Significance
Epstein-Barr virus (EBV) accounts for 200,000 new epithelial and B cell malignancy cases and 140,000 deaths annually. Glycoprotein B (gB) is the sole fusogen that is highly conserved and essential for all herpesvirus entry into target cells and thus, is attracting attention to identify potent antibodies to neutralize viral infection. Here, we discovered two anti-EBV gB neutralizing antibodies, 3A3 and 3A5, that effectively neutralized EBV infection of both B and epithelial cells. They also potently protected against EBV-induced lymphoproliferative disorders in humanized mice. Importantly, the 3A3 and 3A5 epitopes identified here represent the neutralizing antigenic sites to block EBV infection and membrane fusion. They are major targets of protective gB-specific neutralizing antibodies elicited by natural EBV infection in humans.
Keywords: Epstein–Barr virus, glycoprotein B, neutralizing antibody, viral membrane fusion, lymphoproliferative disorder
Abstract
Epstein-Barr virus (EBV) infects more than 90% of the world’s adult population and accounts for a significant cancer burden of epithelial and B cell origins. Glycoprotein B (gB) is the primary fusogen essential for EBV entry into host cells. Here, we isolated two EBV gB-specific neutralizing antibodies, 3A3 and 3A5; both effectively neutralized the dual-tropic EBV infection of B and epithelial cells. In humanized mice, both antibodies showed effective protection from EBV-induced lymphoproliferative disorders. Cryoelectron microscopy analyses identified that 3A3 and 3A5 bind to nonoverlapping sites on domains D-II and D-IV, respectively. Structure-based mutagenesis revealed that 3A3 and 3A5 inhibit membrane fusion through different mechanisms involving the interference with gB-cell interaction and gB activation. Importantly, the 3A3 and 3A5 epitopes are major targets of protective gB-specific neutralizing antibodies elicited by natural EBV infection in humans, providing potential targets for antiviral therapies and vaccines.
Epstein-Barr virus (EBV), a human γ-herpesvirus, establishes persistent infection in ∼95% of adults worldwide (1). EBV is a causative agent for infectious mononucleosis and is closely associated with several lymphomas and epithelial malignancies, including Hodgkin’s lymphoma, Burkitt’s lymphoma, natural killer (NK)/T cell lymphoma, nasopharyngeal carcinoma (NPC), and gastric carcinoma (2). Moreover, EBV reactivation sometimes causes fatal lymphoproliferative disorders (LPDs) in immunocompromised patients with AIDS or posttransplantation (3). However, there are no specific therapeutics or effective vaccines against EBV infection, largely due to the lack of knowledge regarding the immunity to EBV infection and related diseases (2). EBV-related diseases pinpoint two predominant host cells targeted by a viral infection, B cells and epithelial cells (4). EBV encodes multiple envelope glycoproteins that determine cell tropism and accomplish host cell attachment and entry (5). Glycoprotein B (gB), functionally conserved throughout all herpesviruses, is the fusogen protein essential for viral entry into both B cells and epithelial cells (6).
EBV gB forms the core membrane fusion machinery with the glycoprotein H and glycoprotein L heterodimer (gHgL) and drives membrane fusion through conformational change. The activation of gB is triggered stepwise upon gHgL binding to a receptor on the target cell surface (7). The gHgL heterodimer binds to the receptor to trigger epithelial cell entry or human leukocyte antigen class II by forming a complex with glycoprotein gp42 for B cell entry (8–11). Previous studies have shown that the gHgL D-I and D-I/D-II linker regions are involved in gB binding and activation (12, 13). Furthermore, EBV gB interacts directly with neuropilin 1 (NRP1) to promote the EBV infection of epithelial cells through NRP1-facilitated internalization and fusion (14). Despite these advances, the molecular mechanisms and critical domains underlying gB activation remain largely unclear.
As the core components of the fusion machinery, gHgL and gB have drawn increasing attention as targets of potent neutralizing antibodies against EBV infection of both B cells and epithelial cells. Among the anti-gHgL antibodies, E1D1, CL59, and CL40 block epithelial cell infection but not B cell infection (15, 16). Two human anti-gHgL neutralizing antibodies, AMMO1 and 1D8, show dual-tropic inhibition of both B cell and epithelial cell infection (17, 18). AMMO1 protects against EBV challenges in humanized mice and a homologous rhesus lymphocryptovirus in the rhesus monkey (19). It is supposed that AMMO1 likely interferes with gB activation and inhibits membrane fusion. However, for the EBV fusogen protein gB, two antibodies, CL55 and AMMO5, were isolated. CL55, derived from mouse hybridoma, shows no neutralizing activity, while AMMO5, a human antibody, only inhibits epithelial cell infection but not B cell infection (17, 20). Currently, no structural information is available for either anti-gB monoclonal antibody (mAb).
To better characterize the neutralizing antibodies targeting EBV gB, rabbits were immunized with EBVgB to develop anti-EBV gB mAbs. The uniqueness of rabbit antibody repertoires affords an efficient screening of mAbs with high diversity, affinity, and specificity (21). We successfully isolated two gB-specific antibodies, 3A3 and 3A5, that neutralized EBV infection of both B cells and epithelial cells and conferred potent protective activities against EBV-induced LPD in a humanized mouse model. Using cryoelectron microscopy (cryo-EM), we determined that 3A3 and 3A5 recognized two epitopes located at gB D-II and D-IV, respectively. We demonstrated that both epitopes are critical for gB-mediated membrane fusion through different mechanisms. Furthermore, 3A3 and 3A5 epitopes represent the major targets of anti-gB neutralizing antibodies elicited during natural EBV infection. The dual-tropic inhibition and in vivo protection highlight that gB D-II and D-IV are promising targets for anti-EBV therapies and vaccines.
Results
3A3 and 3A5 Specifically Bind to Recombinant and Native gB.
The ectodomain of glycoprotein B (ect-gB) was expressed in 293F cells and purified. The trimeric form of ect-gB was confirmed by native polyacrylamide gel electrophoresis (PAGE), and the monomeric form was confirmed by reducing PAGE (SI Appendix, Fig. S1A). When treated with sodium dodecyl sulfate (SDS) and β-mercaptoethanol, the monomer was further disrupted into two bands of appropriate size corresponding to the two fragments cleaved by furin protease during expression (SI Appendix, Fig. S1A). gB-specific memory B cells were isolated from rabbits immunized with ect-gB by the procedure shown in SI Appendix, Fig. S1B to obtain anti-EBV-gB mAbs.
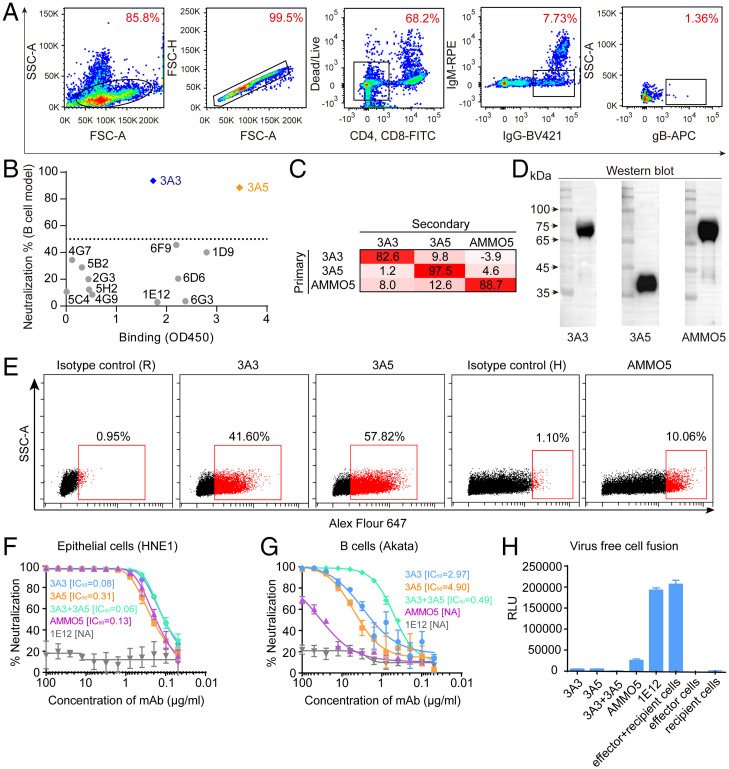
Allophycocyanin-conjugated ect-gB was used as bait to screen gB-specific B cells (Fig. 1A). The sorted B cells were cultured in vitro, and the supernatants were tested by enzyme-linked immunosorbent assay (ELISA) for their reactivities with ect-gB. Then, the gene sequences encoding the paired variable heavy chain (VH) and variable light chain (VL) for 13 gB-specific mAbs were successfully cloned into a recombinant expression vector (SI Appendix, Table S1). Among the 13 mAbs, 3A3 and 3A5 were further verified as neutralizing antibodies using a B cell-based neutralizing assay in vitro and a gB-based ELISA (Fig. 1B and SI Appendix, Fig. S2).
Fig. 1.
Isolation and evaluation of the binding and neutralizing abilities of 3A3 and 3A5. (A) FACS-based staining and gating strategy to sort gB-specific B cells from peripheral blood mononuclear cells derived from a gB-immunized rabbit. (B) Binding and neutralizing activities of 13 mAbs cloned from B cells sorted using the approach in A. The binding activities (x axis; optical density at 450 nm (OD450)) with the recombinant ect-gB were evaluated by ELISA. The neutralizing activities (y axis; percentage neutralization) of 13 mAbs were evaluated by the B cell infection model. The neutralization percentage indicates the neutralizing potency of mAbs blocking the EBV infection of Akata B cells. The formula is neutralization percentage = 1 − the percentage of infected cells with serum or mAb/the percentage of infected cells without serum or mAb × 100%. (C) Cross-competition of mAbs 3A3, 3A5, and AMMO5 was measured by competitive ELISA. The inhibition of the binding of secondary antibody to gB by the primary antibody is shown. In the competitive ELISA, immobilized recombinant gB was presaturated with the primary antibodies, and then, the binding of secondary antibodies, 3A3-HRP (horseradish peroxidase), 3A5-HRP, and AMMO5-HRP, was measured. (D) WB analysis of 3A3, 3A5, and AMMO5 for their activities to ect-gB under reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). 3A3 and AMMO5 bound the ∼70-kDa fragment, and 3A5 bound the ∼40-kDa fragment. (E) Detection of 3A3 or 3A5 binding to total gB of EBV-positive Akata B cells postinduction of the viral lytic cycle using flow cytometry. AMMO5 was used as a control. Flow cytometry staining was performed with cell membrane permeabilization. (F and G) Neutralization of Akata-EBV infection of HNE1 epithelial cells (F) and CNE2-EBV infection of Akata B cells (G) by serial dilutions of anti-gB antibodies, 3A3, 3A5, the combination of 3A3 + 3A5, AMMO5, and 1E12 (nonneutralizing antibody, negative control). Half maximal inhibitory concentration (IC50) values were calculated by sigmoid trend fitting. Data points are shown as the mean of two replicates ± SEM. (H) Blockade of cell-cell fusion by anti-gB antibodies. The nonneutralizing anti-gB antibody 1E12 was used as a negative control. HEK-293T effector cells were transfected with expression plasmids encoding gB, gHgL, and T7 polymerase. HEK-293T recipient cells were transfected with the pT7EMCLuc plasmid expressing luciferase under the control of T7 polymerase. Effector and recipient cells were mixed in the presence or absence of antibodies, and luciferase activity was measured. Data points are shown as the mean of two independent replicates ± SEM. APC, allophycocyanin. SSC-A, side scatter area. FSC-A, forward scatter area. RPE, R.Phycoerythrin. FITC, fluorescein Isothiocyanate. NA, not available. RLU, relative light unit.
We then used a gB-based competitive ELISA to analyze the epitope competition of 3A3, 3A5, and a previously reported human anti-gB mAb, AMMO5, that blocked epithelial cell infection but not B cell infection (17). The results showed that the binding sites of these three mAbs did not overlap with each other (Fig. 1C). Western blot (WB) analysis also confirmed that these two neutralizing mAbs recognized different gB fragments (Fig. 1D).
3A3 and 3A5 showed similar binding activities to ect-gB, with mean half-maximal effective concentrations of 7.0 and 5.2 ng/mL, respectively, which were lower than that of AMMO5 (14.3 ng/mL) (SI Appendix, Fig. S3A). The affinities of antigen binding fragments of 3A3 (3A3Fab) and 3A5Fab against gB were further confirmed with a surface plasmon resonance (SPR) assay, where two antibodies demonstrated nanomolar binding affinities (∼3.49 nM for 3A3 and ∼3.93 nM for 3A5 vs. ∼21.15 nM for AMMO5) (SI Appendix, Fig. S3B).
Next, we investigated the binding activities of the three mAbs to endogenously expressed gB by immunofluorescence assay. All three mAbs specifically recognized native gB expressed in COS7 cells (SI Appendix, Fig. S3C). The binding activities of 3A3, 3A5, and AMMO5 with the virion-encoded gB expressed on EBV-positive Akata cells were confirmed by flow cytometry assay (Fig. 1E). Notably, 3A3 and 3A5 bound to a higher proportion of Akata cells induced for EBV lytic production than AMMO5 (Fig. 1E). These results demonstrated that mAbs 3A3 and 3A5 efficiently bind to recombinant and native gB.
3A3 and 3A5 Effectively Inhibit EBV Infection of Both Epithelial Cells and B Cells.
We then evaluated the neutralizing abilities of 3A3 and 3A5 against EBV infection of epithelial cells (Fig. 1F) and B cells (Fig. 1G), respectively. For comparison, AMMO5 and a nonneutralizing anti-gB mAb 1E12 were also included. In the epithelial cell infection assay, 3A3 and 3A5 showed potent neutralizing potencies with half maximal inhibitory concentration (IC50) values of 0.08 and 0.31 μg/mL, respectively, comparable with AMMO5 of 0.13 μg/mL (Fig. 1F). However, in the B cell infection assay, 3A3 and 3A5 showed at least 10-fold higher neutralizing activities than AMMO5, with IC50 values of 2.97 and 4.90 μg/mL, respectively (Fig. 1G).
We further explored the synergistic neutralizing potency of 3A3 and 3A5. In the epithelial cell model, the 3A3/3A5 combination (1:1 mixture) neutralized EBV infection with a lower IC50 of 0.06 μg/mL (Fig. 1F). A stronger synergistic effect was observed in the B cell infection model. The IC50 of the 3A3 and 3A5 mixture to block B cell infection was 0.49 μg/mL, 5 to 10 times lower than that of the individual neutralizing antibodies (Fig. 1G). For EBV infection of epithelial cells, 3A3 or 3A5 alone showed potent neutralizing activity, which may result in the very weak synergistic effect observed in the epithelial infection model.
In addition, we investigated the interference of 3A3 and 3A5 with membrane fusion using a virus-free epithelial cell fusion model. Specifically, the core fusion machinery, gHgL, and gB, were expressed in the effector cells to drive membrane fusion with recipient cells. Meanwhile, T7 polymerase and a luciferase reporter under the control of the T7 promoter were separately expressed in the effector and recipient cells. 3A3, 3A5, or the control antibody was incubated with effector cells before the effector and recipient cell coculture. Then, these antibodies’ inhibition of cell membrane fusion was measured with luciferase activity in cell lysates after coculture. Of note, 3A3 and 3A5 can inhibit membrane fusion by 95% compared with the nonneutralizing mAb 1E12 as the control without inhibition (Fig. 1H). AMMO5 also blocked membrane fusion by 87% (Fig. 1H). These results demonstrated that 3A3 and 3A5 potently block gB-mediated membrane fusion and inhibit virion entry into B cells and epithelial cells in vitro.
3A3 and 3A5 Confer Protection against Lethal EBV Challenges in Humanized Mice.
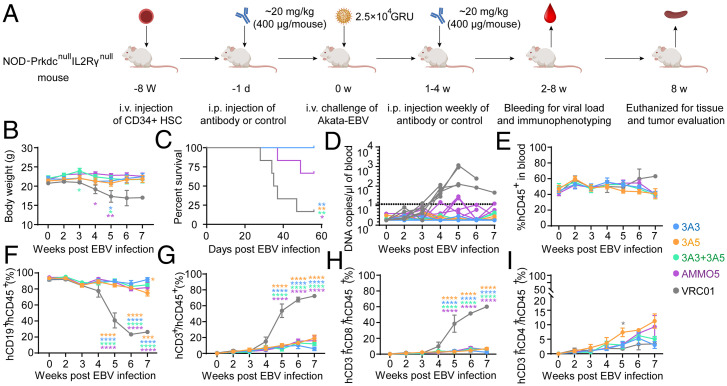
A humanized mouse model was used to test the protective potential of 3A3 and 3A5 in vivo. Humanized mice were established by engrafting CD34+ human hematopoietic stem cells obtained from umbilical cord blood into NOD-Prkdcnull IL2Rγnull (NPI) mice (22). In the humanized mouse experiments, the constant rabbit regions of 3A3 and 3A5 were replaced by the human Immunoglobulin G1 (IgG1) constant region. VRC01, a human anti-HIV-1 antibody, was used as a negative control (23). We adopted a protocol similar to the previous study (18) and chose an antibody dose comparable with the doses used in several studies exploring the mAb protection against EBV challenge in mouse models (19, 24). Briefly, the humanized mice received an intraperitoneal (i.p.) injection containing 400 μg of antibodies (∼20 mg/kg) and were challenged via intravenous (i.v.) injection by Akata-EBV equivalent to ∼25,000 green Raji units 24 h after the first dose of antibody treatment (Fig. 2A). In the following 4 wk postchallenge, all animals received the testing antibodies with the same dose of 400 μg/mouse (∼20 mg/kg) via i.p. injection. All animals were evaluated for body weight, survival, EBV viremia, and blood and tissue pathology.
Fig. 2.
3A3 and 3A5 conferred protection against lethal EBV challenge in humanized mice. (A) Experimental time line for antibody administration, EBV challenge, and monitoring for various biological and clinical outcomes. A total of 400 μg of 3A3, 3A5, 3A3 + 3A5, AMMO5, and VRC01 (negative control; n = 6 for each group) were administered to the NPI mice via i.p. injection 24 h prior to i.v. challenge with 25,000 GRU (green raji units) of Akata-EBV and followed by an additional weekly antibody treatment of the same dose for 4 wk. (B–D) Body weight (B), survival (C), and EBV DNA copies (D) in the peripheral blood of mice were monitored during the experiment. Each line in D represents an individual mouse, and the dashed line indicates the detection limit. (E-I) The percentage changes of hCD45+ (E), hCD19+ (F), hCD3+ (G), hCD8+ (H), and hCD4+ (I) cells in the peripheral blood during the experiment. Data points and error bars in B and D-I represent the mean values and SEMs of the data from the surviving mice at that time point. Data schematics for 3A3, 3A5, 3A3 + 3A5, AMMO5, and VRC01 are colored blue, yellow, green, purple, and black, respectively. Statistical analyses were performed using one-way ANOVA. The color of the asterisks denotes the group with which there is a significant difference from the VRC01 control group determined by a Sidak multiple comparison test. The yellow asterisk in F indicates a significant difference between the 3A3-treated and 3A5-treated groups determined by a Sidak multiple comparison test. *P ≤ 0.0332; **P ≤ 0.0021; ***P ≤ 0.0002; ****P ≤ 0.0001. HSC, hematopoietic stem cells. Schematic diagram was made with BioRender.com.
Compared with the anti-gB antibody-treated groups, the mice in the VRC01-treated group showed significant weight loss beginning at week 3 postchallenge, and only one of six (16.7%) mice survived by 7 wk (Fig. 2 B and C). All mice receiving 3A3, 3A5, and 3A3 + 3A5 survived after the EBV challenge, whereas four (66.7%) of the AMMO5-treated mice survived (Fig. 2C). EBV DNA increased in the peripheral blood until week 5 post-EBV challenge in the VRC01-treated mice. EBV viremia (>10 copies/μL) was observed in all six VRC01-treated mice and two AMMO5-treated mice 4 wk postchallenge (Fig. 2D). In contrast, the mice treated with 3A3, 3A5, and 3A3 + 3A5 remained aviremic (<10 copies/μL) (Fig. 2D).
All of the mice retained similar proportions (40 to 60%) of human CD45+ (hCD45+) lymphocytes in the peripheral blood during the experiments (Fig. 2E). Consistent with EBV viremia, the proportion of human B cells and T cells differed between treatment groups (Fig. 2 F and G). In the VRC01-treated control group, the percentage of hCD19+ B cells in the peripheral blood was significantly decreased from ∼90% at week 2 to ∼30% at week 7, accompanied by an increase of hCD3+ total T cells (Fig. 2 F and G). hCD8+ T cells contributed to the increase of hCD3+ total T cells in VRC01-treated mice (Fig. 2H), while only a mild increase in hCD4+ cells was observed in the peripheral blood (Fig. 2I). In contrast to the VRC01-treated control group, in the 3A3, 3A5, 3A3 + 3A5, and AMMO5-treated mice, the percentage of hCD19+ B cells was maintained at the level of 80 to 90% (Fig. 2F), and the percentage of hCD3+ and hCD8+ T cells remained at a low level of <10% during the protection experiment (Fig. 2 G and H). Altogether, these results demonstrate that 3A3 and 3A5 as well as AMMO5 control EBV infection and confer protection against EBV viremia after lethal EBV challenge in vivo.
Prevention of EBV-Induced LPD in Protected Humanized Mice.
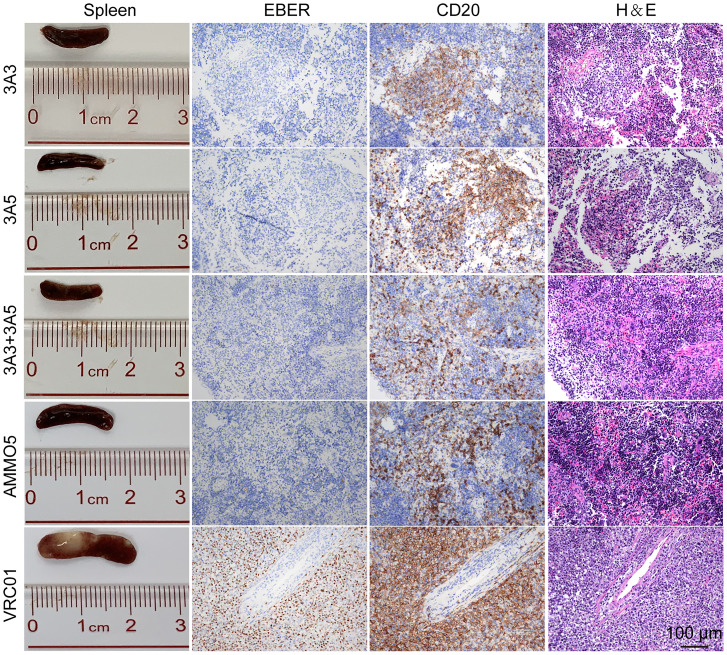
Next, we evaluated the pathological changes of EBV-induced LPD that often occur concurrently with EBV viremia at necropsy. The spleens were significantly enlarged, and tumors were visible in the VRC01-treated mice (Fig. 3 and SI Appendix, Fig. S4A). In contrast, the spleens from the mice treated with 3A3, 3A5, 3A3 + 3A5, and AMMO5 were normal in morphology, without visible tumors (Fig. 3). The in situ hybridization of Epstein-Barr virus-encoded RNAs (EBERs) and hCD20 staining confirmed the development of typical LPDs as a result of the outgrowth of EBV-infected B cells (CD20+ and EBER+) in the spleens of the VRC01-treated mice (Fig. 3). In the mice treated with 3A3, 3A5, 3A3 + 3A5, and AMMO5, although infiltration of CD20+ B cells in spleens was observed, EBV+ B cells were much fewer than in the VRC01-treated group. Consistently, mice from the VRC01-treated group also showed >1,000-fold higher levels of EBV DNA in spleens than the anti-gB antibody-treated mice (SI Appendix, Fig. S4B). The lowest level of EBV DNA was observed in the spleens of mice treated with 3A3 + 3A5 (SI Appendix, Fig. S4B).
Fig. 3.
3A3 and 3A5 protected humanized mice from EBV-induced LPDs. Representative macroscopic spleen and spleen tissue stained for H&E, EBER, and hCD20+ at necropsy. Each image is representative of the experimental group. (Scale bar: 100 μm.)
Furthermore, splenic lymphocytes were analyzed at necropsy. Different antibody-treated groups showed similar proportions of hCD45+ cells (SI Appendix, Fig. S4C). Consistent with the pattern of hCD19+ B cells in peripheral blood, the percentage of hCD19+ B cells in the VRC01-treated group was significantly lower than that in the anti-gB antibody-treated groups (SI Appendix, Fig. S4D). However, a group of highly proliferating memory B cells, the hCD19+hCD24−hCD38+ lymphocytes, showed a dramatic elevation only in the VRC01-treated group (SI Appendix, Fig. S4E). The visible tumor in spleens might be formed by these highly proliferating B cells. Concurrent with the decrease of hCD19+ B cells, significant increases in the proportions of hCD3+ (SI Appendix, Fig. S4F), hCD8+ (SI Appendix, Fig. S4H), and activated hCD137+hCD69+hCD8+ (SI Appendix, Fig. S4I) T lymphocytes were also observed in the VRC01-treated group compared with the anti-gB antibody-treated groups. However, the proportions of hCD4+ cells in the spleen remained similar for all groups (SI Appendix, Fig. S4G). The reduction of hCD19+ B cells in the VRC01-treated group might result from T cell-mediated killing of the EBV-infected human B cells (25).
Collectively, these results demonstrated that 3A3 and 3A5 significantly reduced the viral load and prevented EBV-induced LPD in humanized mice. Although less potent than two neutralizing antibodies, 3A3 and 3A5, AMMO5 also effectively prevented lethal challenges with EBV, possibly through fragment crystallizable-mediated antibody effector functions.
3A3 and 3A5 Epitopes Are Major Targets of Naturally Acquired Anti–gB Neutralizing Antibodies.
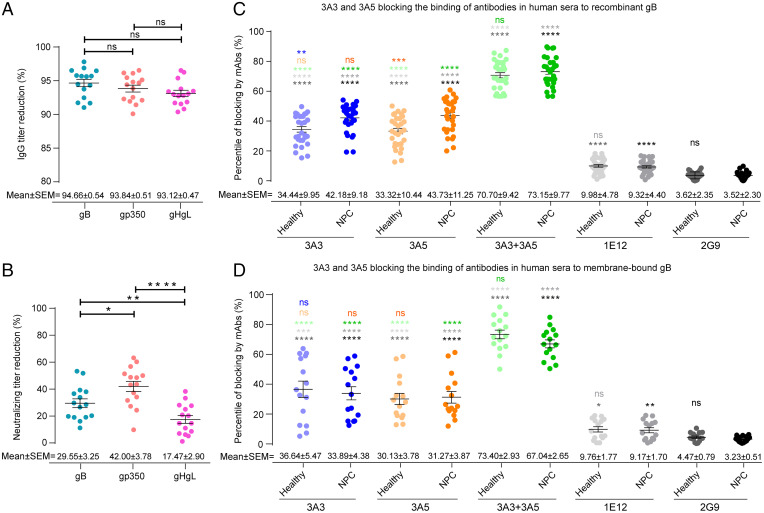
Next, we sought to determine the contribution of gB-specific antibodies to the neutralization of EBV infection and the predominance of 3A3- and 3A5-like antibodies elicited by natural EBV infection in human sera. We depleted serum antibodies against gp350, gB, and gHgL and examined the B cell neutralization efficiency before and after depletion for each serum. The 293T cell lines that separately expressed viral full-length envelope proteins, gp350, gHgL, and gB, were used to deplete antibodies against the corresponding glycoproteins in the sera from 15 healthy adult individuals who were asymptomatic EBV carriers, and the 293T cells transfected with the empty vector were used as the negative control (SI Appendix, SI Materials and Methods). After 15 rounds of depletion, the IgG titers against gB, gp350, and gHgL were decreased by more than 90% (Fig. 4A and SI Appendix, Fig. S5 A–C), whereas the depletion using 293T cells transfected with the empty vector did not reduce the IgG titers against either of the three glycoproteins (SI Appendix, Fig. S5 D–F). Using the B cell infection model, we measured the neutralizing titer of each serum before and after the depletion of over 90% of each glycoprotein-specific antibody. Compared with the undepleted sera, the neutralizing activity decreased on average by 29.55% (±3.25%, SEM), 42.00% (±3.78%, SEM), and 17.47% (±2.90%, SEM) for the gB-, gp350-, and gHgL-specific antibody depletion, respectively (Fig. 4B and SI Appendix, Fig. S5 G–I). These results indicated that, together with anti-gp350 and anti-gHgL antibodies, anti-gB antibodies elicited by natural EBV infection also play a significant role in blocking the EBV infection of B cells.
Fig. 4.
Evaluation of 3A3- or 3A5-like antibodies in human sera. (A) Reduction in glycoprotein-specific immunoglobulin G (IgG) titers after the specific depletion of antibodies against gB, gp350, or gHgL in sera from 15 healthy adult individuals was evaluated by each glycoprotein-based ELISA. The IgG titers were calculated by the end point dilution method, and OD450 = 0.1 was set as the cutoff value. The percentage of IgG titer reduction was calculated by the equation (1 − IgG titer-depleted/IgG titer-before) × 100%, where IgG titer-depleted is each glycoprotein-specific IgG titer of the serum after the specific antibody depletion and IgG titer-before is each glycoprotein-specific serum IgG titer before depletion. (B) Reduction in neutralizing ability after the specific depletion of over 90% of anti-gB, -gp350, or -gHgL IgG antibodies in sera from 15 healthy adult individuals was evaluated by the EBV infection of Akata B cells. The reduction in the neutralizing titer after the specific antibody depletion was calculated by the equation (1 − IC50-depleted/IC50-before) × 100%, where IC50-depleted is the neutralizing titer of the serum depleted by specific glycoprotein-expressed 293T cells and IC50-before is the neutralizing titer of the serum before depletion. (C) Blocking naturally acquired anti-gB antibodies by 3A3 and 3A5 in human sera from binding to recombinant gB was evaluated by competitive ELISA. Sera from 30 healthy adult individuals and 30 patients who were nasopharyngeal carcinoma (NPC) positive for EBV viral capsid antigen (VCA)-IgG, VCA-IgA, early antigen (EA)-IgA, and Epstein-Barr nuclear antigen 1 (EBNA1)-IgA were used. The anti-gB mAbs 3A3, 3A5, 3A3 + 3A5, and 1E12 and an antiinfluenza HA antibody 2G9 (negative control) were used to block the immobilized recombinant gB. The OD values of each serum were determined by ELISA before and after mAb treatment. The blocking ratio of the mAb against each serum was calculated as 1 − (OD value of the serum binding to recombinant gB treated with mAb/OD value of the serum binding to recombinant gB without mAb treatment) × 100%. (D) Blocking naturally acquired anti-gB antibodies in human sera by 3A3 and 3A5 binding to membrane-bound gB expressed by 293T cells was evaluated by flow cytometry. Sera from 15 healthy adult individuals and 15 patients with NPC, tested in C, were used in the antibody blocking assays with membrane-bound gB. The anti-gB mAbs, 3A3, 3A5, their combination, and 1E12, were incubated with gB-expressed 293T cells prior to the incubation with human sera. The anti-influenza HA mAb 2G9 was used as a negative control. After incubation with human sera, cells without membrane permeabilization were stained with the AF647 goat anti-human IgG antibody to detect serum antibody binding to the cell surface by flow cytometry. The blocking ratio of the mAb against each serum was calculated as 1 − (percentage of AF647-positive cells incubated with mAb/percentage of AF647-positive cells without mAb incubation) × 100%. Error bars represent the SEM for each experimental group. P values from the unpaired Welch’s t tests are indicated (significant difference is indicated by asterisks). ns, no significant difference. *P ≤ 0.0332; **P ≤ 0.0021; ***P ≤ 0.0002; ****P ≤ 0.0001.
Moreover, we determined that the correlations between the neutralizing titers and each glycoprotein-specific antibody titer were similar before and after each specific serum antibody depletion. Specifically, the gp350 IgG titers had a moderate and significant correlation with the neutralizing titers in the B cell infection model in both the undepleted and depleted sera, while we did not observe significant correlations between anti-gB or -gHgL IgG titers and neutralization titers in either the undepleted or depleted sera (SI Appendix, Fig. S5 J–O). Together with the results that the neutralization activity decreased most greatly for the gp350-specific antibody depletion (Fig. 4B), these findings suggest that the serum antibodies against gp350 seem to contribute more than the anti-gB or anti-gHgL antibodies to the neutralization of in vitro EBV infection of B cells.
Moreover, to determine whether the 3A3 and 3A5 epitopes represent the major targets of the gB-specific antibody response elicited during EBV infection in humans, we further evaluated the 3A3- and 3A5-like antibodies in the anti-gB antibody pools in sera from healthy donors (asymptomatic EBV carriers) and patients with NPC. 3A3, 3A5, their combination, and the control antibodies (a rabbit anti-gB nonneutralizing mAb 1E12 obtained in this study and a rabbit anti-influenza hemagglutinin (HA) mAb 2G9) were preincubated with the recombinant gB immobilized in the ELISAs. Then, we measured the binding of the serum antibodies to the recombinant gB preincubated with the above mAbs. The irrelevant control mAb 2G9 showed only 3 to 4% nonspecific inhibition of the serum antibodies binding to recombinant gB, whereas 3A3 and 3A5 blocked ∼34.44% (±9.95%, SEM) and 33.32% (±10.44%, SEM) of the sera binding to gB in healthy donors, respectively (Fig. 4C). Blocking was more effective in sera from patients with NPC, with average blocking efficiencies of 42.18% (±9.18%, SEM) and 43.73% (±11.25%, SEM) for 3A3 and 3A5, respectively (Fig. 4C). Furthermore, the combination of 3A3 and 3A5 blocked 70.70% (±9.42%, SEM) and 73.15% (±9.77%, SEM) of anti-gB serum antibodies in healthy and NPC groups from binding to recombinant gB (Fig. 4C), respectively. In contrast, the blocking efficiencies of the nonneutralizing anti-gB mAb 1E12 were 9.98% (±4.78%, SEM) and 9.32% (±4.40%, SEM) for the sera from healthy and NPC groups, respectively, dramatically lower than 3A3, 3A5, and their combination. As the recombinant gB proteins in the antibody blocking assays adopt the postfusion conformation, these findings indicate that 3A3- and 3A5-like antibodies collectively accounted for a major proportion of total anti-gB antibodies recognizing postfusion gB in healthy EBV carriers and patients with NPC.
To further evaluate whether 3A3 and 3A5 could block the serum antibodies targeting gB epitopes in prefusion conformation, we performed the antibody blocking assay with flow cytometry using cell membrane-bound gB proteins expressed by 293T cells that are in the mixture of prefusion and postfusion conformations. Before the incubation with sera, 293T cells that expressed full-length gB were preincubated with 3A3, 3A5, their combination, and the control antibodies, 1E12 and 2G9. We found that 3A3 and 3A5 blocked ∼36.64% (±5.47%, SEM) and 30.13% (±3.78%, SEM) of the serum IgG antibodies binding to membrane-bound gB in healthy donors, respectively, and the irrelevant mAb 2G9 blocked the serum antibody binding at a very low level (∼3 to 4%) (Fig. 4D). The blocking efficiencies in sera from patients with NPC were similar to healthy donors, with average values of 33.89% (±4.38%, SEM) and 31.27% (±3.87%, SEM) for 3A3 and 3A5, respectively. By contrast, the nonneutralizing antibody 1E12 showed much lower blocking efficiency for the sera binding to membrane-bound gB in healthy (9.76 ± 1.77%, SEM) and NPC (9.17 ± 1.70%, SEM) groups compared with 3A3 and 3A5. Furthermore, 3A3 together with 3A5 collectively blocked a major proportion of total anti-gB IgG antibodies in the sera from healthy donors (73.40 ± 2.93%, SEM) and patients with NPC (67.04 ± 2.65%, SEM) from binding to membrane-bound gB (Fig. 4D), consistent with the results observed using recombinant gB (Fig. 4C). Altogether, the blocking assays with the cell membrane-bound gB and the recombinant gB proteins indicate that the 3A3 and 3A5 epitopes constitute the major targets of prefusion and postfusion gB-specific antibodies elicited by natural EBV infection in human sera.
We further compared antibody blocking assays using recombinant gB vs. using cell membrane-bound gB. Interestingly, 3A5 and 3A3 + 3A5 had a higher blocking efficiency for sera from patients with NPC binding to recombinant gB than cell membrane-bound gB (Fig. 4 C and D and SI Appendix, Table S2), indicating that a larger proportion of prefusion than postfusion gB-specific epitopes bound with sera from patients with NPC was not competed by 3A5. These findings suggest that the proportion of serum antibodies targeting the 3A5 epitope of the total antibodies targeting gB in the prefusion conformation is smaller than the proportion of serum antibodies targeting the same 3A5 gB epitope in the postfusion conformation. By contrast, similar proportions of prefusion and postfusion gB-specific epitopes bound with sera from healthy donors were competed by 3A3 and 3A5, implicating similar proportions of serum antibodies targeting 3A3 and 3A5 epitopes in their prefusion and postfusion conformations.
Structural Basis for the Neutralization of EBV by 3A3 and 3A5.
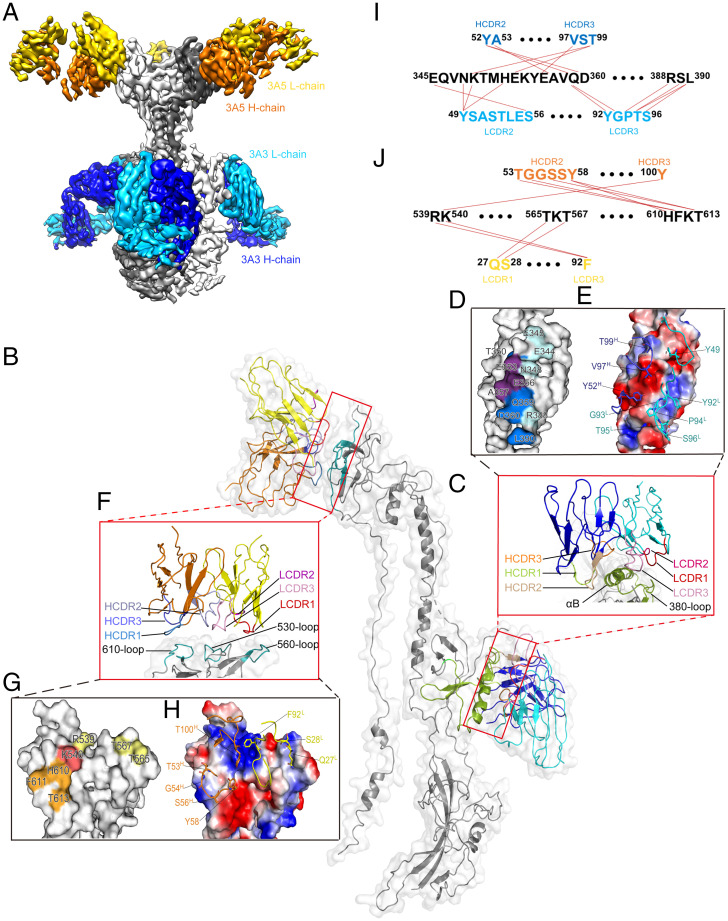
To provide insight into the structural basis for the neutralizing potencies of neutralizing mAbs 3A3 and 3A5, we used cryo-EM to determine the structures of recombinant ect-gB in complex with the Fab fragments of 3A3 and 3A5 at resolutions of 7.1 and 9.0 Å, respectively (SI Appendix, Fig. S6). 3A3Fab binds to D-II at the middle of the three-lobed structure, while 3A5Fab interacts with D-IV at the top of gB. Thus, being distal from each other, 3A3 and 3A5 may bind to gB simultaneously without steric hindrance, consistent with the competitive ELISA (Fig. 1 C and D). Accordingly, we further determined the cryo-EM structure of the ternary gB:3A3Fab:3A5Fab complex at a resolution of 3.9 Å (Fig. 5A and SI Appendix, Fig. S7 and Table S3). In this structure, each protomer of the gB trimer is bound with one 3A3 Fab and one 3A5 Fab, which was in good agreement with the gB:3A3Fab and gB:3A5Fab complex structures (Fig. 5B).
Fig. 5.
Structure determination of gB:3A3Fab:3A5Fab by cryo-EM. (A) Side view of the 3.9-Å cryo-EM structure of gB:3A3Fab:3A5Fab. gB, 3A3Fab, and 3A5Fab are colored gray, blue, and yellow, respectively. (B) Segmentation of monomeric gB binding with one 3A3Fab and one 3A5Fab. The key elements of D-II and D-IV of gB involved in antigen-antibody interactions are colored green and cyan, respectively. 3A3Fab and 3A5Fab are colored blue and yellow, respectively. (C-E) The interface of gB with 3A3Fab. The CDRs of the VH and VL and the gB’s αB-helix and 380 loop are shown differently (C). The residues involved in the 3A3 interactions are mapped on the gB surface, including E344, E345, N348, T350, E353, E356, A357, Q359, D360, R388, and L390 (D). The key residues localized at the VH and VL are labeled, including Y49L, Y92L, G93L, P94L, T95L, S96L, Y52H, V97H, and T99H (E). (F-H) The interface of gB with 3A5Fab. The CDRs of the VH and VL and the 530 loop, 560 loop, and 610 loop of gB are shown with different colors (F). The key residues involved in the 3A5 interactions are mapped on the gB surface, including R539, K540, T565, T567, H610, F611, and T613 (G). The key residues localized at the VH and VL are labeled, including Q27L, S28L, F92L, T53H, G54H, S56H, Y58H, and T100H (H). (I and J) Molecular interactions between gB and the VH and VL of mAbs 3A3 (I) and 3A5 (J).
Four of six complementary determining region (CDR) loops of the 3A3 paratope are involved, with a total buried area at the 3A3 Fab-gB protomer interface of ∼740 Å2 (light chain complementarity determining regions 2 (LCDR2) and -3 accounting for 40%; heavy chain complementarity determining regions 2 (HCDR2) and -3 accounting for 60%) (Fig. 5C and SI Appendix, Fig. S8A). 3A3 recognizes an epitope composed of 11 residues (E344, E345, N348, T350, E353, E356, A357, Q359, D360, R388, and L390) in the D-II of gB (Fig. 5D). The variable domain of the heavy chain contributes to the majority of the interactions, accounting for ∼60% of the buried area, through hydrophobic and hydrophilic contacts (Fig. 5E). Nine of 11 contact residues are located in the αB-helix of D-II (amino acid (aa) 345 to 360), highlighting that this αB-helix is critical for 3A3 binding and viral neutralization (Fig. 5I and SI Appendix, Fig. S8A).
The paratope of 3A5 also constituted four CDR loops, with a total buried area at the 3A5 Fab-gB protomer interface of ∼350 Å2 (LCDR1 and -3 accounting for 46%; HCDR2 and -3 accounting for 54%) (Fig. 5F and SI Appendix, Fig. S8B). The variable domains of the heavy chain and light chain of 3A5 contributed ∼54 and ∼46% of the buried area, respectively (Fig. 5H). The 3A5 epitope located at gB D-IV encompassed six residues: R539/K540 at the loop connecting β27 and β28, T565/T567 at the loop connecting β28 and β29, and H610/T613 in β33 (Fig. 5 G and J).
Residues Critical for the Binding of 3A3 and 3A5.
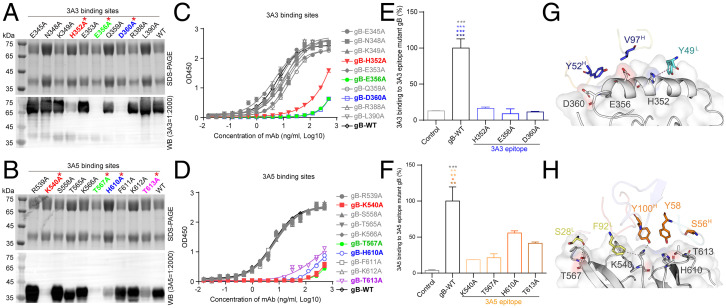
We next sought to identify the key residues mediating the interactions of gB:3A3Fab and gB:3A5Fab using structure-guided mutagenesis. Ten gB residues, E345, N348, K349, H352, E353, E356, Q359, D360, R388, and L390, in gB at the gB:3A3Fab interface were individually mutated to alanine. The mutants were subjected to WB, ELISA, flow cytometry, and analytical ultracentrifuge (AUC) assays to test their interaction with 3A3. We observed that gB with the point mutations of E356A or D360A completely abolished binding with mAb 3A3, while H352A retained a weak interaction with 3A3 (Fig. 6A). In the WB assay, other mutants bound to 3A3 with similar affinities to wild-type glycoprotein B (gB-WT) (Fig. 6A). Consistently, the binding activities of 3A3 to the H352A, E356A, or D360A mutants were significantly decreased compared with gB-WT or other mutant gB in the ELISA (Fig. 6C). We further measured the binding of 3A3 with the membrane-bound mutant gB proteins expressed on 293T cells using flow cytometry. Consistent with the results of the ELISA, the above three 3A3 epitope mutants drastically reduced 3A3 binding to the membrane-bound gB (Fig. 6E). Given that the recombinant gB protein used in the ELISA was in the postfusion conformation and that the membrane-bound gB was likely a mixture of prefusion and postfusion conformations, these results suggest that H352A, E356A, and D360A greatly affect 3A3 binding to these epitope mutations in both the postfusion and prefusion conformation. In addition, the AUC assay confirmed that no complex peak appeared when 3A3Fab was incubated with any of the above three mutants (SI Appendix, Fig. S9A). These three key residues, H352, E356, and D360, in the 3A3 epitope were all located at the αB-helix that contributed to 10 interaction contacts between gB and 3A3 (Fig. 6G).
Fig. 6.
Validation of key residues located at the gB:3A3 and gB:3A5 interfaces. (A and B) Reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and WB of purified recombinant mutant and gB-WT proteins as indicated with anti-gB antibodies 3A3 (A) and 3A5 (B). (C and D) The binding activities of the mutant and gB-WT to 3A3 (C) and 3A5 (D) were determined by ELISA. (E and F) The 293T cells bound with 3A3 (E) or 3A5 (F) were stained with the AF647 goat anti-rabbit IgG secondary antibody and measured by flow cytometry. The 293T cells expressing full-length gB-WT, 3A3, or 3A5 epitope mutants and the control cells transfected with the empty vector are indicated. Antibody staining was performed without cell membrane permeabilization. The level of antibody binding to 293T cells expressing gB-WT and each gB mutant was first normalized to the membrane expression level of gB-WT and each gB mutant, respectively, and then, all values were normalized as a percentage to gB-WT. Statistical analyses were performed using one-way ANOVA. Data in C-F are represented as the mean of two independent replicates ± SEM. (G and H) Key amino acid interactions at the gB:3A3 (E) and gB:3A5 (F) interfaces. The color of the asterisks denotes the group with which there is a significant difference determined by a Sidak multiple comparison test. *P ≤ 0.0332; **P ≤ 0.0021; ***P ≤ 0.0002. Red asterisks represent key residues recognized by 3A3 and 3A5.
For the gB-3A5 interface, 10 epitope residues of R539, K540, S558, T565, K566, T567, H610, F611, K612, and T613 on gB were also individually mutated and screened. Among them, four critical residues, K540, T567, H610, and T613, were verified to affect the binding between 3A5 and gB. 3A5 no longer bound to K540A and T567A mutant gB proteins and bound only weakly to the H610A and T613A mutants in the WB assay (Fig. 6B). Similarly, these four mutations almost abolished the binding of 3A5 in the ELISA (Fig. 6D). Among the four 3A5 epitope mutants of gB, the binding of 3A5 with membrane-bound gB was reduced by K540A and T567A mutants by over 80%, while 3A5 retained 45 and 60% of the binding with membrane-bound H610A and T613A gB mutants, respectively (Fig. 6F). Comparing the results of the ELISA with recombinant gB vs. the results of flow cytometry with the membrane-bound gB, we found that K540A and T567A almost abolished the binding of 3A5 with the mutant epitopes in both the prefusion and postfusion conformations, whereas H610A and T613A had a greater impact on 3A5 binding to these mutations in the postfusion than prefusion conformation. AUC further verified the absence of complex formation between 3A5Fab and gB containing these four mutations (SI Appendix, Fig. S9B). The key residues K540 and T567 formed hydrogen bonds to interact with F92L and Y100H of mAb 3A5, respectively. Altogether, the four key residues, K540, T567, H610, and T613, formed the interaction network with 3A5 and contributed ∼70% (19 of 28) of the interaction contacts (Fig. 6H).
3A3 and 3A5 Inhibit Cell Membrane Fusion via Different Neutralizing Mechanisms.
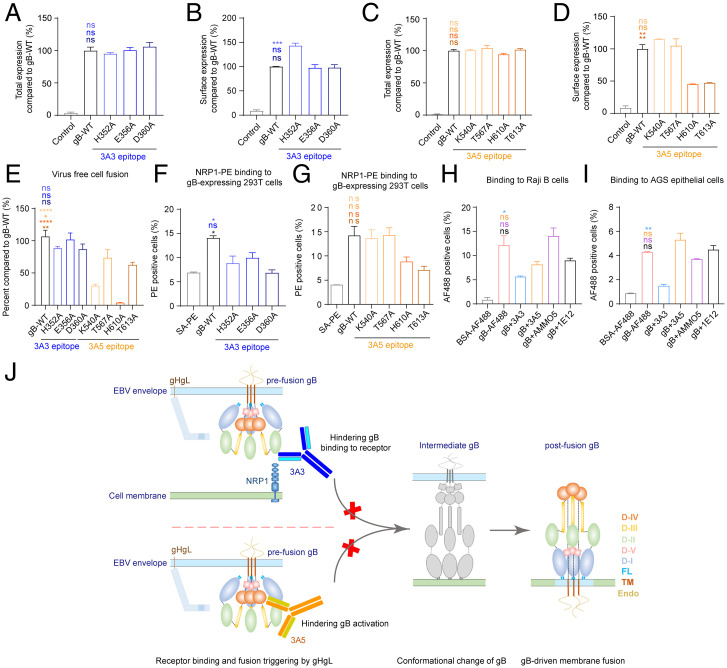
To further determine how 3A3 and 3A5 block EBV infection, we investigated the functional impact of mutations at the key residues involved in the binding interface of 3A3 (H352, E356, and D360) and 3A5 (K540, T567, H610, and T613). The total expression levels of these mutants were comparable with that of gB-WT, as measured by flow cytometry (Fig. 7 A and C). For cell surface expression, E356A, D360A, K540A, and T567A retained similar levels to gB-WT, and H352A increased cell surface-embedded gB, while the H610A and T613A mutations suppressed gB surface expression by ∼30% (Fig. 7 B and D). In the epithelial cell fusion model for mutations located at the 3A3 epitope, including H352A, E356A, and D360A, gB retained its function in mediating cell to cell fusion (Fig. 7E). In contrast, mutations at the 3A5 epitope, including K540A, T567A, and H610A, affected membrane fusion. K540A and T567A dramatically reduced cell fusion without an appreciable change in the cell surface expression of gB, whereas H610A caused a 75% reduction in the cell surface expression of gB but a 90% reduction in membrane fusion (Fig. 7 D and E). T613A showed a greater effect on gB membrane expression (70% reduction) than cell to cell fusion (40% reduction) (Fig. 7E). The significant reduction in membrane expression suggested that the mutated gB harboring H610A and T613A was retained in the cytoplasm, potentially due to suboptimal folding (Fig. 7D). Collectively, these results indicated that the 3A5 epitope is essential for gB to exert membrane fusion function.
Fig. 7.
3A3 and 3A5 binding sites at gB D-II and D-IV are critical for cell binding and fusion. (A-D) The total expression and cell surface expression levels of gB-WT and gB 3A3 mutants (A and B) or 3A5 mutants (C and D) produced by transfected 293T cells were measured with mAb 3A5 (A and B) and mAb 3A3 (C and D) by flow cytometry, respectively. Antibody staining was performed with cell membrane permeabilization when the total expression was evaluated in A and C. Antibody staining was performed without cell membrane permeabilization when cell surface expression was evaluated in B and D. All values were normalized as a percentage to gB-WT. (E) Cell-cell fusion efficiency of gB-WT and gB mutants. All values were normalized as a percentage to gB-WT. (F and G) The binding of NRP1 to 293T cells expressing gB-WT, 3A3 epitope mutants (F), or 3A5 epitope mutants (G) of gB was evaluated by flow cytometry without cell membrane permeabilization. Cells stained with SA-PE alone were used as a negative control. (H and I) The binding of gB to Raji B cells (H) and AGS gastric adenocarcinoma epithelial cells (I) in the presence of anti-gB antibodies was evaluated by flow cytometry without cell membrane permeabilization. Bovine serum albumin (BSA)-AF488 and gB-AF488 were used as negative and positive controls, respectively. (J) Proposed schematic diagram of neutralizing mechanisms of 3A3 and 3A5. 3A3 blocks gB binding to its coreceptor NRP1 by directly restricting access to the interface. The binding of 3A5 to D-IV could restrict the movement of the gB trimer during conformational changes from pre- to postfusion by steric hindrance. It is also possible that the binding of 3A3 and 3A5 could bring steric hindrance that inhibits the triggering of gB activation by gHgL. Data are represented as the mean of two independent replicates ± SEMs. Statistical analyses were performed using one-way ANOVA. The color of the asterisks denotes the group with which there is a significant difference determined by a Sidak multiple comparison test. ns, no significant difference. *P ≤ 0.0332; **P ≤ 0.0021; ***P ≤ 0.0002; ****P ≤ 0.0001. SA-PE, streptavidin-phycoerythrin. PE, phycoerythrin. FL, fusion loop. TM, transmembrane domain. Endo, endodomain.
The interactions between gB and cell surface proteins play a role in the infection of several herpesviruses, including EBV, varicella-zoster virus (VZV), and herpes simplex virus (HSV) (26–30). NRP1 has been reported as the coreceptor for EBV gB, enhancing EBV entry into epithelial cells (14). Flow cytometry assays showed that the H352A and D360A mutations at the 3A3 epitope both significantly reduced the attachment of NRP1 to the gB-overexpressing 293T cells (Fig. 7F), whereas 3A5 epitope mutants (K540A, T567A, H610A, and T613A) did not affect NRP1 binding to the gB-overexpressing 293T cells (Fig. 7G). We observed a trend, although not significant, of reduced binding of NRP1 to H610A and T613A gB mutants (Fig. 7G), which could be explained by the reduced cell surface expression observed with H610A and T613A gB mutants (Fig. 7D). In line with our data, the NRP1 binding site on gB has been reported as aa 23 to 431 encompassing the 3A3 epitope (SI Appendix, Fig. S10) (14), suggesting that 3A3 could directly block gB binding to its putative coreceptor NRP1. We further assessed whether mAbs 3A3 or 3A5 interfered with gB attachment to the cell surface. We demonstrated that 3A3, but not 3A5 and AMMO5, could inhibit soluble gB binding to Raji B cells and AGS gastric cancer cells (Fig. 7 H and I). Similar results were also observed with Akata B cells and HK1 NPC cells (SI Appendix, Fig. S11). These results suggested that 3A3 may occupy the receptor binding site of gB and thus, hinder gB binding to the target cell membrane.
For 3A5, its binding sites may not participate in gB-receptor recognition as 3A5 did not affect gB attachment to the cell membrane of either B cells (Raji or Akata) or epithelial cells (AGS or HK1) (Fig. 7 H and I and SI Appendix, Fig. S11). Together with the 3A5 epitope mutagenesis assays that indicate the 3A5 epitope is essential for fusion function, these results further support that binding of 3A5 is likely to interfere with the necessary interactions required for gB activation or conformational transition, such as the interactions between gB protomers and/or between gB and gHgL, other than blocking gB interaction with its coreceptor. Altogether, we demonstrated that neutralizing mAbs 3A3 and 3A5 block gB-mediated membrane fusion and neutralize EBV infection, possibly through different mechanisms involving gB interaction with its putative coreceptor NRP1 and gB activation (Fig. 7J).
Discussion
Due to its essential role in the viral infection of both B cells and epithelial cells, EBV gB has attracted attention to identifying potent dual-tropic neutralizing antibodies. In this study, we established a panel of specific antibodies against EBV gB, among which 3A3 and 3A5 showed strong neutralizing activities against EBV infection of both B cells and epithelial cells. These two neutralizing mAbs effectively protected humanized mice from EBV viremia and EBV-driven LPD, possibly through controlling infection and thus, reducing EBV loads in vivo. Structural and functional analyses revealed two epitopes in the gB, D-II and D-IV. These two epitopes are critical for EBV fusion. The 3A3 epitope at D-II participates in the interaction with the host cell, and the 3A5 epitope at D-IV is important for the fusion reaction of gB. Importantly, in EBV-infected individuals, 3A3 and 3A5 epitopes represent the major targets of gB-specific immune responses that contribute to the neutralization of EBV infection. The neutralizing antigenic sites defined by 3A3 and 3A5 at gB D-II and D-IV provide potential targets for antiviral vaccines and therapeutics.
Several points need to be discussed here. First, as gB adopts the prefusion to postfusion conformational transition to exert fusion function, it is highly advantageous to isolate neutralizing antibodies capable of blocking the prefusion gB. Although in the present study, we did not resolve the prefusion structure, according to a homology modeling structure of EBV prefusion gB based on the structure of HSV-1 prefusion gB (31), we found that gB D-II and D-IV remain largely intact and exposed in the prefusion conformation (SI Appendix, Fig. S12). Consistently, our results found that 3A3 and 3A5 bound to nascent gB expressed by EBV-positive cells after induction of viral lytic production and potently neutralize EBV infection of B cells and epithelial cells, highlighting that the epitopes of 3A3 and 3A5 are accessible in the prefusion state. Second, our study indicates that the epitopes of mAbs 3A3 and 3A5 represent two major immunogenic sites for neutralization in naturally infected individuals. We show that 3A3- and 3A5-like antibodies account for ∼70% of total anti-gB antibodies elicited by EBV infection in healthy individuals and patients with NPC. Importantly, previous and our current studies show that in infected individuals, EBV elicits broad antibody responses against gp350, gHgL, gp42, and gB, which all contribute to the inhibition of viral infection in B cells, although the levels of these serum antibodies vary greatly among individuals (32). Notably, we observed differences in the blocking efficiency of 3A3 and 3A5 against the binding of antibodies in sera from healthy donors vs. NPC to recombinant gB vs. membrane-bound gB (Fig. 4 C and D and SI Appendix, Table S2), implicating that the antibody responses against the prefusion and postfusion gB differ between healthy individuals and patients with NPC and warrant further investigation.
Since multiple EBV infections are common in healthy individuals (33, 34), it seems reasonable to expect a vaccine that elicits a stronger immune response than naturally acquired EBV infection to provide sufficient protection. Given that EBV elicits broad antibody responses against its glycoproteins in humans, the gB epitopes recognized by 3A3 and 3A5 could be exploited in combination with the other antigenic sites on gp350 and gHgL recognized by the neutralizing mAbs 72A1, AMMO1, 769B10, and 1D8 (18, 24, 32) for the design of potent vaccines. The recent exciting studies reported the design of nanoparticles displaying gp350 and/or gHgL, which elicited potent neutralizing antibodies against EBV infection (32, 35). Carefully designed approaches that combine multiple EBV immunogens and properly expose these neutralizing antigenic sites could be further explored to induce broader and stronger immunity against EBV infection.
Despite recent advances, the critical molecular determinants of EBV gB involved in membrane fusion have not been entirely identified (6). Our findings pinpoint that the critical sites recognized by 3A3 and 3A5 participate the steps along with gB-mediated fusion which underlie the neutralizing ability of 3A3 and 3A5. This study reveals that the 3A3-gB interface at D-II participates in the interaction of gB with its putative coreceptor NRP1, implicating that it might have important functional roles in gB-host cell interaction (Fig. 7J). We show that mutations at the 3A3 epitope (E352A and D360A) lead to a significant loss of NRP1 binding to gB-overexpressed cells and that 3A3 blocks gB attachment to both epithelial and B cells. Meanwhile, the 3A3 binding sites at D-II coincide with the gB domain that interacts with NRP1 (SI Appendix, Fig. S10), further suggesting the involvement of D-II in the virus-host interactions (14). In addition to NRP1 for EBV, cell surface proteins that interact with gB and facilitate entry have been reported to be involved in VZV (MAG) and HSV-1 (PILRα) infection (26–30). Notably, we observed that 3A3 could only reduce but not abolish the attachment of gB to the cell surface. The inhibition of gB attachment to B cells by 3A3 was less obvious than to epithelial cells, which may partly explain why 3A3 was more effective in blocking the EBV infection of epithelial cells than B cells. It is plausible that other gB coreceptors remain identified, especially for B cell entry.
Here, we show that the 3A5-gB interaction interface at D-IV, specifically the crown structure, is important for the fulfilling gB-mediated membrane fusion. Our point mutagenesis assays at residues K540, T567, and H610 in the 3A5 epitope were all expressed on the cell surface at detectable levels but were deficient in gB-mediated cell-cell fusion in the epithelial cell fusion model. Among the point mutations in the 3A5 epitope at D-IV, gB with the mutations of H610A and T613A displayed a reduction of more than 50% in cell surface expression, suggesting that these gB mutants may be less folded and retained within the cytoplasm. A previous EBV gB functional mutagenesis study also showed that insertional mutations at F561, Y597, E602, and A620 encompassing the 3A5 epitope affected gB cell surface expression and/or abolished the formation of complex oligomers (36). The homologous regions in D-IV of HSV-1 and VZV gB have been reported to be important for oligomerization and cell surface expression (37–39). Direct evidence that D-IV contributed to the formation of the gB trimer was obtained from the cryo-ET structure of HSV-1 prefusion gB in which D-IV, cyclically swapped within the trimer, makes contacts with neighboring protomers (31). Together, these studies suggest that D-IV, encompassing the 3A5 epitope, participates in the formation of trimer interfaces and is important for maintaining the prefusion conformation for membrane fusion. Furthermore, comparisons of the prefusion and postfusion conformations of HSV-1 gB indicated that D-I, D-II, and D-IV flip around the central part containing D-IV and that the C-terminal regions of D-III and D-IV remain rather intact during the transition in gB (31). Through binding to D-IV, 3A5 may hinder the transition of gB from the prefusion to postfusion state (Fig. 7J).
Despite structural and functional insights, we are still uncertain about the exact mechanism of gB-mediated membrane fusion. EBV gB functional domains interacting with 3A3 and 3A5 may participate in the unexplored gB-gHgL and/or other gB-protein interactions required for membrane fusion. Future studies on the structures of prefusion EBV gB and gB in complex with gHgL and/or other proteins will help to determine the interactions and conformational changes required for gB-mediated membrane fusion.
Various gB-specific mAbs have been demonstrated to effectively neutralize infection with other herpesviruses, including cytomegalovirus (CMV), HSV-1, and VZV (40–43). Structurally, these neutralizing mAbs were mapped to three major antigenic sites on postfusion gB: D-I (1G2 for human cytomegalovirus (HCMV)), D-II (SM5-1 and -3 to -25 for CMV), and D-IV (93k for VZV and 7B11 for pseudorabies (PRV)) (SI Appendix, Fig. S13) (43–47). Two of them, 93k (VZV) and 7B11 (PRV), bind to D-IV as 3A5, but the binding footprints and orientations of 93k and 7B11 are different from that of 3A5. The binding site of SM5-1 is out of the αB-helix in the D-II, different from 3A3. Consistent with the different binding footprints among these antibodies, the mAbs against HCMVgB (3–25) or VZVgB (93k) showed no cross-reactivity to EBVgB (SI Appendix, Fig. S14). The epitopes of 3A3 and 3A5 defined in this study represent the major immunogenic sites specific for EBV gB in human sera. This study highlights the importance of the neutralizing antigenic sites at gB D-II and D-IV together with those at gp350 and gHgL recognized by neutralizing antibodies (18, 24, 32, 35) for the rational design of successful anti-EBV vaccines and therapeutics.
Materials and Methods
Experimental details on cloning and mutagenesis, expression and purification of proteins and antibodies, rabbit B cell isolation and recombinant antibody cloning, indirect ELISA, competitive ELISA and antibody blocking assay, the antibody blocking assay by flow cytometry, immunofluorescence assay, antibody depletion from human sera, binding to induced Akata-EBV-GFP (green fluorescent protein) cells, cryo-EM sample preparation and data collection, image processing and three-dimensional reconstruction, model building and refinement, virus production, the neutralization assay, SPR, AUC, the virus-free epithelial cell fusion assay, quantification of the cell surface and total expression of gB mutants, the cell surface binding assay, EBV infection in humanized mice, detection of EBV DNA in blood and tissues, hematoxylin and eosin (H&E) staining, immunohistochemistry, in situ hybridization, the flow cytometry assay of human cells in humanized mice, rabbit immunization and statistical analysis are described in the SI Appendix, SI Materials and Methods.
Human Specimens.
Sera from 38- to 64-y-old healthy individuals and patients with NPC were collected from Sun Yat-sen University Cancer Center. This study was approved by the Institutional Ethics Committee of the Sun Yat-sen University Cancer Center. Written informed consent was obtained from all participants.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 82030046 (to M.-S.Z.), 82073756 (to Yixin Chen), and 81872228 (to M.X.); Science and Technology Major Projects of Xiamen Grant 3502Z20203023 (to Q. Zheng); Guangdong Basic and Applied Basic Research Foundation Grant 2020B1515020002 (to M.X.); and Fundamental Research Funds for the Central Universities (to M.X.). We thank Prof. Richard Longnecker for providing the plasmids pCAGGS-T7, pCAGGS-gH, pCAGGS-gL, pCAGGS-gB, and pT7EMCLuc and Prof. Shoudeng Chen and Prof. Yaoqing Chen from Sun Yat-sen University for their suggestions on the experiments and manuscript writing.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202371119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Shannon-Lowe C., Rickinson A., The global landscape of EBV-associated tumors. Front. Oncol. 9, 713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J. I., Fauci A. S., Varmus H., Nabel G. J., Epstein-Barr virus: An important vaccine target for cancer prevention. Sci. Transl. Med. 3, 107fs7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shindiapina P., Ahmed E. H., Mozhenkova A., Abebe T., Baiocchi R. A., Immunology of EBV-related lymphoproliferative disease in HIV-positive individuals. Front. Oncol. 10, 1723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Longnecker R., Epithelial cell infection by Epstein-Barr virus. FEMS Microbiol. Rev. 43, 674–683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutt-Fletcher L. M., EBV glycoproteins: Where are we now? Future Virol. 10, 1155–1162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connolly S. A., Jardetzky T. S., Longnecker R., The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 19, 110–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sathiyamoorthy K., et al. , Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog. 10, e1004309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., et al. , Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71, 4657–4662 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., et al. , Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat. Microbiol. 3, 172–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chesnokova L. S., Nishimura S. L., Hutt-Fletcher L. M., Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc. Natl. Acad. Sci. U.S.A. 106, 20464–20469 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H., et al. , Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat. Microbiol. 3, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Plate A. E., Smajlović J., Jardetzky T. S., Longnecker R., Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J. Virol. 83, 7678–7689 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandramouli S., et al. , Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci. Immunol. 2, eaan1457 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Wang H. B., et al. , Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 6, 6240 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sathiyamoorthy K., et al. , Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc. Natl. Acad. Sci. U.S.A. 114, E8703–E8710 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sathiyamoorthy K., et al. , Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat. Commun. 7, 13557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijder J., et al. , An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein-Barr virus. Immunity 48, 799–811.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Q. Y., et al. , A potent and protective human neutralizing antibody targeting a novel vulnerable site of Epstein-Barr virus. Nat. Commun. 12, 6624 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh S., et al. , Neutralizing antibodies protect against oral transmission of lymphocryptovirus. Cell Rep. Med. 1, 100033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L., Borza C. M., Hutt-Fletcher L. M., Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J. Virol. 79, 10923–10930 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber J., Peng H., Rader C., From rabbit antibody repertoires to rabbit monoclonal antibodies. Exp. Mol. Med. 49, e305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo S., et al. , Oncological and genetic factors impacting PDX model construction with NSG mice in pancreatic cancer. FASEB J. 33, 873–884 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Mgodi N. M., et al. ; HVTN 703/HPTN 081 Team, A phase 2b study to evaluate the safety and efficacy of VRC01 broadly neutralizing monoclonal antibody in reducing acquisition of HIV-1 infection in women in sub-Saharan Africa: Baseline findings. J. Acquir. Immune Defic. Syndr. 87, 680–687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque T., et al. , A mouse monoclonal antibody against Epstein-Barr virus envelope glycoprotein 350 prevents infection both in vitro and in vivo. J. Infect. Dis. 194, 584–587 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Strowig T., et al. , Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J. Exp. Med. 206, 1423–1434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh T., et al. , PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132, 935–944 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., et al. , Binding of herpes simplex virus glycoprotein B (gB) to paired immunoglobulin-like type 2 receptor alpha depends on specific sialylated O-linked glycans on gB. J. Virol. 83, 13042–13045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suenaga T., et al. , Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses. Proc. Natl. Acad. Sci. U.S.A. 107, 866–871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Q., et al. , PILRα and PILRβ have a siglec fold and provide the basis of binding to sialic acid. Proc. Natl. Acad. Sci. U.S.A. 111, 8221–8226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suenaga T., et al. , Sialic acids on varicella-zoster virus glycoprotein B are required for cell-cell fusion. J. Biol. Chem. 290, 19833–19843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vollmer B., et al. , The prefusion structure of herpes simplex virus glycoprotein B. Sci. Adv. 6, eabc1726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu W., et al. , Immunization with components of the viral fusion apparatus elicits antibodies that neutralize Epstein-Barr virus in B cells and epithelial cells. Immunity 50, 1305–1316.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwok H., Chan K. W., Chan K. H., Chiang A. K., Distribution, persistence and interchange of Epstein-Barr virus strains among PBMC, plasma and saliva of primary infection subjects. PLoS One 10, e0120710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walling D. M., Brown A. L., Etienne W., Keitel W. A., Ling P. D., Multiple Epstein-Barr virus infections in healthy individuals. J. Virol. 77, 6546–6550 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei C. J., et al. , A bivalent Epstein-Barr virus vaccine induces neutralizing antibodies that block infection and confer immunity in humanized mice. Sci. Transl. Med. 14, eabf3685 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reimer J. J., Backovic M., Deshpande C. G., Jardetzky T., Longnecker R., Analysis of Epstein-Barr virus glycoprotein B functional domains via linker insertion mutagenesis. J. Virol. 83, 734–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin E., Spear P. G., Random linker-insertion mutagenesis to identify functional domains of herpes simplex virus type 1 glycoprotein B. Proc. Natl. Acad. Sci. U.S.A. 104, 13140–13145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Britt W. J., Jarvis M. A., Drummond D. D., Mach M., Antigenic domain 1 is required for oligomerization of human cytomegalovirus glycoprotein B. J. Virol. 79, 4066–4079 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speckner A., Kropff B., Knör S., Mach M., The antigenic domain 1 of human cytomegalovirus glycoprotein B contains an intramolecular disulphide bond. J. Gen. Virol. 81, 2659–2663 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Pötzsch S., et al. , B cell repertoire analysis identifies new antigenic domains on glycoprotein B of human cytomegalovirus which are target of neutralizing antibodies. PLoS Pathog. 7, e1002172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns T. M., et al. , Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J. Virol. 88, 2677–2689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atanasiu D., et al. , Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84, 3825–3834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliver S. L., et al. , A glycoprotein B-neutralizing antibody structure at 2.8 Å uncovers a critical domain for herpesvirus fusion initiation. Nat. Commun. 11, 4141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye X., et al. , Recognition of a highly conserved glycoprotein B epitope by a bivalent antibody neutralizing HCMV at a post-attachment step. PLoS Pathog. 16, e1008736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spindler N., et al. , Structural basis for the recognition of human cytomegalovirus glycoprotein B by a neutralizing human antibody. PLoS Pathog. 10, e1004377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandramouli S., et al. , Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat. Commun. 6, 8176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., et al. , Two classes of protective antibodies against Pseudorabies virus variant glycoprotein B: Implications for vaccine design. PLoS Pathog. 13, e1006777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.