Significance
G protein–coupled receptors (GPCRs) constitute the largest and pharmacologically most important family of cell-surface receptors. Some GPCRs interact specifically with receptor-activity-modifying proteins (RAMPs), but the consequences of this interaction for the receptor activation mechanism are not well understood. Using a set of fluorescent biosensors for the parathyroid hormone 1 receptor (PTH1R) and its downstream signaling partners, we show here that RAMP2 induces a unique, preactivated receptor state that shows faster activation and altered downstream signaling. This type of GPCR modulation may open new methods of drug design.
Keywords: GPCR, RAMP, BRET, FRET
Abstract
Receptor-activity-modifying proteins (RAMPs) are ubiquitously expressed membrane proteins that associate with different G protein–coupled receptors (GPCRs), including the parathyroid hormone 1 receptor (PTH1R), a class B GPCR and an important modulator of mineral ion homeostasis and bone metabolism. However, it is unknown whether and how RAMP proteins may affect PTH1R function. Using different optical biosensors to measure the activation of PTH1R and its downstream signaling, we describe here that RAMP2 acts as a specific allosteric modulator of PTH1R, shifting PTH1R to a unique preactivated state that permits faster activation in a ligand-specific manner. Moreover, RAMP2 modulates PTH1R downstream signaling in an agonist-dependent manner, most notably increasing the PTH-mediated Gi3 signaling sensitivity. Additionally, RAMP2 increases both PTH- and PTHrP-triggered β-arrestin2 recruitment to PTH1R. Employing homology modeling, we describe the putative structural molecular basis underlying our functional findings. These data uncover a critical role of RAMPs in the activation and signaling of a GPCR that may provide a new venue for highly specific modulation of GPCR function and advanced drug design.
G protein–coupled receptors(GPCRs) represent the largest class of membrane-bound proteins and are involved in a multitude of biological processes (1). They are characterized by a seven-transmembrane helix structure, which undergoes a characteristic rearrangement upon binding of agonists. Agonist binding to its cognate receptor induces conformational changes in the transmembrane helices, which are transmitted to the cytosolic face of the receptors and ultimately result in receptor activation, which represents the key step of signal transduction. The combination of crystallographic and cryogenic electron microscopy studies and the employment of optical biosensors to study the reorganization of the seven transmembrane domains has allowed a detailed understanding of the general mechanisms of GPCR activation (2–5).
Earlier structural studies suggest that GPCRs undergo similar conformational changes upon activation, including, most prominently, an outward movement of the transmembrane helix 6 at the cytosolic face, thereby creating a pocket to which the G protein α-subunit can couple (5). More recent studies, however, have revealed that the exact type of changes may depend on the receptor class and the specific receptor (6–8). Class- and receptor-specific differences may also exist in the interaction of receptors not only with downstream G proteins and β-arrestins but also with accessory and modulatory proteins (9).
Studies of the kinetic steps that govern the structural rearrangements which underlie receptor activation (10) showed that its speed might depend on the receptor class and the specific receptor. For example, when exposed to saturating agonist concentrations, most class A GPCRs switch into the active state within tens of milliseconds. The same process takes 1 to 2 ms for a class C GPCR and may take up to a second for class B receptors (11–15). Little is known whether the activation kinetics of GPCRs can be modulated by their cellular context and whether proteins other than the receptors themselves might play a role in shaping signaling kinetics and specificity.
Here, we study the parathyroid hormone 1 receptor (PTH1R), a prototypical member of class B GPCRs characterized by a large N-terminal domain that binds a major part of their cognate peptide agonists (16, 17). Compared to class A GPCRs, PTH1R activation is relatively slow and occurs in a two-step process: The initial N-terminal binding step has a time constant of ∼140 ms, followed by an interaction of the ligand with the transmembrane core, which changes into its active conformation with a time constant of ∼1 s (11, 14). Pleiotropic in its downstream coupling, PTH1R signals primarily via Gs but can also couple to Gq (18), G12/13 (19), and Gi (20) and interacts with and signals via β-arrestins (21, 22). The two endogenous agonists, parathyroid hormone (PTH) and parathyroid hormone-related peptide (PTHrP), trigger PTH1R activation with similar kinetics and specificity for the various intracellular pathways (23–25). However, PTH can induce prolonged signaling from intracellular sites, while PTHrP signals exclusively from the cell surface (26).
PTH1R has been reported to interact with modulatory proteins of the receptor-activity-modifying protein (RAMP) family (27–29). RAMPs constitute a family of single transmembrane helix proteins with three members: RAMP1, RAMP2, and RAMP3.
It is controversial whether PTH1R interacts only or preferentially with RAMP2 (28) or all three RAMPs (28, 29). In RAMP2 knock-out mice, PTH1R function is deregulated, and placental dysfunction is observed (30), suggesting a major physiological role of the PTH1R/RAMP2 interaction. Yet, the molecular mechanisms of how RAMPs may modulate the activation dynamics of PTH1R and their signaling properties remain to be elucidated.
To address these questions, we develop and employ biosensors for PTH1R activation and investigate an array of downstream signaling pathways to assess the effects of RAMPs on the activation dynamics and signaling properties of PTH1R in response to its two endogenous ligands, PTH and PTHrP. We observe that RAMP2 specifically interacts with PTH1R and modulates its activation kinetics as well as signaling dynamics in an agonist-dependent manner.
Results
Analysis of PTH1R/RAMP Interactions at the Cell Surface.
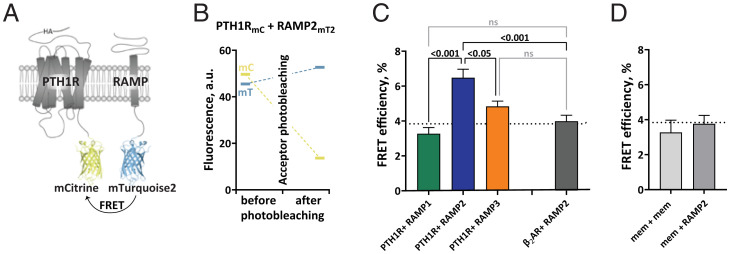
First, we investigated the interactions of PTH1R with the three RAMPs at the surface of intact cells. To do so, we performed acceptor photobleaching experiments to quantify fluorescence resonance energy transfer (FRET) efficiencies between PTH1R, labeled with a C-terminal mCitrine (31) (mC), and the three different RAMPs, labeled at their intracellular C terminus with mTurquoise2 (32) (mT2) (Fig. 1A). Experiments were conducted in human embryonic kidney (HEK293) cells that express negligible amounts of endogenously PTH1R and have only low levels of RAMPs. FRET efficiencies were quantified by measuring the recovery of donor emission after photobleaching of the acceptor (Fig. 1B) in HEK293 cells transiently coexpressing comparable levels of tagged PTH1R in combination with tagged RAMP1, 2, or 3 (SI Appendix, Fig. S1A).
Fig. 1.
Intermolecular FRET reveals RAMP2 as an interaction partner of PTH1R. (A) Schematic representation of the constructs for FRET acceptor photobleaching experiments between PTH1R and RAMP2. Photobleaching experiments were conducted in HEK293 cells, transiently cotransfected with a combination of donor- and acceptor-tagged constructs. The acceptor fluorophore (mCitrine, mC) was fused to the C-terminus of the PTH1R (PTH1RmC), to the control β2-adrenergic receptor (β2ARmC) or targeted to the plasma membrane via a -CAAX sequence (memmC). The donor fluorophore (mTurquoise2, mT2) was fused to the C-terminal of RAMPs (RAMP1/2/3mT2) or targeted to the plasma membrane via a -CAAX sequence (memmT2). (B) Representative experiment showing photobleaching efficiency in cells expressing PTH1RmC and RAMP2mT2. Fluorescence emission of both donor (mC) and acceptor (mT2) was recorded before and after acceptor photobleaching. (C and D) FRET efficiencies from photobleaching experiments recorded with a confocal microscope. The data are expressed as percent of donor emission increase after photobleaching for each experimental group. The dotted line indicates the average FRET efficiency of negative control groups (gray bars). The data are derived from at least three independent experiments and following numbers of cells: PTH1RmC + RAMP1mT2 (n = 46), PTH1RmC + RAMP2mT2 (n = 70), PTH1RmC + RAMP3mT2 (n = 71), β2ARmC + RAMP2mT2 (n = 51), memmC + RAMP2mT2 (n = 37), and memmC + memmT2 (n = 9); bars represent means ± SEM. Significance between the groups was assessed by Brown–Forsythe ANOVA, followed by Dunnett’s multiple comparisons test; not significant (ns): P > 0.05.
The FRET efficiency was significantly higher for cells expressing PTH1RmC with RAMP2mT2 than for combinations with either RAMP1mT2 or RAMP3mT2 (Fig. 1C). In fact, the FRET efficiencies for the latter two were not significantly different from background FRET (P > 0.05; dotted line in Fig. 1 C and D), which is determined by either nonspecific FRET between two membrane tags or between a membrane tag and RAMP2mT2 (Fig. 1D), or by FRET between RAMP2mT2 and the β2-adrenergic receptor (β2ARmC), a GPCR shown not to interact with RAMP2 (29, 33). These data indicate that PTH1R forms complexes with RAMP2 at the cell surface but very little or none with RAMP3 or RAMP1.
RAMP2 Expression Modulates PTH1R Basal and PTH-Bound Conformations.
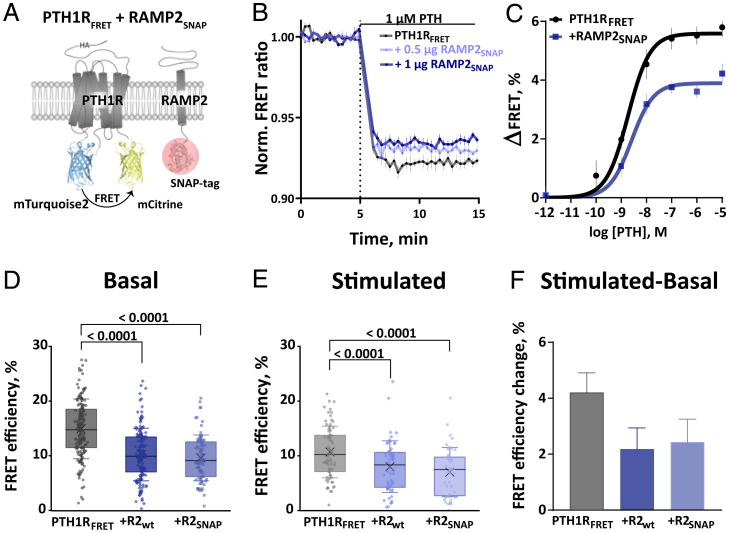
We then aimed to investigate whether RAMP2 regulates PTH1R activation dynamics. Based on previously reported PTH1R biosensors with donor and acceptor fluorophores fused to conformationally sensitive sites (11, 34), we generated an improved conformational biosensor, PTH1RFRET. Preserving insertion sites in the third intracellular loop and at the C terminus, we exchanged the fluorophores with brighter and more photostable fluorophores, namely mT2 and mC (Fig. 2A).
Fig. 2.
RAMP2 modulates PTH1R basal and PTH-induced conformations. (A) Schematic representation of intramolecular PTH1RFRET biosensor. To control RAMP2 expression, its C-terminally fused SNAP-tag was labeled with 1 µM SNAP-Cell SiR-647 (red circle). (B) Average time course of PTH-induced FRET changes recorded in a plate reader from HEK293 cells transiently expressing PTH1RFRET (black) alone or together with RAMP2SNAP (0.5 or 1 µg cDNA, blue). The data represents the mean ± SEM of five independent experiments normalized to the initial FRET value (set to 1). (C) Concentration–response curves obtained from control and 1 µg RAMP2SNAP traces as in B. ΔFRET values are expressed as percent maximal change from the initial FRET value. Curve fitting gave pEC50 values (means ± SEM) of PTH1RFRET = 8.73 ± 0.12 and PTH1RFRET + RAMP2SNAP = 8.59 ± 0.17. (D and E) FRET efficiencies from photobleaching experiments were recorded with a confocal microscope. The data are expressed as percent of donor emission increase after photobleaching for each experimental group. FRET efficiencies of basal (D) and 100 µM PTH-stimulated (E) HEK293 cells transiently expressing PTH1RFRET (black) alone or together with RAMP2wt (dark blue) or RAMP2SNAP (light blue). The data are from the following numbers of cells obtained in six (basal) and three (stimulated) independent experiments. Basal: PTH1RFRET (n = 153), PTH1RFRET + RAMP2wt (n = 130), PTH1RFRET + RAMP2SNAP (n = 82). Stimulated PTH1RFRET (n = 73), PTH1RFRET + RAMP2wt (n = 56), PTH1RFRET + RAMP2SNAP (n = 44). Data show values from individual cells; boxes represent the first and third interquartile range, and whiskers indicate SD. Significance between the groups was tested with one-way ANOVA followed by Dunnett‘s multiple comparisons test; not significant (ns): P > 0.05. (F) FRET efficiency changes calculated from D and E, represented as percent change ± SEM.
To measure PTH1R activation dynamics, we cotransfected HEK293 cells with PTH1RFRET biosensor with and without RAMP2SNAP, mimicking physiological contexts in which both are expressed abundantly (i.e., lungs, kidneys, and placenta). To estimate the expression level of RAMP2 in these experiments, we tagged the C terminus of RAMP2 with a SNAP-tag, which was labeled with the permeable fluorescent dye SNAP-Cell SiR-647 (35). This allowed us to determine that PTH1R expression was not affected by up to 2 µg of RAMP2SNAP complementary DNA (cDNA) (corresponding to a 1:2 PTH1R:RAMP2 transfection ratio), and a 1:1 transfection ratio (PTH1R:RAMP2) was used for all subsequent experiments unless otherwise noted (SI Appendix, Fig. S2 A and B). Further control experiments showed that neither cell-surface expression of PTH1RFRET (measured with enzyme-linked immunosorbent assay [ELISA] via detection of an HA-tag in the PTH1RFRET; SI Appendix, Fig. S2C) nor the total expression of PTH1RFRET (measured by direct excitation of mC in the PTH1RFRET; SI Appendix, Fig. S2D) was affected by the expression of RAMP2, either in its wild-type (WT) or in its SNAP-tagged form.
We then measured the changes in FRET of PTH1RFRET evoked by different concentrations of PTH, initially by manual addition in a microtiter plate format. As in similar GPCR biosensors, agonists evoked a decrease of FRET, presumably induced by a movement of the third intracellular loop away from the C terminus, which is thought to cause an increased distance between the two fluorophores in the biosensor (11, 15–17). Fig. 2B shows the time courses of the PTH-induced decrease in the FRET ratio in control (black) and cells transfected with 0.5 or 1 µg of cDNA encoding for RAMP2SNAP (blue). The amplitude of this decrease was smaller in RAMP2SNAP-expressing cells than in control cells at all concentrations of PTH (Fig. 2 B and C), while the potencies of PTH were not different between the two conditions (Fig. 2C). We observed that higher expression levels of RAMP2SNAP (0.5 vs. 1 µg) caused a more significant reduction of the amplitude of PTH-induced PTH1R activation (Fig. 2B and SI Appendix, Fig. S2E). These effects were the same whether SNAP-tagged or WT RAMP2 was used (SI Appendix, Fig. S2F).
Additionally, we evaluated whether also RAMP1 and RAMP3 might modulate PTH1RFRET ligand-induced activation, despite their poor interaction with the PTH1R (Fig. 1C). We therefore transfected a stable cell line expressing PTH1RFRET biosensor with 1 µg of cDNA encoding for the different RAMPSNAP isoforms. We found that neither RAMP1SNAP nor RAMP3SNAP was prominently modulating the amplitude of the PTH1RFRET activation induced by PTH or PTHrP (SI Appendix, Fig. S3 B and C). However, in the same experimental setup, RAMP2SNAP moderately but significantly decreased the PTH- but not the PTHrP-induced activation of PTH1RFRET, suggesting an agonist-specific effect of RAMP2 on PTH1R modulation. Control experiments showed that PTH1RFRET biosensor expression was comparable across all tested groups (SI Appendix, Fig. S3A)
To assess whether the decrease in the amplitude of the PTH-induced FRET signal by RAMP2 might be caused by a change in the initial conformation and, hence, basal FRET of the biosensor, we performed photobleaching experiments of PTH1RFRET in the absence or presence of RAMP2SNAP. We observed that under basal conditions the FRET efficiency was significantly higher in the absence than in the presence of RAMP2, with no difference between C-terminally labeled RAMP2SNAP or WT RAMP2 (Fig. 2D).
After 5 min of stimulation with a high concentration (100 µM) of PTH, a similar pattern was observed, i.e., the FRET efficiency was higher in the absence than in the presence of RAMP2 (Fig. 2E). Again, there was no difference between WT RAMP2 and RAMP2SNAP, indicating that the two could be used interchangeably and that a SNAP-tag on the C terminus did not affect the effect of RAMP2 on PTH1R. Interestingly, the RAMP2-induced decrease in FRET efficiency was smaller in the PTH-activated state than under basal conditions (Fig. 2 E vs. D). Similarly, the PTH-induced decrease in FRET was smaller in the presence of RAMP2 than in its absence (Fig. 2F).
Control experiments showed that PTH1RFRET biosensor expression was comparable across all tested groups (SI Appendix, Fig. S4A), as was the amount of bleaching in each group (SI Appendix, Fig. S4B). Additionally, a hyperbolic increase of FRET efficiencies at increasing acceptor concentrations as determined by prebleached emissions demonstrated that FRET was specific (15) (SI Appendix, Fig. S4C).
Taken together, these data indicate that RAMP2 modulates the conformation of the PTH1RFRET biosensor: It decreases FRET in the basal state and, less so, in the PTH-activated state, and it decreases the PTH-induced FRET signal. A possible explanation for these findings is that RAMP2 induces a kind of preactivation of PTH1RFRET, characterized by decreased basal FRET.
RAMP2 Modulates the Activation Speed and the Amplitude of the PTH1RFRET Biosensor.
To assess whether the interaction of RAMP2 with PTH1R might modulate the activation kinetics, we performed experiments with a rapid superfusion system (11), using a stable cell line expressing PTH1RFRET (Fig. 2A). Transient coexpression and visualization of RAMP2SNAP were performed as described above.
Stimulation with a saturating concentration of PTH (10 µM), in the presence and absence of RAMP2SNAP, resulted in a rapid decrease of the FRET ratio (Fig. 3 A and B), characterized by antiparallel changes of donor and acceptor emission channels (SI Appendix, Fig. S5). The amplitude of the FRET change, similar to microtiter plate and photobleaching experiments (Fig. 2), was about twofold higher in the absence than in the presence of RAMP2SNAP (Fig. 3D).
Fig. 3.
Modulatory effects of RAMP2SNAP coexpression on PTH1RFRET and PTH1RcpGFP biosensor activation. (A) Schematic representation of intramolecular PTH1RFRET biosensor. To control RAMP2 expression, its C-terminally fused SNAP-tag was labeled with 1 µM SNAP-Cell SiR-647 (red circle). (B) Representative ratio traces of PTH-mediated FRET changes in single HEK293 cells stably expressing PTH1RFRET and in the presence of transiently coexpressed RAMP2SNAP, recorded in a microscopic FRET setup. To analyze cells that expressed both, cells were labeled with 1 µM SNAP-Cell SiR-647, and regions of interest were selected where PTH1RFRET and RAMP2SNAP were coexpressed. Horizontal lines indicate the application of 10 μM PTH with a rapid superfusion system. Traces were normalized to the baseline (set to 1) and plateau after stimulation (set to 0). FRET ratio traces are raw (gray and light blue) and Fourier low-passed (black, dark blue). Traces are representative of n = 41 cells (PTH1RFRET) and n = 46 cells (+RAMP2SNAP), acquired in at least five independent experiments. (C) Time constants τ of PTH-induced FRET changes derived from traces as in B, calculated from monoexponential curve fitting. The data were fitted with a lognormal distribution, and the dashed line indicates mode, global maximum of the distribution for PTH1RFRET = 410 ms; +RAMP2SNAP = 50 ms. Median value and 95% CIs for τ were PTH1RFRET = 710 ms [516, 946 CI], n = 41 cells; RAMP2SNAP = 330 ms [144, 416 CI], n = 46 cells. A Mann–Whitney U test was used to assess a significant difference between the groups (P < 0.001). (D) Amplitude of FRET changes induced by PTH obtained from traces as in B. Bars represent means ± SEM, percent of the FRET amplitudes from all examined cells: PTH1RFRET = 7.5 ± 0.5%, PTH1RFRET+RAMP2SNAP = 3.8 ± 0.3%. A t test was used to assess a significant difference between the groups (P < 0.001). (E) Schematic representation of the single-color biosensor to monitor PTH1R activity in single-cell experiments. Receptor activation upon agonist stimulation was monitored by recording fluorescence at 516 nm. (F and I) Representative fluorescence traces of PTH (F) and PTHrP (I) mediated changes in ΔF/F0 recorded in a microscopic FRET setup in single HEK293 cells transiently expressing PTH1RcpGFP alone or with RAMP2SNAP, labeled with 1 µM SNAP-Cell SiR-647. Horizontal lines indicate the application of 10 μM PTH or PTHrP with a rapid superfusion system. (G and J) Time constants τ of PTH-induced and PTHrP-induced activation derived from traces as in F and I. The data were analyzed as described in C. Dashed line indicates mode, global maximum of the distribution: PTH1RcpGFP = 760 ms and PTH1RcpGFP+RAMP2SNAP = 190 ms. Median value and 95% CIs for τ for PTH: PTH1RcpGFP = 950 ms [817, 1057 CI], n = 78 cells; PTH1RcpGFP+RAMP2SNAP = 400 ms [322, 448 CI], n = 75 cells. A Mann–Whitney U test was used to assess a significant difference between the groups (P < 0.001). PTHrP: PTH1RcpGFP = 1,960 ms [1,770, 2,660 CI], n = 38; PTH1RcpGFP+RAMP2SNAP = 1,910 ms [1,670, 2,100 CI], n = 41. (H and K) Effects of RAMP2 on the amplitude of the ΔF/F0 signals induced by PTH (F) and PTHrP (H). Bars represent means ± SEM in percent of the ΔF/F0 amplitudes from all cells examined: PTH: PTH1RcpGFP = 15.3 ± 1.1% (n = 78 cells); PTH1RcpGFP+RAMP2SNAP = 9.1 ± 1.1%, (n = 77 cells), from at least five independent experiments. PTHrP: PTH1RcpGFP = 17.5 ± 1.8% (n = 38 cells); PTH1RcpGFP + RAMP2SNAP = 13.6 ± 1.4% (n = 41 cells), from at least two independent experiments. A t test was used to assess a significant difference between the groups (P < 0.001).
Activation time constants (τ) were calculated by monoexponential curve fitting. In accordance with earlier data (11), PTH activated PTH1RFRET with a median time constant of 710 ms (Fig. 3C). However, when RAMP2SNAP was coexpressed, the PTH-induced activation was twice as fast with a median τ value of only 330 ms (Fig. 3C). Fig. 3C shows the distribution of the time constants under the two conditions, which peak at 410 ms and 50 ms, respectively.
Control experiments showed that neither membrane expression of the PTH1RFRET biosensor measured with ELISA via detection of the HA-tag in PTH1RFRET (SI Appendix, Fig. S2C) nor the total expression of PTH1RFRET measured as direct emission of biosensor’s acceptor was affected by RAMP2 (SI Appendix, Fig. S2D). This excluded the possibility that differences between the control and RAMP2 group were due to different expression levels of the biosensor.
Overall, these results indicate that RAMP2 has two distinct effects on receptor activation as observed with PTH1RFRET: It increases the speed severalfold and reduces the amplitude (from a lower starting value) by approximately twofold.
A Single-Color PTH1R Biosensor Confirms the Modulatory Role of RAMP2.
To further substantiate our results, we generated an orthogonal PTH1R biosensor based on a single fluorophore, circularly permuted green fluorescent protein (cpGFP). This approach was initially developed to visualize fast calcium dynamics (36) and neurotransmitter release—processes with subsecond time courses (37). We generated a PTH1RcpGFP biosensor by inserting a cpGFP module with linkers into the third intracellular loop (Fig. 3E), similar to the donor insertion position in PTH1RFRET. In preliminary experiments in microtiter plate format, PTH1RcpGFP transiently expressed in HEK293 cells showed a marked increase in fluorescence in response to agonist activation, which occurred with potency (SI Appendix, Fig. S6B) similar to that in WT PTH1R (11) or PTH1RFRET (Fig. 2C and SI Appendix, Fig. S2F). Again, we monitored coexpression of RAMP2SNAP via a C-terminal SNAP-tag and analyzed only cells that expressed both PTH1RcpGFP and RAMP2SNAP (Fig. 3E).
We then performed single-cell experiments with these cells, applying agonists via a fast perfusion system. Ten micromolar PTH evoked an increase in fluorescence in control and RAMP2-expressing cells (Fig. 3F). In agreement with results obtained with the PTH1RFRET biosensor, the amplitude of the signal and also the speed of the activation process were affected by RAMP2; in particular, RAMP2 decreased the amplitude (ΔF/F0) and increased the speed of activation by PTH (Fig. 3 G and H). The time constant (τ) of activation was decreased from 950 ms to 390 ms and the peak of the τ-value distribution from 760 ms to 190 ms by the presence of RAMP2.
Interestingly, similar experiments with the second endogenous agonist, PTHrP, revealed that these effects were agonist-specific: Using PTHrP (10 µM) as the agonist, no significant differences in the amplitude (ΔF/F0) or the time constant (τ) were detected between control and RAMP2-coexpressing cells (Fig. 3 I–K and SI Appendix, Fig. S3C). These results suggest that RAMP2 modulation of PTH1R is agonist-specific.
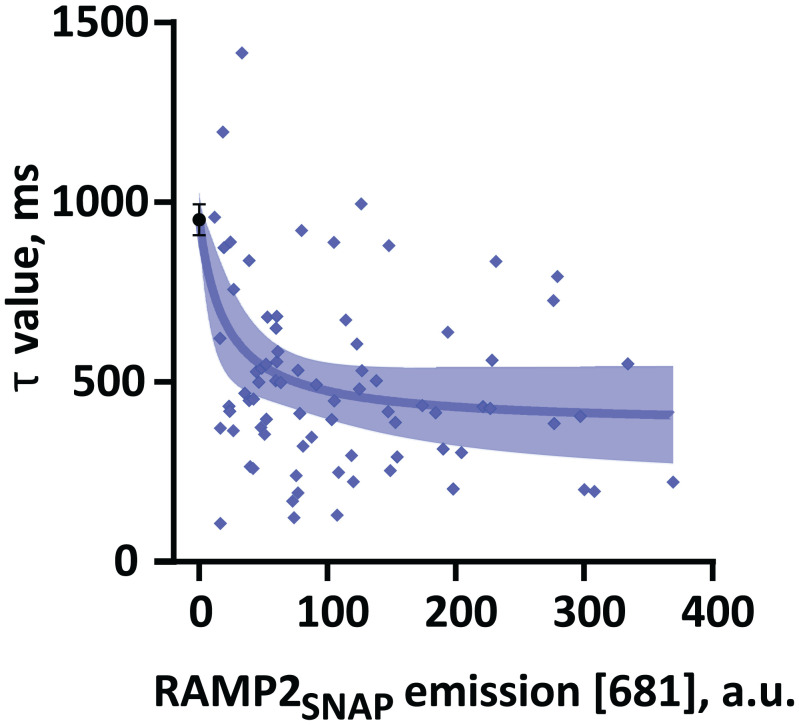
To quantitatively analyze the effect of RAMP2 expression on PTH-induced PTH1R kinetics, we plotted the time constant τ for each cell as a function of RAMP2 expression visualized via SNAP labeling. Fig. 4 shows that the time constant τ of PTH-induced PTH1R activation decreases with the expression level of RAMP2SNAP. These data suggest that modulation of PTH1R amplitude of activation (Fig. 2B and SI Appendix, Fig. S2E) and time constant (Fig. 4) by RAMP2 depends on its expression level (Fig. 4) in a saturable manner.
Fig. 4.
Effects of RAMP2 expression levels on the modulation of PTH1R activation dynamics. Time constants τ of PTH-induced ΔF/F0 changes in single HEK293 cells transiently expressing PTH1RcpGFP alone (black) or with RAMP2SNAP (blue). Data from Fig. 3G were analyzed for the time constant τ as a function of RAMP2SNAP expression, quantified by labeling with 1 µM SNAP-Cell SiR-647. Data were fitted to a three-parameter logistic function. Shown is fit ± 95% CI. a.u., arbitrary units.
Ligand-Specific Effects of RAMP2 on G Protein Activation by PTH1R.
Since RAMP2 appears to change the PTH1R conformation, resulting in faster agonist-specific activation, we wondered whether it might also affect downstream signaling by the PTH1R. To test this hypothesis, we used a suite of bioluminescence resonance energy transfer (BRET)- and FRET-based biosensors to quantify the effect of PTH1R activation on different signaling pathways, using both PTH and PTHrP.
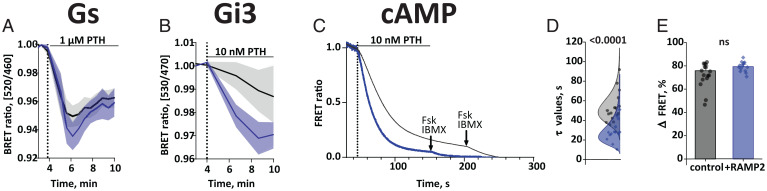
We first compared the ability of PTH1R to activate different G proteins, using BRET-based biosensors (38), which respond to PTH1R activation with a decrease in BRET between their Gγ subunit labeled with the bioluminescent donor NanoLuc and their Gα subunits tagged with the acceptor cpVenus. HEK293 cells were transiently transfected with the specific BRET biosensor along with PTH1Rwt with or without RAMP2wt, as described above. Experiments were conducted in microtiter plates, and BRET signals were recorded over time until they reached their maximal response.
The values of basal BRET ratio of all four G protein biosensors were similar in control and RAMP2-expressing cells, indicating that the RAMP2-induced changes of the PTH1R did not translate into a direct activation of G proteins (SI Appendix, Fig. S7 A–D). Significant PTH-induced changes in BRET were observed with all four G protein biosensors. They were all concentration-dependent with EC50 (half maximal effective concentration, 50%) values in a range reflecting G protein preferences of this receptor in the order Gs > Gq > G13 > Gi3 (SI Appendix, Fig. S8, Upper and Table S1).
Two changes in the G protein activation patterns were notable. First, the presence of RAMP2wt caused more rapid activation of Gs by PTH with a brief initial overshoot peaking at 2 min after receptor activation (Fig. 5A); when measured at this time point, the presence of RAMP2wt increased the PTH-induced BRET change (Fig. 5A). Second, RAMP2wt caused a specific increase in potency for PTH-triggered Gi3 activation, resulting in a significant difference of Gi3 activation by low concentrations of PTH (Fig. 5B and SI Appendix, Fig. S8G and Table S1); at 10 nM PTH, the presence of RAMP2wt markedly accelerated Gi3 activation (Fig. 5B). In contrast, the potencies, and efficacies of PTH-stimulated activation for Gq and G13 were not affected by RAMP2 (SI Appendix, Fig. S8, Upper and Table S1).
Fig. 5.
RAMP2 effects on PTH-stimulated G protein activation and cAMP accumulation. (A and B) HEK293 cells transiently transfected with cDNA encoding for BRET biosensors of G proteins: Gs (A) and Gi3 (B) along with PTH1Rwt, with or without RAMP2wt. BRET signals were recorded in a plate reader from cells stimulated with the indicated concentrations of PTH. Shown are time courses of agonist stimulation. Data are means ± SEM of at least three independent experiments performed in duplicates or more. For further statistics and concentration response curves see SI Appendix, Fig. S6 and Tables S1 and S2. (C) HEK293 cells transiently transfected with cDNA encoding for the cAMP-based FRET biosensor (Epac-SH187), along with PTH1Rwt, with or without RAMP2wt. Shown are representative ratio traces of PTH-mediated FRET changes in single HEK293 cells, recorded in a microscopic FRET setup. Horizontal line indicates application of 10 nM PTH with a rapid superfusion system. The arrow indicates addition of 10 µM forskolin and 100 µM IBMX after signal saturation. Traces were normalized to the baseline (set to 1) and plateau after stimulation with forskolin and IBMX (set to 0). Traces are representative of n = 16 cells (control) and n = 14 cells (+RAMP2wt), acquired in two independent experiments. (D) Time constants τ of PTH-induced FRET changes derived from traces as in C, calculated from monoexponential curve fitting. The data were fitted with a lognormal distribution. Median value and 95% CIs were: PTH1RFRET = 49 s [45, 62 CI], n = 16 cells; RAMP2SNAP = 28 s [23, 35 CI], n = 14 cells. A Mann–Whitney U test was used to assess a significant difference between the groups (P < 0.001). (E) Effects of RAMP2 on the amplitude of the FRET signals induced by PTH. Bars represent means ± SEM in percent of the ΔFRET amplitudes from all cells examined: Epac-SH187+PTH1Rwt = 75.8 ± 4.1% (n = 16 cells); Epac-SH187+PTH1Rwt +RAMP2wt = 79.3 ± 2.1%, (n = 14 cells). A t test was used to assess a significant difference between the groups (not significant [ns]: P > 0.05).
PTHrP elicited similar decreases in BRET for all G protein biosensors (SI Appendix, Fig. S8, Lower), which occurred with time courses similar to those for PTH. However, in contrast to PTH, the coexpression of RAMP2wt did not significantly change the potency or efficacy of PTHrP stimulation for any of the G proteins analyzed (SI Appendix, Fig. S8, Lower and Table S2).
To assess whether downstream effects corresponded to those seen at the G protein level, we also measured cAMP accumulation using Epac-SH187 cAMP biosensor (39). The amplitude of cAMP accumulation recorded in microtiter plates was similar in RAMP2wt-expressing cells and in control cells at all concentrations of PTH (Fig. 5E and SI Appendix, Fig. S8I). Strikingly, in line with the accelerated Gs activation, and in agreement with results obtained with the PTH1RFRET biosensor, the speed of cAMP accumulation measured at a single-cell level with a rapid superfusion system (11) was affected by RAMP2; in particular, RAMP2 accelerated the PTH-induced cAMP accumulation (Fig. 5 C and D). The time constant (τ) of activation was decreased from the median time constant 45 s to 25 s by the presence of RAMP2 (Fig. 5D).
RAMP2 Effects on Non-G-Protein Signaling.
In addition to the activation of G proteins, agonist-activated PTH1R is phosphorylated by G protein–coupled receptor kinases (GRKs) and then binds β-arrestins, thereby triggering receptor internalization and signaling by extracellular signal-regulated kinases (ERKs). The latter process appears to have different conformational requirements compared to G protein activation (21–23, 40). We therefore set out to also assess the effects of RAMP2 on these signaling mechanisms, employing various BRET and FRET biosensors to quantify GRK2 and β-arrestin2 recruitment to the PTH1R along with ERK activation. To monitor the recruitment of GRK2 and or β-arrestin2, we used BRET assays, in which PTH1R was tagged with the donor NanoLuc (PTH1RNanoLuc) and GRK2 and β-arrestin2 were tagged with yellow fluorescent protein (YFP) and mVenus, respectively. We measured BRET signals at the maximal response time after full agonist occupancy (Fig. 6 A, C, and E).
Fig. 6.
RAMP2 effects on β-arrestin recruitment. HEK293 cells were transiently transfected with cDNA encoding for β-arrestin2mVenus along with PTH1RNanoLuc (A–D) or β2ARNanoLuc (E and F), with or without RAMP2wt. BRET signals were recorded in a plate reader from cells stimulated with PTH (A and B), PTHrP (C and D), or isoprenaline (E and F). Shown are averaged time courses of agonist stimulation (A, C, and E) and corresponding concentration-response curves (B, D, and F), fitted with a three-parameter concentration–response curve fit. ΔBRET values were calculated in saturation and represent maximal change in response from the initial BRET value. Data are means ± SEM of at least n = 3 independent experiments performed in quadruplicates or more. For further statistics and results see SI Appendix, Tables S3 and S4.
The main change induced by RAMP2 in this series of experiments was a marked increase in β-arrestin2 recruitment, visualized as a significant increase in the amplitude of BRET ratio for both PTH and PTHrP (Fig. 6 A and C and SI Appendix, Tables S3 and S4). This increase was visible at all concentrations of PTH and PTHrP (Fig. 6 B and D). Control experiments showed that RAMP2 did not affect β-arrestin2 recruitment in the absence of receptor stimulation (SI Appendix, Fig. S9J). Further control experiments indicated the specificity of the effects of RAMP2 because it did not alter β-arrestin2 recruitment to the β2-adrenergic receptor (Fig. 6 E and F and SI Appendix, Table S5), which does not interact with RAMP2 (29, 33) (Fig. 1C).
In contrast to these major and very robust effects on β-arrestin2 recruitment, there were only minor or no effects on GRK2 recruitment and ERK activation (SI Appendix, Fig. S10 and Tables S3 and S4).
Taking all data on PTH1R signaling together, we demonstrate two significant effects of RAMP2: 1) a PTH-selective increase in the speed of stimulating Gs and potency of Gi3 activation (Fig. 5 and SI Appendix, Fig. S8) and 2) an increase in β-arrestin2 recruitment which is seen for both agonists (Fig. 6). Interestingly, the latter effects are not translated into increased nuclear ERK signaling by PTH1R. However, the speed of the increased recruitment of β-arrestin2 corresponds to the kinetics of the overshoot in Gs activation, in line with the role of β-arrestins to limit G protein activation.
Structural Models of Putative PTH1R–Ligand–RAMP2–Gs Complexes.
The modeling approach carried out here resulted in two different proposals for the complex formation of RAMP2 with PTH1R and PTH ligand variants (SI Appendix, Fig. S14A). In the model version I, the RAMP2–ECD (Extra-cellular domain) is bound to the PTH1R–ECD, as suggested by the known CLR–CGRP–RAMP1 complexes (41), but the overall receptor–ECD orientation to the transmembrane (TM) region is maintained according to the known PTH1R–LA–PTH–Gs complex (16). In this scenario—with unchanged complex arrangement and ligand conformation, but additionally bound RAMP2—no significant changes in ligand binding or inter- or intramolecular interactions are apparent despite a few receptor–RAMP2 interactions mainly in the TM region.
In contrast, taking the previously solved CLR–CGRP–RAMP1–Gs complex (41) as a structural template for modeling a putative complex between PTH1R–PTH/PTHrP–Gs with RAMP2 (SI Appendix, Fig. S14B), several highly relevant structural parts are potentially altered with modified interaction patterns both intra- and intermolecular compared to the known PTH1R–LA–PTH complex (16). The linker region of RAMP2 (e.g., F138 and D140) would interact with the C terminus of PTH1R EL2, in addition to specific RAMP2–ECD to PTH1R–ECD contacts (e.g., receptor–RAMP2: Q45–R97). The PTH1R ECD can contact EL3 (e.g., E431), which is expected to directly affect neighboring helices TM6 and TM7, which are known to be important for signal transduction and regulation. Furthermore, the ligand–receptor interaction pattern is modified in this model (Fig. 7), assuming additional contacts (e.g., receptor–PTH: D133–K13).
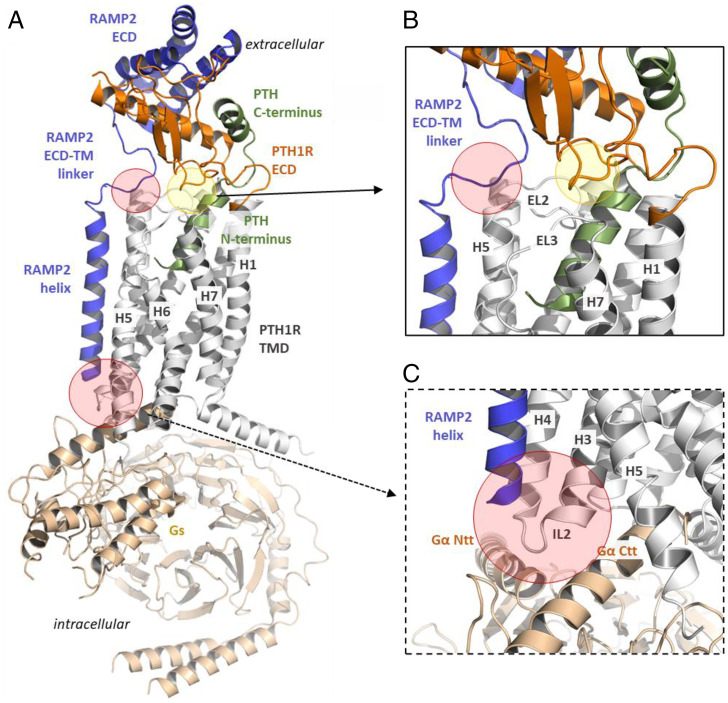
Fig. 7.
Putative RAMP2 binding mode in a PTH1R–PTH–Gs complex model. (A) A homology model between PTH1R–PTH–Gs and RAMP2 suggests several specific contacts between the components of this complex. RAMP2 should interact at the extracellular region with the EL2 of the receptor (B) but also at the intracellular site with the IL2 and adjacent transitions to receptor helices 3 and 4 (C) (red translucent circles), a region that is associated with G protein binding (Ntt, N-terminal tail; Ctt, C-terminal tail). There are also new intramolecular contacts from the PTH1R ECD to the TMD (transparent green circle), which are not observable in the recently determined PTH1R complex structure (16) (Protein Data Bank ID code: 6nbh). In this model the receptor ECD would interact directly with the EL3 but potentially also with EL1. In addition, several new receptor ECD–ligand contacts are feasible, such as K13 (PTH) and D133 of the receptor.
Discussion
RAMPs have coevolved and are coexpressed with several GPCRs (28, 33, 42, 43). More than 40 partner proteins have been described to interact with them (29, 44–46). The effects of such interactions have been studied in most detail for their prototypical interaction partners—the class B GPCRs calcitonin and calcitonin-like receptors (CTR and CTRL) (41). RAMP’s binding to these receptors has been described to facilitate the transport of the receptors to the cell surface, to change ligand specificity, and also to alter their downstream signaling cascades (47). However, little is known about how RAMPs might affect the activation process of a GPCR itself.
Here, we investigated possible modulatory effects using the class B PTH1R as a model system. Coexpression of RAMP isoforms with PTH1R in HEK293 cells mimics the physiological context in which both receptor and RAMPs are abundantly expressed (30); these conditions make a confounding contribution of the low levels of endogenous RAMPs in HEK293 cells negligible. Among the three RAMPs, this receptor showed a clear preference for RAMP2. Here, RAMP2 increased the activation speed of the PTH1R severalfold, and it reduced the amplitude of the activation signal by approximately twofold. Such effects are dependent on the RAMP2 expression level in a saturable manner. Both effects were seen with a FRET sensor as well as a new cpGFP-based biosensor with similar effect sizes. In line with the smaller signal amplitude, basal FRET of the FRET biosensor was decreased by RAMP2, i.e., altered in a way similar to the effects of (partial) agonists. This might suggest that RAMP2 induced a partially preactivated state, from which agonist-induced activation may proceed with much greater speed. Based on the various fluorescence and FRET readouts, this preactivated state appears to be distinct both from the inactive and from the fully active state. RAMP2 changed the basal and stimulated conformations of the PTH1R. However, it did not evoke any detectable change in the basal BRET ratio of G protein biosensors or other downstream effectors. We interpret this lack of change in basal activity of G protein as evidence for a RAMP2-induced shift of the PTH1R into a conformation that is more easily converted into an active state, but which per se is not recognized as “active” by any of the G proteins analyzed with our currently available biosensors.
Interestingly, observed effects were ligand-specific: They were prominent for PTH but essentially absent for PTHrP. This was true for both the increase in activation speed and the decrease in amplitude of the activation signals. It suggests, first, that the two endogenous ligands can be regulated in a differential manner and, second, that RAMP2 can exert very specific and subtle effects on PTH1R. Such specific conformational changes are somewhat reminiscent of analogous kinetic effects that have been observed in homo- and heterodimeric GPCRs (13, 15). Ligand-dependent effects were previously described for some with RAMPs-interacting GPCRs (41, 48).
RAMP1–CTRL heterooligomers have been shown to RAMP-specifically propagate extracellular dynamics to the cell interior and, through that, to control receptor phenotypes (49). Structurally, all RAMPs are tightly packed with their interacting GPCR partner, being placed between transmembrane domains 3, 4, and 5 and making contacts with the second extracellular loop (EL2) (41, 49). Thus, RAMPs are placed near structural motifs that govern GPCR activation.
In an attempt to interpret our data in a structural manner, we performed structural homology modeling of PTH1R–ligand–RAMP2–Gs complexes to suppose potential intermolecular interactions, preactivation, and possible impact on the intracellular signaling of PTH1R–RAMP2 oligomer (Fig. 6A). Starting from available structural information (16, 41, 49) (SI Appendix, Figs. S12–S14), our model provides indications that RAMP2-mediated preactivation might originate from the interaction of the RAMP2 linker (Fig. 7B) with the receptor EL2 (upper part of TM5) and ECD (red circle), which is additionally in contact with the EL3 (green circle) that connects TM6 and TM7. Mutations in this interaction may offer a model-based approach to assess further the contribution of these postulated interactions for preactivation of the PTH1R and their impact on downstream signaling. In addition, presumed intramolecular interactions in the ECD and EL3 receptors (Fig. 7B, green circle) that may contribute to preactivation can also be tested by exchanging contacting amino acids, which could lead to “silencing” effects or an increase in preactivation. The extracellular contributions of intra- and intermolecular contacts might cause preactivation or stabilize a preactivated conformation of the receptor, comparable to scenarios known from several GPCRs with “intramolecular agonists” (50). In this case, the binding of RAMP2 would act as an initial (partial) activation trigger. Moreover, the EL2 of PTH1R has been recognized as an allosteric hotspot, where selective modulation or mutational alterations can affect the balance between G protein coupling and β-arrestin–driven signaling (51, 52). In addition, the RAMP2 helix contacts the receptor IL2 (Fig. 6C, red circle) and the adjacent connections to TM3 and TM4 intracellularly. This receptor part is highly interrelated with G protein binding, and thus any modification by RAMP2 binding should result in altered functional receptor properties even in the basal state. IL2 has been shown to predispose to constitutive receptor activation in several class A GPCRs, as demonstrated by mutagenesis studies (53, 54) and may also be highly relevant in class B.
In line with the predicted altered G protein function, we observed a number of remarkably specific changes on PTH1R downstream signaling by RAMP2. First, we found that RAMP2 caused a specific and selective increase in Gs and Gi3 activation kinetics by PTH, suggesting selectivity in the modulation of G protein coupling. Again, this effect was ligand-specific, being much more pronounced for PTH than for PTHrP.
Among the non-G-protein interactions of the PTH1R, we observed a substantial and specific increase in β-arrestin2 recruitment by RAMP2; this recruitment paralleled the overshoot in Gs activation, suggesting that it might limit Gs activation and signaling. In contrast, increased β-arrestin2 recruitment was not translated to other β-arrestin–dependent cascades such as nuclear ERK activation. It remains to be seen whether the increased β-arrestin2 recruitment BRET signal is due to an increased amount of β-arrestin2 recruited, or to a different state of β-arrestin2 induced by the PTH1R/RAMP complex compared to PTH1R alone (55, 56), and whether β-arrestins are somehow shielded from their downstream interaction partners by RAMP2. Since specific β-arrestin–dependent transcription programs have been identified for the PTH1R, which might provide a therapeutically interesting pathway to increase bone mass (22, 57), RAMP2-dependent modulation of these pathways may provide a new type of pharmacological target. There are several tissues in which both PTH1R and RAMP2 are highly coexpressed (i.e., lungs, kidneys, and placenta)—mimicked by our experimental setup—however, there are also tissues in which PTH1R is expressed without RAMP2 (i.e., pancreas). Since PTH1R/RAMP2 complexation is not obligate but tissue-dependent, this mechanism might be pharmacologically attractive and might represent a source of unique, tissue-specific biased signaling patterns. Such a unique pharmacological targeting approach was used during the development of erenumab, an antibody that specifically targets the CTRL/RAMP interface and indicates that a GPCR/RAMP interface can be exploited as a pharmacological target (47, 57, 58).
In summary, our data, together with recent structural insights, highlight a unique conformation of the PTH1R when interacting with its regulator RAMP2. This specific conformation is not recognized as active by G protein but promotes faster and ligand-specific activation of PTH1R and controls its signaling specificity. These data illustrate the critical role of RAMP in PTH1R activation and signaling.
Materials and Methods
Chemicals.
The peptide ligands parathyroid hormone PTH (1–34) (human, H-4835-GMP, 4033364) and parathyroid hormone-related peptide (PTHrP) (1–34) (human, mouse, rat; 4017147) were from Bachem. SNAP-Cell 647-SiR (S9102S) was from New England Biosciences. Anti–HA-tag antibody (ab9110) was from Abcam and anti-rabbit IgG, horseradish peroxidase (HRP)-linked antibody (7074P2) was from Cell Signaling. The 3,3′,5,5′-tetramethylbenzidine (TMB, T8665) was from Sigma-Aldrich. NanoBRET Nano-Glo Substrate–furimazine (N1663) and HaloTag NanoBRET 618 Ligand (G9801) were from Promega. Dimethyl sulfoxide (A994.2) for cell culture was from Carl Roth GmbH & Co. KG. Bovine serum albumin (BSA, SAFSA7030) was from VWR International.
Molecular Cloning.
All PTH1R-based constructs were cloned from human full-length PTH1R. Plasmids were either created by molecular restriction cloning or by the Gibson Assembly technique (New England Biolabs, Inc.).
HA-PTH1RmTurquoise/mCitrine (PTH1RFRET) and HA-PTH1RmCitrine were modified from previously described biosensors (11, 34). For HA-PTH1RNanoLuc, NanoLuc was fused to the C-terminal of HA-PTH1Rwt (59).
PTH1RcpGFP biosensor was cloned into pCMV Twist vector and designed according to the previously described dLight1 cpGFP biosensor (36) and synthesized by Twist Bioscience. Influenza A signaling peptide (MKTIIALSYIFCLVFADYKDDDDA) was fused to the N terminus of PTH1R, and LSSLI-cpGFP-NHDQL was inserted between Lys388 and Arg400 in the third intracellular loop.
WT RAMP constructs were a gift from Annette Beck-Sickinger, University of Leipzig, Leipzig, Germany. RAMPmCitrine and RAMPSNAP were generated by fusing mCitrine or SNAP-tag to the C-termini of RAMPs. SNAP-tag sequence was amplified from a SNAP-GABAB1 receptor template, kindly provided by Jean-Philippe Pin, Institut de Génomique Fonctionnelle, Montpellier, France. The C terminus of the CAAX sequence was tagged with mCitrine or mTurquoise2. Nuclear EKAR (Cerulean-Venus) was a gift from Karel Svoboda, Janelia Research Campus, Ashburn, VA; Addgene plasmid 18682) (60); biosensor Epac-SH187 was a gift from Kees Jalink, The Netherlands Cancer Institute, Amsterdam, The Netherlands (39); and GRK2YFP (61) was described previously. For β2ARmCitrine and β2ARNanoLuc, mCitrine and NanoLuc were fused to the C terminus of β2AR, respectively. β-arrestin2mVenus was modified from previously described β-arrestin2EYFP (62) by exchanging EYFP for mVenus.
The expression vector in all plasmids was pcDNA3(+) unless otherwise noted.
All constructs were verified by sequencing by Eurofins or LGC genomics.
Cell Culture.
Different clones of human embryonic kidney cells (HEK293) were employed. HEK293 (ECACC 85120602, CRL-1573, ATCC) was used for the generation of a stable cell line, HEK293T for most experiments (ECACC 96121229, Sigma-Aldrich) and HEK293A (R70507, Thermo Fisher) for plate reader experiments of G protein activation. We refer to all three clones of HEK293 cells as HEK293 in the main text.
Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Pan Biotech) supplemented with 2 mM l-glutamine (Pan Biotech), 10% fetal calf serum (Biochrome), 100 μg/mL streptomycin, and 100 U/mL penicillin (Gibco) at 37 °C with 5% CO2. Cells were washed with phosphate-buffered saline (PBS, Sigma-Aldrich) and passaged with 0.05%/0.02% trypsin/ethylenediaminetetraacetic acid (Pan Biotech) every 2 to 3 d when reaching 80% confluency. Cells were routinely tested for mycoplasma infection using MycoAlert Mycoplasma Detection Kit (Lonza). Cells were not contaminated with mycoplasma.
Creation of the Stable Cell Line.
HEK293 cells were used to develop a stable cell line of PTH1RFRET biosensor. Cells seeded into 100-mm dishes were transfected at a confluence of 60% with 2 µg of cDNA encoding PTH1RFRET with Lipofectamine 3000 Transfection Reagent Kit (Qiagen), according to the manufacturer’s protocol. Transfected clones were selected with 600 μg/mL G-418 (VWR International) and sorted with a flow cytometer. Monoclonal single clones were grown in DMEM supplemented with 200 μg/mL G-418. The best clone was selected for further experiments based on the brightness and amplitude of the saturating PTH stimulation of the PTH1RFRET biosensor in plate reader experiments.
Seeding and Transfection.
Coverslips or microtiter plates were covered with poly-d-lysine (PDL) for 30 min, washed two times with PBS, and left to dry before seeding. For microscopy experiments, 2 × 105 cells were seeded onto 25-mm coverslips (Sigma-Aldrich) into a six-well plate. After 24 h, cells were transfected with Lipofectamine 3000 (Qiagen), according to the manufacturer’s protocol. For all transfections, PTH1R:pcDNA3/RAMP ratio was 1:1, unless otherwise noted. The empty backbone of pcDNA3 was used throughout to maintain a consistent level of total cDNA. Each methods section contains a detailed description of seeding and transfection protocol for plate reader experiments.
FRET Acceptor Photobleaching (FRET-AB) in Confocal Microscopy.
Cells were imaged 36 h after transfection. Coverslips were mounted onto Attofluor chamber (Fisher Scientific) and washed once with FRET buffer (137 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 20 mM Hepes, pH 7.4) containing 0.1% (wt/vol) BSA (AppliChem). Cells were kept in the FRET buffer at room temperature throughout the experiment.
The chamber was mounted onto a Leica SP8 confocal laser-scanning microscope, equipped with an oil-immersion objective (HC PL APO CS2 40×/1.3 numerical aperture). LAS X microscope control software and the Leica FRET-AB wizard tool were used to perform experiments. A 1.5-mW white-light laser was set to 1%, and a 431-nm laser line was used at 1% power for donor imaging. For acceptor imaging, a 512-nm laser line at 1% power was used, and for the bleaching step increased to 100% for 10 frames. The 512- × 512-pixel images were acquired with a hybrid detector in standard mode. Emission of donor channel was recorded within 440 to 512 nm, and emission of acceptor channel was recorded within 517 to 620 nm. The zoom factor was set to 5.5×, resulting in a pixel size of 103 nm, and the laser scanning speed was set to 400 Hz. Fixed-size regions of interest (ROIs) were selected on the cell membrane. For intramolecular FRET-AB experiments, ROIs expressing both PTH1RFRET and RAMP2SNAP were selected.
SNAP-Tag Labeling.
Before FRET experiments, coverslips expressing a combination of PTH1RFRET and RAMP2SNAP were labeled with 1 µM SNAP-Cell 647-SiR in serum-free Fluorobrite DMEM (Gibco) for 30 min and kept in the incubator at 37 °C and 5% CO2. Excessive dye was washed by exchanging medium three times every 10 min.
Single-Cell PTH1R Biosensor Experiments in Intact Cells.
Cells were imaged 36 h after the transfection. Coverslips were mounted onto an Attofluor chamber and washed once with FRET buffer. Cells were kept in FRET buffer at room temperature throughout the experiment. The chamber was mounted onto an inverted microscope (DMi8, Leica Microsystems), equipped with an oil-immersion objective (HC PL APO 63×/1.40–0.60 oil, Leica Microsystems), dichroic beamsplitter T505lpxr (Visitron Systems), and xenon lamp coupled with a continuously tunable Visichrome high-speed polychromator (Visitron Systems). Images were acquired with a scientific complementary metal–oxide–semiconductor camera (Prime 95B, Teledyne Photometrics) using a dual image splitter (OptoSplit II, Cairn Research). Image sequences had 40-ms (PTH1RFRET) or 100-ms (Epac-SH187) acquisition intervals and were recorded with the VisiView 4.0 software (Visitron Systems). Ligand application was performed using a solenoid valves perfusion system with a 200-µm inner diameter manifold-tip (Octaflow II, ALA Scientific Instruments).
To check for RAMP2SNAP-expressing cells, cells were excited at 640 nm for 100 ms and fluorescence emission was recorded at 690/50 nm.
For FRET experiments, cells expressing PTH1RFRET or Epac-SH187 were excited with 445 nm, and fluorescence emission was simultaneously recorded at 470/24 nm and 535/30 nm. Cells expressing PTH1RcpGFP were excited at 483 nm and fluorescence emission was recorded at 506 nm.
Fluorescence Spectrum and Fluorescence Experiments in the Plate Reader.
HEK293T cells (3 × 106) were seeded into 100-mm dishes and transfected after 24 h with the combination of PTH1RFRET/PTH1RcpGFP and pcDNA3/RAMP2wt/RAMP2SNAP or PTH1Rwt, Epac-SH187/EKAR biosensor, and pcDNA3/RAMP2wt/RAMP2SNAP. Combinations were transfected at a ratio of 1:1 or 1:1:1, respectively. Twenty-four hours after the transfection, cells were transferred to PDL-precoated black-wall, black-bottomed 96-well plates (Brand) at a density of 70,000 cells per well. Thirty-six hours after the transfection, cells expressing biosensors were washed and medium was substituted with FRET buffer. Plate reader experiments were conducted at 37 °C using a Synergy Neo2 plate reader (BioTek) equipped with a monochromator and filter optics. Ten excitation flashes were applied per data point.
For the fluorescence emission spectrum of PTH1RcpGFP, cells were excited at 455/10 nm and fluorescence emission was recorded with 1-nm resolution within 500 to 660 nm.
For amplitude experiments and generation of concentration-response curves, basal reads for 5 min were recorded in 90 µL FRET buffer. Subsequently, 10 µL of 10-fold ligand solution or FRET buffer was applied to each well and the stimulated reads were recorded for a further 10 min. For FRET experiments, a 420/50-nm excitation filter and 485/20-nm and 540/25 nm dual-emission filter were used. For PTH1RcpGFP-expressing cells, 485/20-nm excitation and 516/20-nm emission filters were used.
For PTH1RFRET, EKAR or Epac-SH187 biosensor-expressing cells, expression levels were measured with monochromator optics. Cells were excited at 510/20 nm and fluorescence emission was recorded at 560/20 nm.
Live-Cell ELISA.
HEK293T cells (3 × 106) were seeded into 100-mm dishes and transfected 24 h later with a combination of PTH1RFRET and pcDNA3/RAMP2wt/RAMP2SNAP or PTH1RcpGFP and pcDNA3 (no HA-tag control) at a ratio of 1:1. The medium was exchanged after 12 h and 24 h after the transfection and the cells were transferred to PDL-precoated transparent 96-well plates (Brand) at a density of 70,000 cells per well. Forty-eight hours later, cells were washed two times with 0.5% BSA/PBS. Subsequently, cells were incubated for 1 h at 4 °C with rabbit anti-HA tag antibody (1:1,000) in 1% BSA/PBS. Following incubation, cells were washed four times with 0.5% BSA/PBS and incubated with goat anti-rabbit IgG, HRP-linked antibody (1:4,000) in 1% BSA/PBS for 1 h at 4 °C. Finally, cells were washed three times with 0.5% BSA/PBS, and 50 µL of the peroxidase substrate TMB was added. Following a 30-min incubation and development of a blue product, absorbance was recorded at 665 nm using a Neo2 plate reader.
BRET-Based G Protein Activation Assay.
HEK293A cells were transfected with PTH1Rwt, G protein BRET biosensor (63) and pcDNA3/RAMP2wt at a ratio of 1:1:1. Constructs were transfected in suspension with Lipofectamine 2000 (2 µL transfection reagent/1 µg total cDNA) according to the manufacturer’s protocol and seeded into a PDL-precoated, white-wall, white-bottomed 96-well microtiter plate (30,000 cells per well). Forty-eight hours after the transfection, cells were washed with Hanks’ balanced salt solution (HBSS) and incubated with 90 µL of a 1:1,000 (vol:vol) stock solution of furimazine in HBSS. Five minutes later, three consecutive reads were recorded as basal reads. Subsequently, 10 µL of a 10-fold ligand solution or HBSS was applied to each well and the stimulated reads were recorded.
All experiments were conducted using a CLARIOstar plate reader (BMG Labtech) recording NanoLuc and cpVenus emission with 450/80-nm (gain 3600) and 530/30-nm (gain 4000) monochromator settings, respectively, and an integration time of 0.3 s.
BRET-Based GRK2 Recruitment, β-Arrestin2 Recruitment, and Gs Protein Activation Assay.
HEK293T cells were transfected with GRK2EYFP, PTH1RNanoLuc and pcDNA3/RAMP2wt; β-arrestin2mVenus, PTH1RNanoLuc and pcDNA3/RAMP2wt or Gs protein BRET biosensor, PTH1Rwt and pcDNA3/RAMP2wt at a of ratio 1:1:1. Combinations were transfected with Lipofectamine 3000 according to the manufacturer’s protocol with a total of 6 µg of cDNA. After 12 h medium was exchanged and after 24 h cells were transferred into a white-wall, white-bottomed, 96-well microtiter plate, at a density of 60,000 cells per well. Twenty-four hours after the reseeding, the medium was removed, and cells were washed once with FRET buffer and incubated with 90 μL of a 1:1,000 (vol:vol) stock solution of furimazine in FRET buffer. Five minutes later, basal reads were recorded for 4 min and subsequently 10 µL of 10-fold ligand solution or FRET buffer was applied to each well and the stimulated reads were further recorded.
Measurements were performed at 37 °C using a Synergy Neo2 Plate Reader with the NanoBRET filter set, integration time per data point was set to 0.3 s and gain to 100/120 (GRK2 recruitment) or 90/110 (β-arrestin2 recruitment, Gs activation).
GRK2YFP- and βarr2mVenus-expressing cells were excited at 510/20-nm and fluorescence emission was recorded at 560/20 nm for quantification of expression level.
Data Analysis and Statistics.
For microscopic FRET experiments, fluorescence emission time courses of both FRET donor and acceptor were routinely corrected for background and spectral bleedthrough, and the FRET ratio was calculated as described earlier (11, 63). For calculating the time constant (τ), agonist-independent changes in FRET due to photobleaching were subtracted. The decrease in FRET ratio was fitted to the one-phase decay equation r(t) = A × (1 − e−t/τ), where τ is the time constant (s) and A is the amplitude. X0 was constrained to the time when the decay began. Δ FRET values were calculated as normalized differences between basal and stimulated FRET ratios.
For FRET acceptor photobleaching experiments, FRET efficiencies were calculated with the manufacturer’s Wizard tool, based on the provided Eq. 1 and previously described method (64), where I denotes the fluorescence emission intensity:
| [1] |
A maximum of four cells was taken for analysis per image. To ensure coexpression integrity and enough bleaching of the acceptor only cells with initial emission ratios (mCitrine/mTurquoise2) within 0.25 and 4, and bleaching >20%, were considered for statistical analysis.
For plate reader experiments, the data were analyzed in Microsoft Excel and, if needed, wells out of the fluorescence or bioluminescence range of plate readers were excluded as an outlier. For FRET and BRET experiments, raw RET ratios were defined as acceptor emission/donor emission. RET ratios before ligand/buffer addition were averaged and defined as RETbasal. To quantify ligand-induced RET changes, ΔRET was calculated for each well and time point as percent over basal ([(RETstim− RETbasal)/RETbasal] × 100). Subsequently, the average ΔRET of buffer-treated control wells was subtracted. To reduce the fluctuation of the BRET ratio, three consecutive BRET ratios were averaged before and after ligand addition (65). Concentration–response curve experiments were fitted using a three- or four-parameter logistic curve fit as stated in corresponding figure legends.
Statistical differences were evaluated using a one-way ANOVA test followed by Tukey multiple comparisons, Brown–Forsythe ANOVA, followed by Dunnett’s T3 multiple comparisons test, Student’s t test, Mann–Whitney U test, or extrasum-of-squares F test. Each figure legend contains a description of statistical treatment. Differences were considered significant for values of P < 0.05. The data were analyzed and visualized using Microsoft Excel 2016 (Microsoft), GraphPad Prism software 8.1.2 (GraphPad Software), and OriginPro 2018 software (OriginLab).
Supplementary Material
Acknowledgments
We thank all members of the M.J.L. laboratory, especially Ali Isbilir and Jan Möller, for valuable discussions on the manuscript. G.K. and P.S. were supported by the DFG - German Research Foundation through CRC 1423, project number 421152132, subprojects A01/A05/Z03; through CRC 1365, project number 394046635, subproject A03; through Germany’s Excellence Strategies − EXC2008/1 (UniSysCat) – 390540038, and the European Union’s Horizon 2020 MSCA Program under grant agreement 956314 [ALLODD]. This work was supported by the German Research Foundation (SFB688-B08 and 427840891) and Elitenetzwerk Bayern, Receptor Dynamics program and Federal Ministry of Research (BMBF; 03V0830) to M.J.L. and University of Nottingham Anne McLaren Research Fellowship to I.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122037119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter B., Tate C. G., Active state structures of G protein-coupled receptors highlight the similarities and differences in the G protein and arrestin coupling interfaces. Curr. Opin. Struct. Biol. 45, 124–132 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wacker D., Stevens R. C., Roth B. L., How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao Z., Kobilka B., Using synthetic lipids to stabilize purified beta2 adrenoceptor in detergent micelles. Anal. Biochem. 343, 344–346 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Weis W. I., Kobilka B. K., The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velazhahan V., et al. , Structure of the class D GPCR Ste2 dimer coupled to two G proteins. Nature 589, 148–153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao C., et al. , Cryo-EM structures of inactive and active GABAB receptor. Cell Res. 30, 564–573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilger D., et al. , Structural insights into differences in G protein activation by family A and family B GPCRs. Science 369, eaba3373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wootten D., Christopoulos A., Marti-Solano M., Babu M. M., Sexton P. M., Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 19, 638–653 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Lohse M. J., Nuber S., Hoffmann C., Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol. Rev. 64, 299–336 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Vilardaga J. P., Bünemann M., Krasel C., Castro M., Lohse M. J., Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat. Biotechnol. 21, 807–812 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Hlavackova V., et al. , Sequential inter- and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci. Signal. 5, ra59 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Grushevskyi E. O., et al. , Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. Proc. Natl. Acad. Sci. U.S.A. 116, 10150–10155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro M., Nikolaev V. O., Palm D., Lohse M. J., Vilardaga J.-P., Turn-on switch in parathyroid hormone receptor by a two-step parathyroid hormone binding mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 16084–16089 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilardaga J.-P., et al. , Conformational cross-talk between α2A-adrenergic and μ-opioid receptors controls cell signaling. Nat. Chem. Biol. 4, 126–131 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Zhao L.-H., et al. , Structure and dynamics of the active human parathyroid hormone receptor-1. Science 364, 148–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrenmann J., et al. , High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. Nat. Struct. Mol. Biol. 25, 1086–1092 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Abou-Samra A. B., et al. , Expression cloning of a common receptor for parathyroid hormone and parathyroid hormone-related peptide from rat osteoblast-like cells: A single receptor stimulates intracellular accumulation of both cAMP and inositol trisphosphates and increases intracellular free calcium. Proc. Natl. Acad. Sci. U.S.A. 89, 2732–2736 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh A. T., Gilchrist A., Voyno-Yasenetskaya T., Radeff-Huang J. M., Stern P. H., G alpha12/G alpha13 subunits of heterotrimeric G proteins mediate parathyroid hormone activation of phospholipase D in UMR-106 osteoblastic cells. Endocrinology 146, 2171–2175 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Miyauchi A., et al. , Stimulation of transient elevations in cytosolic Ca2+ is related to inhibition of Pi transport in OK cells. Am. J. Physiol. 259, F485–F493 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Syme C. A., Friedman P. A., Bisello A., Parathyroid hormone receptor trafficking contributes to the activation of extracellular signal-regulated kinases but is not required for regulation of cAMP signaling. J. Biol. Chem. 280, 11281–11288 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Gesty-Palmer D., et al. , Distinct beta-arrestin- and G protein-dependent pathways for parathyroid hormone receptor-stimulated ERK1/2 activation. J. Biol. Chem. 281, 10856–10864 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Cupp M. E., Nayak S. K., Adem A. S., Thomsen W. J., Parathyroid hormone (PTH) and PTH-related peptide domains contributing to activation of different PTH receptor-mediated signaling pathways. J. Pharmacol. Exp. Ther. 345, 404–418 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Dean T., Vilardaga J.-P., J. T. Potts, Jr, Gardella T. J., Altered selectivity of parathyroid hormone (PTH) and PTH-related protein (PTHrP) for distinct conformations of the PTH/PTHrP receptor. Mol. Endocrinol. 22, 156–166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T., et al. , Comparable initial engagement of intracellular signaling pathways by parathyroid hormone receptor ligands teriparatide, abaloparatide, and long-acting PTH. JBMR Plus 5, e10441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilardaga J. P., Romero G., Feinstein T. N., Wehbi V. L., Kinetics and dynamics in the G protein-coupled receptor signaling cascade. Methods Enzymol. 522, 337–363 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Christopoulos A., et al. , Novel receptor partners and function of receptor activity-modifying proteins. J. Biol. Chem. 278, 3293–3297 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Lorenzen E., et al. , Multiplexed analysis of the secretin-like GPCR-RAMP interactome.Sci. Adv. 5, eaaw2778 (2019). [DOI] [PMC free article] [PubMed]
- 29.Matthew Harris D. I. M., et al. , RAMPs regulate signalling bias and internalisation of the GIPR. bioRxiv [Preprint] (2021). 10.1101/2021.04.08.436756. Accessed 30 November 2021.
- 30.Kadmiel M., Matson B. C., Espenschied S. T., Lenhart P. M., Caron K. M., Loss of receptor activity-modifying protein 2 in mice causes placental dysfunction and alters PTH1R regulation. PLoS One 12, e0181597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., Tsien R. Y., Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J. Biol. Chem. 276, 29188–29194 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Goedhart J., et al. , Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3, 751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackie D. I., et al. , RAMP3 determines rapid recycling of atypical chemokine receptor-3 for guided angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 116, 24093–24099 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schihada H., et al. , A universal bioluminescence resonance energy transfer sensor design enables high-sensitivity screening of GPCR activation dynamics. Commun. Biol. 1, 105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keppler A., et al. , A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Chen T. W., et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patriarchi T., et al. , Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schihada H., Shekhani R., Schulte G., Quantitative assessment of constitutive G protein-coupled receptor activity with BRET-based G protein biosensors. Sci. Signal. 14, eabf1653 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Klarenbeek J., Goedhart J., van Batenburg A., Groenewald D., Jalink K., Fourth-generation epac-based FRET sensors for cAMP feature exceptional brightness, photostability and dynamic range: Characterization of dedicated sensors for FLIM, for ratiometry and with high affinity. PLoS One 10, e0122513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilardaga J.-P., et al. , Differential conformational requirements for activation of G proteins and the regulatory proteins arrestin and G protein-coupled receptor kinase in the G protein-coupled receptor for parathyroid hormone (PTH)/PTH-related protein. J. Biol. Chem. 276, 33435–33443 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Liang Y.-L., et al. , Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor. Nature 561, 492–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbash S., Lorenzen E., Persson T., Huber T., Sakmar T. P., GPCRs globally coevolved with receptor activity-modifying proteins, RAMPs. Proc. Natl. Acad. Sci. U.S.A. 114, 12015–12020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbash S., et al. , Detection of concordance between transcriptional levels of GPCRs and receptor-activity-modifying proteins. iScience 11, 366–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGlone E. R., et al. , Receptor activity-modifying protein 2 (RAMP2) alters glucagon receptor trafficking in hepatocytes with functional effects on receptor signalling. Mol. Metab. 53, 101296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cegla J., et al. , RAMP2 influences glucagon receptor pharmacology via trafficking and signaling. Endocrinology 158, 2680–2693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey S., et al. , Interactions between RAMP2 and CRF receptors: The effect of receptor subtypes, splice variants and cell context. Biochim. Biophys. Acta Biomembr. 1861, 997–1003 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Serafin D. S., Harris N. R., Nielsen N. R., Mackie D. I., Caron K. M., Dawn of a New RAMPage. Trends Pharmacol. Sci. 41, 249–265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weston C., et al. , Modulation of glucagon receptor pharmacology by receptor activity-modifying protein-2 (RAMP2). J. Biol. Chem. 290, 23009–23022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang Y.-L., et al. , Structure and dynamics of adrenomedullin receptors AM1 and AM2 reveal key mechanisms in the control of receptor phenotype by receptor activity-modifying proteins. ACS Pharmacol. Transl. Sci. 3, 263–284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schöneberg T., Kleinau G., Brüser A., What are they waiting for?-Tethered agonism in G protein-coupled receptors. Pharmacol. Res. 108, 9–15 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Clark L. J., et al. , Allosteric interactions in the parathyroid hormone GPCR-arrestin complex formation. Nat. Chem. Biol. 16, 1096–1104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkar K., et al. , Modulation of PTH1R signaling by an ECD binding antibody results in inhibition of β-arrestin 2 coupling. Sci. Rep. 9, 14432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L.-K., Tao Y.-X., Alanine scanning mutagenesis of the DRYxxI motif and intracellular loop 2 of human melanocortin-4 receptor. Int. J. Mol. Sci. 21, 7611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng X., Müller T., Mizrachi D., Fanelli F., Segaloff D. L., An intracellular loop (IL2) residue confers different basal constitutive activities to the human lutropin receptor and human thyrotropin receptor through structural communication between IL2 and helix 6, via helix 3. Endocrinology 149, 1705–1717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nuber S., et al. , β-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature 531, 661–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M.-H., et al. , The conformational signature of β-arrestin2 predicts its trafficking and signalling functions. Nature 531, 665–668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gesty-Palmer D., Luttrell L. M., ‘Biasing’ the parathyroid hormone receptor: A novel anabolic approach to increasing bone mass? Br. J. Pharmacol. 164, 59–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolgin E., First GPCR-directed antibody passes approval milestone. Nat. Rev. Drug Discov. 17, 457–459 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Dicker F., Quitterer U., Winstel R., Honold K., Lohse M. J., Phosphorylation-independent inhibition of parathyroid hormone receptor signaling by G protein-coupled receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 96, 5476–5481 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harvey C. D., et al. , A genetically encoded fluorescent sensor of ERK activity. Proc. Natl. Acad. Sci. U.S.A. 105, 19264–19269 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolters V., Krasel C., Brockmann J., Bünemann M., Influence of gαq on the dynamics of m3-acetylcholine receptor-g-protein-coupled receptor kinase 2 interaction. Mol. Pharmacol. 87, 9–17 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Krasel C., Bünemann M., Lorenz K., Lohse M. J., Beta-arrestin binding to the beta2-adrenergic receptor requires both receptor phosphorylation and receptor activation. J. Biol. Chem. 280, 9528–9535 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Wesslowski J., et al. , eGFP-tagged Wnt-3a enables functional analysis of Wnt trafficking and signaling and kinetic assessment of Wnt binding to full-length Frizzled. J. Biol. Chem. 295, 8759–8774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Işbilir A., et al. , Advanced fluorescence microscopy reveals disruption of dynamic CXCR4 dimerization by subpocket-specific inverse agonists. Proc. Natl. Acad. Sci. U.S.A. 117, 29144–29154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schihada H., Nemec K., Lohse M. J., Maiellaro I., Bioluminescence in G protein-coupled receptors drug screening using nanoluciferase and halo-tag technology. Methods Mol. Biol. 2268, 137–147 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.