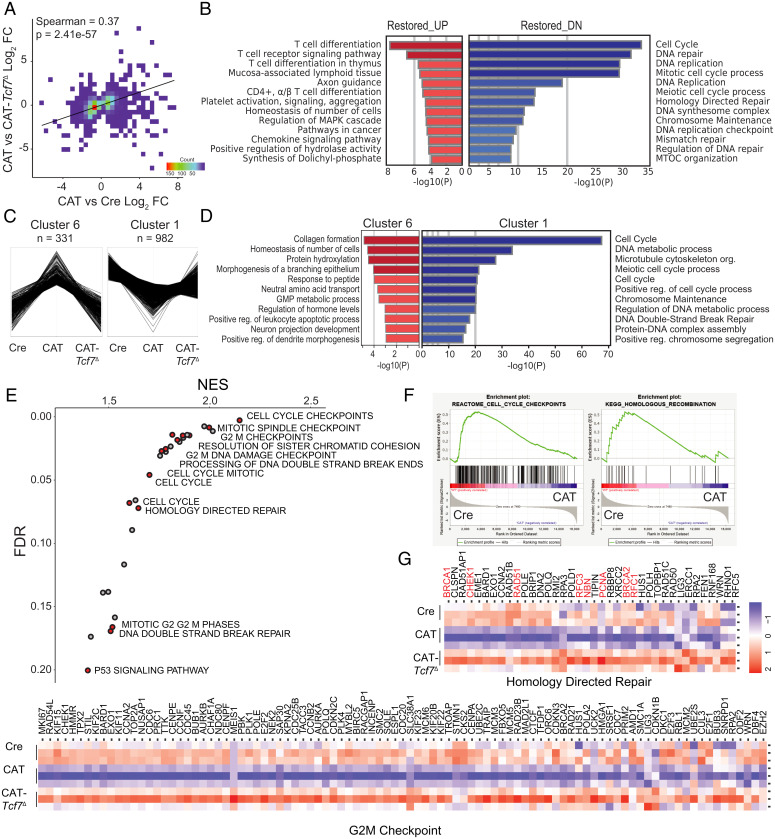
To identify the Tcf-1–mediated processes involved in the transformation of CAT thymocytes by β-catenin, we performed RNA sequencing in pretransformed (6 wk) sorted DPs. Pairwise comparisons of gene expression in the three genotypes, represented in volcano plots, exhibited altered transcriptional programs with both up- and down-regulated genes (SI Appendix, Fig. S3A). To determine the extent of rescue in CAT-Tcf7Δ thymocytes at the transcriptional level, we identified the differentially expressed genes (DEGs, P < 0.05) between CAT and either Cre or CAT-Tcf7Δ cells, finding 1,181 and 3,735 DEGs, respectively. As expected, Spearman rank correlation of these two sets of DEGs established that deletion of Tcf7 in CAT thymocytes significantly reverts transcriptional programs to those in Cre thymocytes (Spearman = 0.37, P = 2.41 × 10−57; Fig. 3A). Importantly, the restored DEGs that were down-regulated by β-catenin and returned by ablation of Tcf-1 show strong enrichment of cell cycle, DNA repair, and DNA replication pathways, specifically implicating homologous recombination, the error-free program for DSB repair (Fig. 3B, blue). Interestingly, the up-regulated DEGs showed significant enrichment in T cell development pathways and are likely contributing to the DP block in CAT thymocytes (Fig. 3B, red). Nevertheless, restoration of these genes upon the deletion of Tcf-7 is insufficient to overcome the developmental stalling in β-catenin stabilized thymocytes.
Fig. 3.
β-Catenin uses Tcf-1 to down-regulate replication, HR repair, and checkpoint control pathway genes. (A) Spearman correlation of differentially expressed genes (P < 0.05) between CAT and either Cre or CAT-Tcf7Δ. (B) Pathway analysis of restored DEGs that were up-regulated (red) or down-regulated (DN, blue) in CAT versus Cre and CAT-Tcf7Δ (Metascape, www.metascape.org). (C) Unsupervised clustering of triplicate RNA-seq profiles (FPKMs) for Cre (n = 3), CAT (n = 3), and CAT-Tcf7Δ (n = 3) DP (CD4+CD8+) thymocytes; gene clusters associated with altered expression in CAT thymocytes that are restored to Cre levels in CAT-Tcf7Δ are shown. (D) Functional pathways enriched in cluster 1 and cluster 6 gene sets (Metascape). (E) GSEA/MSigDB of cluster 1 gene expression showing significant signatures (normalized enrichment score (NES) > 1 Faulse Discovery Rate (FDR) < 0.2) with relevant genome maintenance programs labeled and highlighted in red. (F) Representative GSEA profiles and (G) heatmaps of core enrichment genes of the indicated pathways; central replication and repair genes are highlighted (red).
To specifically identify gene sets that corelate with the lymphomagenesis phenotype in CAT mice, we performed unbiased clustering analysis (ExpressCluster, www.GenePattern.org) of expression levels (FPKM = Fragments Per Kilobase of transcript per Million mapped reads) across all genes in Cre, CAT, and CAT-Tcf7Δ thymocytes. Two out of eight gene clusters were up-regulated (cluster 6, n = 331) or down-regulated (cluster 1, n = 982) in CAT thymocytes and restored in the absence of Tcf-1 (Fig. 3C and SI Appendix, Fig. S3B). Interestingly, pathway analysis of the down-regulated genes (www.metascape.org) also identified strong enrichment of genome maintenance pathways, including DSB repair (cluster 1, Fig. 3D). In contrast, up-regulated genes did not show strong enrichment for functionally relevant pathways (cluster 6, Fig. 3D). We also investigated two gene clusters that were altered in CAT thymocytes and remained at similar levels in CAT-Tcf7Δ cells; these genes exhibited mild enrichment for some basic T cell and homeostatic functions (SI Appendix, Fig. S3 C and D). Taken together, this analysis suggests that down-regulated genes in CAT thymocytes include those responsible for dysregulated genome maintenance pathways that may contribute to transformation. However, genes that are not restored could contribute to the continued developmental defects.
Finally, we performed gene set enrichment analysis (GSEA) (42) on cluster 1 genes, which accounts for magnitude and rank order of gene expression changes associated with specific cellular functions. The most significantly enriched pathways confirmed the Tcf-1–mediated dysregulation of cell cycle checkpoints and genome maintenance pathways in CAT thymocytes (Fig. 3E). We further identified the core enrichment genes at the leading edge of the GSEA profiles in five relevant pathway associations (Fig. 3 F and G and SI Appendix, Fig. S3D). Importantly, core regulators of HR-mediated repair, including Brca1, Brca2, Rad51, and Rbbp8, and replication or cell cycle checkpoint engagement, including Pcna, Rfc1, Chek1, and Chek2, were down-regulated in CAT thymocytes. Pathways involving chromosome maintenance and processes necessary for mitosis were also overrepresented. Taken together, these data provide strong transcriptional evidence that β-catenin contributes to the genomic instability of thymocytes through a Tcf-1–mediated down-regulation of genes required for cell cycle regulation, faithful DNA replication, and DNA repair of DSBs, whereas a set of genes that are positively regulated by Tcf-1 are more likely involved in the developmental effects of β-catenin.
Dominant Active β-Catenin Directs Novel Tcf-1 Binding to HR Repair and Checkpoint Control Genes.
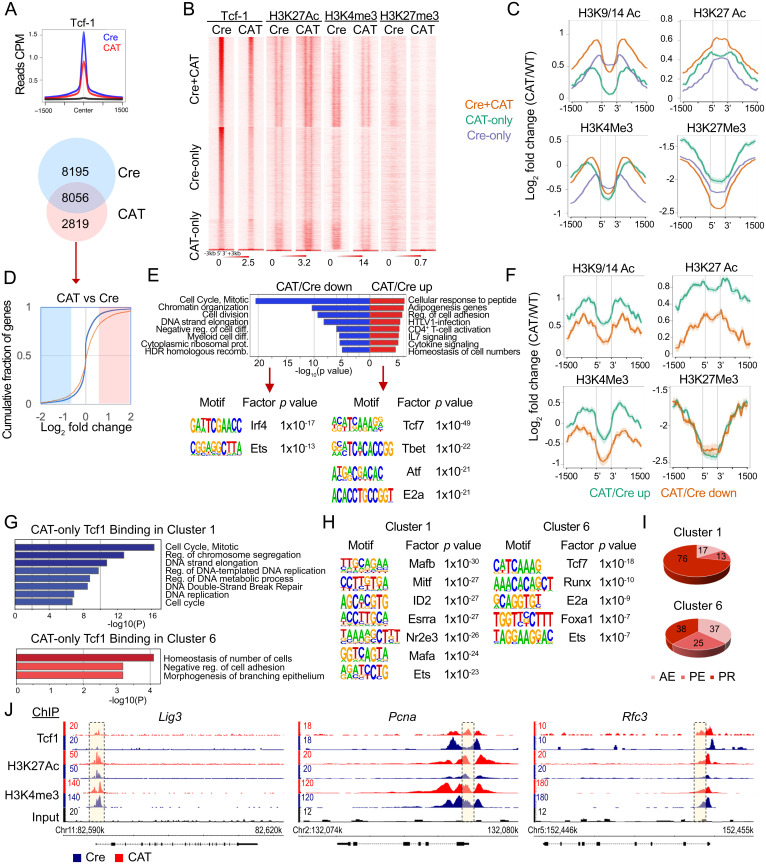
We considered that expression changes in CAT thymocytes may reflect β-catenin–mediated redirection of Tcf-1 binding and/or changes in chromatin accessibility. Therefore, we mapped the distribution of Tcf-1 binding in CAT DP thymocytes using ChIP-seq. We identified 10,875 high-confidence Tcf-1 peaks (P = 10e-4) in these cells compared to 16,251 we had previously detected in Cre DP thymocytes (33). Tcf-1 peaks in CAT were overall less enriched compared to Cre thymocytes (Fig. 4A). Although the majority of Tcf-1 peaks (8,056) in CAT were shared with those in Cre DP thymocytes, there were 2,819 new Tcf-1 peaks that are unique to CAT thymocytes (Fig. 4A).
Fig. 4.
Dominant active β-catenin alters the epigenetic and transcription profile of DP thymocytes by redirecting Tcf-1 binding. (A) Histogram overlays of Tcf-1 binding centered on Tcf-1–bound regions in Cre DP thymocytes (±1.5 kb, Top) and Venn diagram of overlapping Tcf-1 peaks between Cre and CAT DP thymocytes. (B) Heat maps of ChIP-seq peaks for Tcf-1 and indicated histone marks, centered on Tcf-1 binding (±3 kb) at shared or unique sites as indicated. (C) Comparative enrichment histograms of histone modifications associated with active promoters (H3K9/14Ac), active enhancers (H3K27ac), transcription (H3K4Me3), and closed/poised chromatin (H3K27me3). (D) CDF plot of expression changes in genes uniquely bound by Tcf-1 in CAT DP thymocytes (n = 2819, red line) compared to all genes (blue line). (E) Pathway analysis of down-regulated (blue) and up-regulated (red) genes uniquely bound by Tcf-1 in CAT (Top) and the most significantly enriched transcription factor binding motifs (HOMER, Bottom). (F) Comparative enrichment histograms for histone modifications (H3K9/14Ac, H3K27ac, H3K4Me3, and H3K27Me3) centered on CAT-unique Tcf-1 binding sites corresponding to up- or down-regulated genes in CAT versus Cre as indicated. (G) Pathway analysis of cluster 1 (blue, Top) and cluster 6 (red, Bottom) genes uniquely bound by Tcf-1 in CAT. (H) The most significantly enriched transcription factor binding motifs (HOMER) of cluster 1 (Left) and cluster 6 (Right) genes uniquely bound by Tcf-1 in CAT. (I) Pie charts show the distrubution of CAT unique Tcf-1 binding sites to PR, PE, or AE of genes associated with cluster 1 and cluster 6. (J) Representative enrichment tracks (Integrated Genome Browser), of Tcf-1, H3K27Ac, and H3K4me3 ChIP-seq in Cre or CAT DP thymocytes as indicated, showing key replication and repair genes that are differentially expressed and have novel Tcf-1 sites in CAT (yellow boxes).
To assess the effect of β-catenin stabilization on the chromatin landscape, we determined the distribution of histone marks in CAT DP thymocytes by ChIP-seq and compared it to our data from Cre DP thymocytes (33). We mapped histone 3 acetylation at lysines 9/14 (H3K9/14Ac), which marks active promoters, H3K27Ac, which marks active enhancers, H3K4me3, which is associated with activation of transcription, and H3K27me3, which marks poised/closed chromatin. Overall, near Tcf-1 peaks, enrichment for H3K9/14Ac and H3K27Ac marks increased while enrichment for H3K27me3 marks decreased in CAT compared to Cre DP thymocytes (Fig. 4 B and C). The degree of changes in histone mark enrichment (log2 fold change [FC]) varied depending on whether the Tcf-1 peaks were unique to Cre or CAT or shared between the two genotypes. The greatest increase of the active chromatin histone marks H3K9/14Ac and H3K27Ac and reduction of the poised/inactive H3K27me3 histone mark was detected in shared Cre and CAT Tcf-1 sites compared to the novel CAT Tcf-1 sites. In contrast to the dynamic changes in these histone marks, H3K4me3 showed little change in Cre versus CAT for shared or CAT-only Tcf-1 peaks and showed reduced enrichment in Cre-only Tcf-1 peaks. Thus, β-catenin stabilization enhances the presence of active chromatin marks in Tcf-1–bound sites consistent with the classical understanding of its epigenetic functions.
We found no bias in the overall Tcf-1 redistribution in CAT DP thymocytes when separating genomic regions by promoters (PRs), active enhancers (AEs), or poised enhancers (PEs), as we previously defined in Cre DP thymocytes (33) (SI Appendix, Fig. S4 B and C). The novel Tcf-1 sites in CAT mice also showed similar distribution to Tcf-1 sites we previously reported for Cre mice (33), with 57% versus 43% in PRs, 18% versus 24% in AEs, and 25% versus 33% in PEs (SI Appendix, Fig. S4D). The novel Tcf-1 peaks in AEs and PEs were highly enriched for the conserved Tcf-1 motif, whereas peaks at PR sites lacked such enrichment (SI Appendix, Fig. S4F). Annotation of genes associated with novel Tcf-1 binding at PRs, AEs, or PEs (www.metascape.org/) revealed that the PR-bound group was enriched in genes involved in chromatin organization, cellular response to stress, and DNA damage stimulus. The AE-bound group was enriched in genes associated with T cell–related processes and the PE group showed no specific strong enrichment. The distribution of the novel Tcf-1 sites in PRs, AEs, or PEs, and the corresponding chromatin changes, suggest that these sites may selectively affect the expression of the associated genes.
To address this possibility, we compared the expression changes in Cre versus CAT of the genes associated with novel Tcf-1 sites to all genes, using cumulative distribution function (CDF) analysis (Fig. 4D). Consistent with the finding that stabilization of β-catenin results in an overall increase in active chromatin marks, the genes associated with novel Tcf-1 binding were significantly more likely to become up-regulated in CAT thymocytes(Kolmogorov–Smirnov (K&S) P < 0.0001). We next identified two groups of significantly up-(Log2FC > 0.56) or down-regulated (Log2FC < −0.56) genes with novel Tcf-1 peaks in CAT thymocytes. Pathway enrichment analysis (www.metascape.org/) revealed that the group of up-regulated genes with new Tcf-1 peaks was enriched in genes involved in general T cell functions, whereas the group of down-regulated genes was enriched in HR DNA repair and cell cycle pathways (Fig. 4E). We also investigated the Tcf-1 peaks that were unique to Cre thymocytes, which did not appear to contribute to genome maintenance pathways and were also enriched for homeostatic or basic T cell functions (SI Appendix, Fig. S4A). We further characterized the chromatin changes specifically at these CAT-unique, Tcf-1–bound DEGs. Novel Tcf-1 binding led to greater increases in H3K9/14Ac and H3K27Ac active chromatin marks in up-regulated compared to down-regulated genes (Fig. 4F). Importantly, enrichment of H3K4me3 only increased in novel Tcf-1 sites associated with up-regulated genes but decreased in sites associated with down-regulated genes, in line with the association of this histone mark with activation of transcription. To determine how Tcf-1 coordinates these opposing expression programs, we performed motif enrichment analysis on novel Tcf-1 binding sites associated with genes that were up- or down-regulated in CAT thymocytes. Tcf-1 peaks in up-regulated genes were enriched for the Tcf-1 binding motif (Tcf7) in addition to other key transcriptional regulators in T cell development such as Tbet and E2A. In contrast, novel Tcf-1 peaks in down-regulated genes were not enriched for the Tcf-1 consensus motif, suggesting that Tcf-1 acts in distinct complexes with other transcriptional regulators to enforce these different outcomes on chromatin and transcription.
To test whether the regulatory changes we detected were reflected in the cluster analysis that identified CAT lymphoma–associated gene sets, we selected the genes in cluster 1 (144 genes) or cluster 6 (65 genes) that acquired CAT unique Tcf1 binding sites (Fig. 3C). Pathway analysis of cluster 1 genes with novel Tcf-1 binding sites again enriched for DNA replication and DSB repair functions, while the equivalent cluster 6 genes were enriched for basic cell functions (Fig. 4G). Furthermore, Tcf-1 binding in down-regulated cluster 1 genes also lacked its own consensus binding motif, while cluster 6 genes contained Tcf7 and other similar transcription factor (TF) motifs to those in all up-regulated genes (Fig. 4H). Compared to all new Tcf-1 peaks, the peaks associated with cluster 1 genes showed an increased bias for PR sites (76% versus 57%) (Fig. 4I) while the peaks associated with cluster 6 genes showed an increased bias for AE sites (37% versus 18%) (Fig. 4I). Finally, representative tracks show new Tcf-1 binding sites and the corresponding H3K27Ac and H3K4me3 profiles in promoters of important genome maintenance genes Lig3, Pcna, and Rfc3 (Fig. 4I) among cluster 1 genes that acquired new Tcf-1 sites in CAT. Altogether these findings provide a mechanistic explanation for the down-regulation of HR repair and cell cycle genes in CAT thymocytes and directly link these expression changes to Tcf-1 binding. This outcome also explains why ablation of Tcf-1 eliminates these expression changes and highlights their direct mechanistic link to transformation.
Altered Replication Process and Response to Replication Stress in CAT Thymocytes.
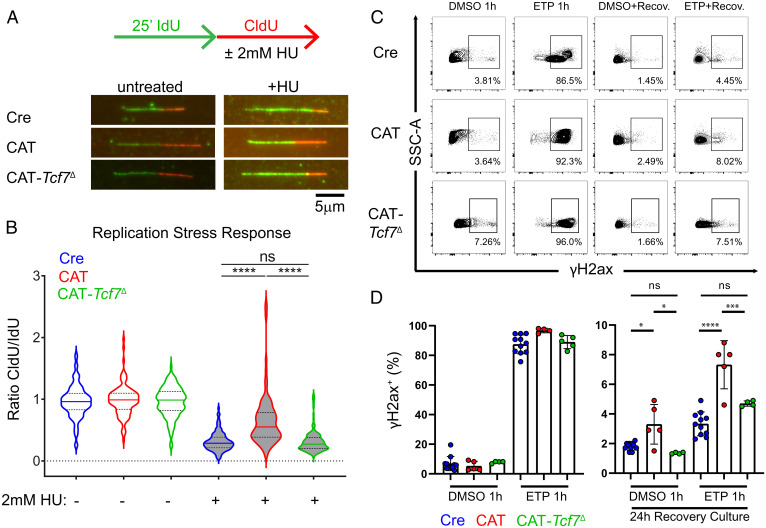
Our studies above (Fig. 1) established that the DSBs in the Myc-Pvt1 locus were Rag independent. However, the transcriptional evidence suggested that replication checkpoints may be impaired in pretransformed CAT thymocytes. Moreover, failures in replication and repair programs are increasingly appreciated as a source of genomic instability in cancer (8). Therefore, to directly test replication fork fidelity, we performed nascent DNA fiber assays in DP thymocytes cultured with the replication stressor, hydroxyurea (HU). In these experiments, newly replicated DNA is pulse labeled by two successive cocultures with nucleotide analogs 5-Iodo-2′deoxyuridine (IdU) and 5-Chloro-2′deoxyuridine (CldU). The fidelity of replication fork arrest due to replication stress checkpoints is tested through the addition of 2 mM HU during the CldU pulse (Fig. 5A). Compared to the equal IdU and CldU track lengths in untreated thymocytes, both Cre and CAT-Tcf7Δ DPs exhibit shortening of CldU tracks during HU coculture conditions (Fig. 5B). CAT thymocytes, however, continue to replicate under HU stress, leading to significantly higher ratios of CldU to IdU track lengths, suggesting that replication fork arrest is impaired.
Fig. 5.
Response to replication stress is impaired in pretransformed CAT thymocytes. (A) Schematic of assay and representative nascent DNA fibers from Cre, CAT, and CAT-Tcf7Δ CD4+CD8+ (DP) thymocytes cultured in successive 25-min pulses of media containing nucleotide analog IdU (green) followed by CldU (red) ± 2 mM HU during the CldU pulse. (B) Ratios of CldU:IdU tract lengths from untreated (vehicle) and HU-treated thymocytes in the replication stress assay. Data are representative of three biological replicates per condition in two independent experiments (n > 100 fibers/condition); results depicted as violin plots with median (solid) and quartile (dotted) lines, with statistics from two-sided, unpaired t tests; ****P ≤ 0.0001. (C) Representative flow cytometric contour plots of intracellular staining for DNA damage marker γH2ax in live DP thymocytes cultured ex vivo for 1 h in 25 mM etoposide (ETP) or vehicle control (Dimethylsulfoxide (DMSO)), followed by 24-h recovery culture in fresh media. (D) Cumulative analysis of Cre (n = 11), CAT (n = 5), and CAT-Tcf7Δ (n = 4) DP cultures showing the percent of live, γH2ax+ cells from two independent experiments in the indicated conditions. Data are represented as the mean ± SEM and statistical testing depicted as two-sided, unpaired t tests; ns > 0.05, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
We then tested whether CAT thymocytes were able to repair DNA damage generated by etoposide (ETP), which freezes topoisomerase II after generating DSBs and prevents religation, with the strongest effects on early replicating genomic regions (8). After a brief 1-h culture in 25 mM ETP, nearly all thymocytes were positive for the early DSB marker, phosphorylated H2ax (γH2ax), suggesting that the detection of DNA breaks and sensitivity to DNA damage induction was normal in CAT thymocytes. However, after ETP washout and 24 h of recovery in fresh media, CAT thymocyte cultures retained significantly more γH2ax+ cells (Fig. 5 C and D). This finding suggests that CAT thymocytes are either less proficient at repairing DSBs and/or they fail to initiate apoptosis to eliminate unrepaired cells. Taken together these findings support a model wherein CAT thymocytes fail to recognize replication stress and allow unrepaired DSBs to escape cell cycle checkpoints that typically restrict replication-mediated damage to the error-free, HR repair in S phase. This would permit Myc-Pvt1 breaks due to impaired replication and/or repair programs to coexist with Rag-mediated breaks in Tcra during G1, providing the conditions for error-prone DSB repair by NHEJ resulting in chromosomal translocations.
β-Catenin Induced Leukemias Are Sensitive to Parp Inhibitors.
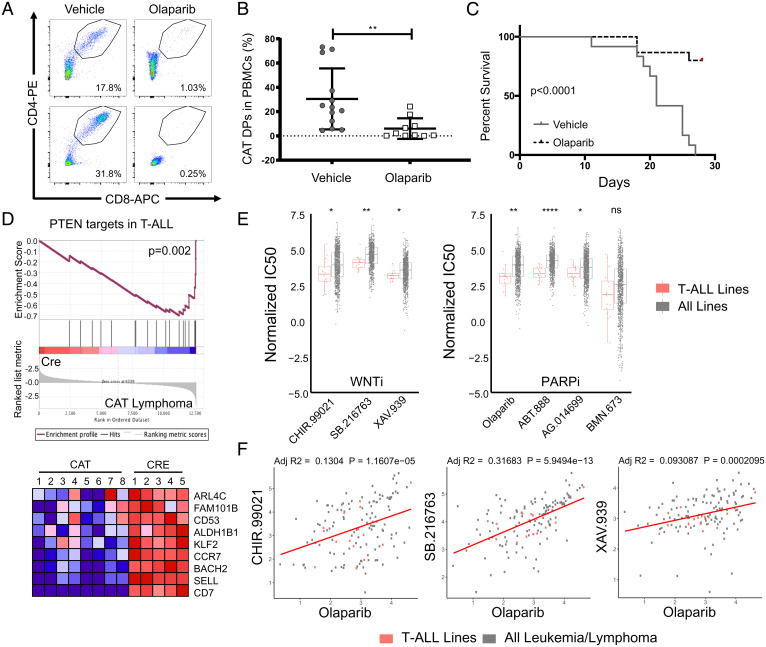
We then asked whether the β-catenin and Tcf1-mediated down-regulation of HR DNA repair genes might also render the resulting lymphomas sensitive to PARPis. Olaparib is a PARPi that was first approved for use in cancer patients that have lost HR activity (most commonly loss of BRCA1/2) due to its synthetic lethal targeting of cancer cells that are now overly dependent on backup DNA repair pathways that require Parp1 (43). To test this hypothesis, we transferred CAT lymphomas into the periphery of Rag−/− mice and treated lymphoma recipients with a 3-wk course of olaparib (50 mg/kg daily for 5 d) or vehicle control. Excitingly, tracking of CD4+CD8+ DPs in peripheral blood showed that olaparib treatment controlled the expansion of CAT lymphomas and prolonged survival of transplant recipients (Fig. 6 A–C).
Fig. 6.
CAT lymphomas and T-ALL cell lines are sensitive to Parp inhibitors. (A) Representative flow cytometry pseudocolor dot plots and (B) cumulative quantification of engraftment flow of live, CD4+CD8+ DPs in peripheral blood (PB) of Rag−/− mice (n = 10/donor) that received CAT lymphomas (n = 3). PB was sampled 2-wk posttransfer during treatment course, with recipients split evenly between olaparib (50 mg/kg/day) or vehicle controls. Data are represented as the mean ± SEM and statistical testing is depicted as two-sided, unpaired t tests; **P ≤ 0.01. (C) Kaplan–Meier survival curve of CAT lymphoma recipients in the indicated conditions. (D) GSEA comparing PTEN targets in T-ALL (44) to RNA expression profile derived from five CAT lymphoma samples. Heat map shows genes at the leading edge of the GSEA. (E) Normalized IC50 data from the GDSC database comparing T-ALL cell line (n = 16) sensitivity to all cancer cell lines (n = 974) for inhibitors targeting Wnt (WNTi) or Parp (PARPi), as indicated. Data are represented as the mean ± SEM, and statistical testing is depicted as two-sided, unpaired t tests; *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. (F) Correlations of olaparib sensitivity (IC50) to WNTi compounds in all leukemia and lymphoma cell lines (n = 150) with T-ALL lines highlighted (n = 16, red). Linear regressions analysis was performed in R, assuming Gaussian distribution and correlations represented as adjusted R-squared values (adj R2).
To directly connect these findings to human leukemia, we next investigated whether PARPi sensitivity might be applicable more broadly in human T-ALL patients. Loss of PTEN, the second most frequent mutation (20%) in T-ALL patients (44), activates AKT and disrupts the GSK3b/CK1 destruction complex, leading to stabilization of β-catenin. Furthermore, elevated β-catenin has been reported in patients with mutations activating NOTCH (>50%), making β-catenin a common feature of T-ALL pathology (45). We therefore compared the transcriptional profiles derived from five CAT lymphomas to PTEN target genes identified in human T-ALL using GSEA (44). Like in PTEN-deficient T-ALL, these genes were also down-regulated in CAT lymphomas, supporting the relevance to these human tumors (Fig. 6D). Furthermore, we extracted drug sensitivity data from the Genomics of Drug Sensitivity in Cancer (GDSC) database, which contains normalized half maximal inhibitory concentration (IC50) data on hundreds of compounds in more than 900 cancer cell lines. In comparison to all lines, T-ALL samples were found to be more sensitive to three separate WNT inhibitors (CHIR-99021, SB-216763, and XAV-939) as well as four compounds targeting AKT, which includes both NOTCH1 and PTEN mutant lines (Fig. 6E and SI Appendix, Fig S5A). Importantly, T-ALL lines were also more sensitive to 3/4 PARPi compounds in this database (olaparib, ABT-88 [veliparib], and AG-014699 [rucaparib]) (Fig. 6E). Analysis of all leukemia and lymphoma cell lines found similar sensitivity profiles (SI Appendix, Fig S5B), suggesting relevance to other hematologic malignancies. Moreover, the degree of drug sensitivity to PARP targeting compounds correlated with that of WNT targeted therapies (Fig. 6F). Taken together, these findings support a paradigm where stabilization of β-catenin signaling leads to dysregulated HR in T-ALL, opening a therapeutic opportunity for drugs that leverage synthetic lethality approaches in a patient population with urgent needs for targeted therapies.
Discussion
Chromosomal translocations involving the immunoglobulin (Ig) loci in B cells and the Tcr loci in T cells have been frequently reported in leukemia and lymphoma patients (46). A common partner to Ig and Tcr interchromosomal fusions is the Myc-Pvt1 locus whose abnormal expression has been causally implicated in a variety of cancer types (46, 47). Here we provide a mechanistic explanation for a recurrent, spontaneous Tcra/Myc-Pvt1 translocation promoted by aberrantly stabilized β-catenin in late thymocyte development. We demonstrate that while DSBs at the Tcra breakpoint site of the translocation are generated by Rag recombinase, DSBs at the Myc-Pvt1 site result from Tcf-1 controlled impairment of the HR repair and cell cycle checkpoint processes. Specifically, stabilized β-catenin redirects Tcf-1 binding to novel sites associated with genes involved in replication-associated DNA damage resulting in their down-regulation. We demonstrate that this HR repair impairment implicated in the etiology of the translocations opens an exciting therapeutic opportunity for PARP inhibitors, with potential for use more widely in Wnt/β-catenin stabilized leukemias and lymphomas.
Tcf-1, a member of the Tcf/Lef family of transcriptional regulators, plays critical roles at multiple stages of T cell development and function, including the DP stage investigated here (30, 48, 49). Tcf-1 has been shown to have a context-dependent tumor suppressor function, since its DP thymocyte deletion did not result in transformation, while its germline or early thymocyte deletion caused malignant transformation (50), (51). Classically, Tcf-1 can both negatively regulate genes when in complex with factors like Groucho, while also positively regulating Wnt-target genes, acting as the canonical DNA-binding partner of β-catenin (29). However, the functions of Tcf-1 are highly context dependent and it can mediate transcriptional repression even in complex with β-catenin (52, 53). Accordingly, here we show widespread and bidirectional alterations in Tcf-1–dependent transcriptional programs when β-catenin is aberrantly stabilized. We show that β-catenin can partially redirect Tcf-1 from its normal role and distribution on chromatin in DP thymocytes. Interestingly, genes in cluster 6 that are directly up-regulated through novel Tcf-1 binding in CAT thymocytes, and their expression is restored upon Tcf-1 loss, contain its consensus motif and show a relative preference for AE regions. By contrast, the down-regulated genes in cluster 1 exhibit new Tcf-1 binding at alternative TF motifs with a preference for PR regions. This is in line with our previous observations in Cre DP thymocytes, indicating that binding of Tcf-1 to its conserved motif in AE sites favors gene up-regulation, while binding to PR sites lacking its conserved motif favors gene down-regulation (33). This may help explain how Tcf-1 can simultaneously coordinate different outcomes on gene regulation, which has been invoked by us and others studying Tcf-1 and its different regulatory outcomes in T cells (30, 33). At present, our analysis cannot define precise rules for secondary factors. Future studies will need to address TF-complex formation and coordination with Tcf-1 throughout thymocyte development.
Tcf-1 has been appreciated as an epigenetic regulator and has been predominately associated with enhancing chromatin accessibility (35). Work from the Gounari laboratory and others has confirmed widespread alterations in chromatin accessibility mediated by Tcf-1 in both DP thymocytes and T regulatory cells (33, 54, 55). Accumulating evidence suggests that Tcf-1 acts in various molecular complexes with other transcriptional and epigenetic regulators to alter expression programs by modifying the chromatin landscape. More recently Tcf-1 has also been found to have intrinsic HDAC activity (56). Here we show that stabilized β-catenin directs Tcf-1 to novel binding sites and significantly enhances active chromatin marks at both existing and novel Tcf-1 binding sites. This fits within the general paradigm of Tcf-1 acting on top of a previously existent T cell chromatin state to control stage-specific programs by fine tuning the chromatin landscape during T cell development and activation (33, 35). In agreement with this idea, novel Tcf-1 binding was frequently found proximal to previously existing Tcf-1 binding sites in Cre thymocytes (Fig. 4G), which could be evidence of alterations in regulatory complexes at these loci. Thus, deregulated activation of β-catenin in DP thymocytes engages Tcf-1 to predominately enhance active chromatin and promote aberrant gene expression programs.
Our present findings highlight a Tcf-1–controlled cluster of down-regulated genes that is significantly enriched in genome maintenance pathways, which we implicate in DSB retention at the Myc-Pvt1 locus. Importantly, this includes core regulators of homologous recombination, such as Brca1 and Brca2, indicating that an impaired error-free DSB repair underlies the translocation formation, as has been shown in other translocation studies (57, 58). Furthermore, pathway analysis strongly implicated altered replication biology, which can also lead to DSBs through a variety of mechanisms. These include replication fork errors or underreplicated DNA leading to chromosome compaction and segregation errors that produce DSBs and translocations when the mitotic G2 checkpoint arrest fails to facilitate repair. Indeed, prior work in APC-mutant colon cancers has shown that overactive Wnt/β-catenin signaling contributes to anaphase bridges that lead to high levels of genomic instability in these tumors (59). Here we provide functional evidence that CAT thymocytes are less efficient at resolving DSBs generated by the replication poison etoposide and are also insensitive to HU stress in nascent fiber assays, which can lead to underreplicated DNA regions that are a danger to genomic stability. Taken together, we suggest a model wherein Tcf-1–mediated down-regulation of the HR and repair machinery produces replication failures that lead to DSBs, which are also not properly recognized for repair and fail to initiate S/G2 arrest. This would allow the coexistence of breaks in the Myc-Pvt1 locus that traverse the cell cycle and are aberrantly joined to rearranging receptor loci in the following G1, placing Myc under the control of highly active regulatory elements.
The present mechanistic study of Tcra/Myc-Pvt1 fusion conditions also uncovered an HR deficiency that renders the subsequent lymphomas sensitive to the Parp inhibitor, olaparib. Human T-ALL patients, particularly adults and pediatric cases after relapse, have poor outcomes and few therapeutic options (6). Although mutations in β-catenin are not directly reported in T-ALL, stabilization and elevated Wnt signaling is seen broadly, including pediatric cases, NOTCH-driven transformation, PTEN loss with or without activating NOTCH, maintenance of leukemic stem cells, as well as several subtypes of adult differentiated T-ALL (17–21). Excitingly, we highlight the potential for a wider application of PARPis in T-ALL due to β-catenin–associated HR deficiency as nearly all T-ALL cell lines exhibit sensitivity compared to other cancer lines. Importantly, sensitivity to WNT pathway inhibitors correlated with PARPi sensitivity within leukemia and lymphoma lines. This finding suggests T-ALL patients may see significant benefits with the addition of PARPis to therapeutic regimens. Future work should address whether β-catenin stabilization can also lead to dysregulation of homologous recombination in non-T cell cancers, particularly B cell leukemias harboring Ig-Myc translocations, and with the potential for use as a biomarker to predict PARPi sensitivity.
Materials and Methods
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods. Mice were maintained on a C57BL/6 background and housed at the University of Chicago animal facility in accordance with protocol #71880, approved by the University of Chicago Institutional Animal Care and Use Committee. Thymi were resected from 6- to 8-wk-old mice and lymphoma samples from 2- to 3-mo-old Cd4-Cre/Ctnnb1ex3fl mice with enlarged thymi. Thymocytes were surface stained for flow cytometry. Data were acquired on an LSRII flow cytometer (Becton Dickinson) and analyzed with FlowJo software (Becton Dickinson). For identifying translocation breakpoints, libraries prepared from four independent CAT leukemia samples were sequenced using HiSeq2500 and processed. Precise breakpoint sequences were determined with Sanger sequencing (SI Appendix, Table S1). Chromatin immunoprecipitation and sequencing, RNA-seq, and analysis of the data are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the University of Chicago Cytometry and Antibody Technology Facility for their assistance with all flow cytometry experiments, RRID: SCR_017760. We also thank the University of Chicago Animal Resources Center, RRID:SCR_021806. This work was supported by NIH Grant AI147652-01A1 (to F.G.). S.A. was supported by T32 CA009594. P.M. was supported by T32 HL07605 and a Leukemia and Lymphoma Society fellowship. J.Q. was supported by an American Association of Immunologists Careers in Immunology fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201493119/-/DCSupplemental.
Data Availability
Raw data and associated analysis files for RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus data repository (ID GSE205453). All other study data are included in the article and/or supporting information. Previously published data sets used for comparative analysis are detailed in SI Appendix, Materials and Methods and are publicly available under IDs GSE46662, GSE32311, and SRP142342.).
References
- 1.Lindahl T., Barnes D. E., Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 65, 127–133 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Vermeer M. H., et al. , Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res. 68, 2689–2698 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Graux C., Cools J., Michaux L., Vandenberghe P., Hagemeijer A., Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: From thymocyte to lymphoblast. Leukemia 20, 1496–1510 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P., Pieters R., Beverloo H. B., Meijerink J. P., Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 143, 153–168 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Teachey D. T., O’Connor D., How I treat newly diagnosed T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma in children. Blood 135, 159–166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samra B., Jabbour E., Ravandi F., Kantarjian H., Short N. J., Evolving therapy of adult acute lymphoblastic leukemia: State-of-the-art treatment and future directions. J. Hematol. Oncol. 13, 70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belver L., Ferrando A., The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 16, 494–507 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tubbs A., et al. , Dual roles of poly(dA:dT) tracts in replication initiation and fork collapse. Cell 174, 1127–1142.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fugmann S. D., Lee A. I., Shockett P. E., Villey I. J., Schatz D. G., The RAG proteins and V(D)J recombination: Complexes, ends, and transposition. Annu. Rev. Immunol. 18, 495–527 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Lieber M. R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jasin M., Rothstein R., Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love P. E., Bhandoola A., Signal integration and crosstalk during thymocyte migration and emigration. Nat. Rev. Immunol. 11, 469–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg E. V., Programming for T-lymphocyte fates: Modularity and mechanisms. Genes Dev. 33, 1117–1135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kastan M. B., Bartek J., Cell-cycle checkpoints and cancer. Nature 432, 316–323 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Larmonie N. S., et al. , Breakpoint sites disclose the role of the V(D)J recombination machinery in the formation of T-cell receptor (TCR) and non-TCR associated aberrations in T-cell acute lymphoblastic leukemia. Haematologica 98, 1173–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lento W., Congdon K., Voermans C., Kritzik M., Reya T., Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb. Perspect. Biol. 5, a008011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groen R. W., et al. , Illegitimate WNT pathway activation by beta-catenin mutation or autocrine stimulation in T-cell malignancies. Cancer Res. 68, 6969–6977 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Bellei B., Cota C., Amantea A., Muscardin L., Picardo M., Association of p53 Arg72Pro polymorphism and beta-catenin accumulation in mycosis fungoides. Br. J. Dermatol. 155, 1223–1229 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Ram-Wolff C., Martin-Garcia N., Bensussan A., Bagot M., Ortonne N., Histopathologic diagnosis of lymphomatous versus inflammatory erythroderma: A morphologic and phenotypic study on 47 skin biopsies. Am. J. Dermatopathol. 32, 755–763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng O. H., et al. , Deregulated WNT signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J. 4, e192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gekas C., et al. , β-Catenin is required for T-cell leukemia initiation and MYC transcription downstream of Notch1. Leukemia 30, 2002–2010 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Gutierrez A., et al. , High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood 114, 647–650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z., et al. , Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood 109, 5463–5472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dose M., et al. , β-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl. Acad. Sci. U.S.A. 111, 391–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaveri D., et al. , β-Catenin activation synergizes with Pten loss and Myc overexpression in Notch-independent T-ALL. Blood 122, 694–704 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Guo W., et al. , Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature 453, 529–533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timakhov R. A., et al. , Recurrent chromosomal rearrangements implicate oncogenes contributing to T-cell lymphomagenesis in Lck-MyrAkt2 transgenic mice. Genes Chromosomes Cancer 48, 786–794 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosimann C., Hausmann G., Basler K., Beta-catenin hits chromatin: Regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10, 276–286 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Zhao X., Shan Q., Xue H. H., TCF1 in T cell immunity: A broadened frontier. Nat. Rev. Immunol. 22, 147–157 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Germar K., et al. , T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 108, 20060–20065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber B. N., et al. , A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emmanuel A. O., et al. , TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4+CD8+ thymocytes. Nat. Immunol. 19, 1366–1378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinke F. C., et al. , TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 15, 646–656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson J. L., et al. , Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48, 243–257.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada N., et al. , Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931–5942 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng G., et al. , RAG represents a widespread threat to the lymphocyte genome. Cell 162, 751–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merelli I., et al. , RSSsite: A reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 38, W262–W267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bredemeyer A. L., et al. , ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 442, 466–470 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Ma A., et al. , Bclx regulates the survival of double-positive thymocytes. Proc. Natl. Acad. Sci. U.S.A. 92, 4763–4767 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walton K. D., et al. , Conditional deletion of the bcl-x gene from mouse mammary epithelium results in accelerated apoptosis during involution but does not compromise cell function during lactation. Mech. Dev. 109, 281–293 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lord C. J., Ashworth A., PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palomero T., et al. , Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13, 1203–1210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girardi T., Vicente C., Cools J., De Keersmaecker K., The genetics and molecular biology of T-ALL. Blood 129, 1113–1123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieber M. R., Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer 16, 387–398 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng Y. Y., et al. , PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mielke L. A., et al. , TCF-1 limits the formation of Tc17 cells via repression of the MAF-RORγt axis. J. Exp. Med. 216, 1682–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escobar G., Mangani D., Anderson A. C., T cell factor 1: A master regulator of the T cell response in disease. Sci. Immunol. 5, eabb9726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu S., et al. , The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F., et al. , Exploring the stage-specific roles of Tcf-1 in T cell development and malignancy at single-cell resolution. Cell. Mol. Immunol. 18, 644–659 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arce L., Yokoyama N. N., Waterman M. L., Diversity of LEF/TCF action in development and disease. Oncogene 25, 7492–7504 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Yu S., Xue H. H., TCF-1 mediates repression of Notch pathway in T lineage-committed early thymocytes. Blood 121, 4008–4009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Veeken J., et al. , The transcription factor Foxp3 shapes regulatory T cell identity by tuning the activity of trans-acting intermediaries. Immunity 53, 971–984.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quandt J., et al. , Wnt-β-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression. Nat. Immunol. 22, 471–484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing S., et al. , Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 17, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scully R., Livingston D. M., In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408, 429–432 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vasanthakumar A., et al. , Brca1 deficiency causes bone marrow failure and spontaneous hematologic malignancies in mice. Blood 127, 310–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aoki K., et al. , Chromosomal instability by beta-catenin/TCF transcription in APC or beta-catenin mutant cells. Oncogene 26, 3511–3520 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and associated analysis files for RNA-seq and ChIP-seq data have been deposited in the Gene Expression Omnibus data repository (ID GSE205453). All other study data are included in the article and/or supporting information. Previously published data sets used for comparative analysis are detailed in SI Appendix, Materials and Methods and are publicly available under IDs GSE46662, GSE32311, and SRP142342.).