Significance
Multipartite viruses package each of their genome segments in different particles, facing a potentially huge cost if the entire genomic information needs to be present concomitantly. Previous work with the octapartite faba bean necrotic stunt virus (FBNSV) showed this issue can be resolved at the within-host level through a supracellular functioning, as follows: all viral segments do not need to be present within the same host cell but may complement each other through intercellular trafficking of their products (protein or messenger RNA [mRNA]). Here, we show that full-genome infections can be reconstituted and function through separate acquisition and/or inoculation of complementary genome segment sets in recipient hosts, thus decreasing the genomic integrity cost during between-host transmission through a suprahost functioning.
Keywords: multipartite virus, virus transmission, genome architecture
Abstract
Because multipartite viruses package their genome segments in different viral particles, they face a potentially huge cost if the entire genomic information, i.e., all genome segments, needs to be present concomitantly for the infection to function. Previous work with the octapartite faba bean necrotic stunt virus (FBNSV; family Nanoviridae, genus Nanovirus) showed that this issue can be resolved at the within-host level through a supracellular functioning; all viral segments do not need to be present within the same host cell but may complement each other through intercellular trafficking of their products (protein or messenger RNA [mRNA]). Here, we report on whether FBNSV can as well decrease the genomic integrity cost during between-host transmission. Using viable infections lacking nonessential virus segments, we show that full-genome infections can be reconstituted and function through separate acquisition and/or inoculation of complementary sets of genome segments in recipient hosts. This separate acquisition/inoculation can occur either through the transmission of different segment sets by different individual aphid vectors or by the sequential acquisition by the same aphid of complementary sets of segments from different hosts. The possibility of a separate between-host transmission of different genome segments thus offers a way to at least partially resolve the genomic maintenance problem faced by multipartite viruses.
About >25% of annotated viral species have their genomic information carried by more than one nucleic acid molecule (1). The most well-known ones are the “segmented viruses” that package all their genome segments in a single virion for transmission, such as the influenza virus A. The “multipartite viruses,” despite being more numerous (∼17% of all viral species), are much less studied, perhaps because their vast majority infects plants or fungi; these viruses package each segment in a distinct viral particle.
Multipartite viruses have important economical and plant health consequences but at present remain an evolutionary puzzle; while none of the proposed advantages is specific to this genomic architecture (all are shared with segmented viruses, but a surprising and idiosyncratic increased lifespan of viral particles has been observed in an artificial system (2)), they face a potentially huge cost if the entire genomic information, i.e., all genome segments, needs to be present concomitantly for the infection to function (1, 3). Indeed, if the different segments were transmitted randomly, an unrealistically large number of viral particles would need to enter each host cell (multiplicity of infection [MOI]), particularly for multipartite viruses with more than four segments (4, 5).
Our work with the aphid-transmitted octapartite faba bean necrotic stunt virus (FBNSV; family Nanoviridae, genus Nanovirus) has added to the puzzle. The FBNSV genome is composed of 8 circular single-stranded DNA segments, each of about 1,000 bases and encoding a single protein (6, 7). Segment C encodes the cell cycle–linked protein (Clink); M encodes the movement protein (MP); N encodes the nuclear shuttle protein (NSP); S encodes the coat protein (CP); R encodes the master replication–associated protein (M-Rep); and U1, U2, and U4 encode proteins of unknown functions. We previously showed that 1) the eight genome segments do not occur at equal frequencies within host plants; the frequency distribution converges to a host plant species–specific distribution, which we termed “genome formula” (8). The discovery of the genome formula, subsequently revealed in another nanovirus species (9) and in two unrelated multipartite viruses (10, 11), other than unveiling another biological weirdness of these viruses begging for an explanation, further questions their existence; if some segments are rare, the MOI needs to be even higher. 2) We subsequently showed that within individual hosts, all viral segments do not need to be present within the same host cell but that they may complement each other through intercellular trafficking of their expression products (messenger RNA and/or protein); this distributed mode of functioning should greatly reduce the within-host putative genomic integrity cost (12). Nevertheless, the potential genomic integrity cost during host-to-host transmission still needs to be addressed. We directly showed that very few copies of each segment were aphid transmitted from plant to plant and that they were two to three orders of magnitude fewer than predicted from evolutionary models (5). This result, which empirically illustrates the between-host cost, was in agreement with the only other report on transmission bottlenecks in multipartite viruses, which found that when an aphid successfully inoculated the tripartite cucumber mosaic virus to a host plant, only one or two copies of each segment were transmitted (13).
A possibility to circumvent the cost of host-to-host transmission would be that the concomitant transmission of all genome segments is not necessary; the segments could be sequentially acquired by the same aphid from different host plants and then transmitted or different aphids could acquire different segments from different plants and then transmit them to the same plant (14). In this paper, we partly tested this hypothesis. We took advantage of the fact that, although all segments were found in the 10 known FBNSV field isolates (15–18), within-host infection can be achieved in the laboratory if some of them are not present (19, 20). We inoculated different sets of plants with incomplete viral genomes, namely, FBNSVC−, FBNSVN−, or FBNSVU4−, with each lacking the genome segment indicated in superscript. It has been previously shown that the absence of these segments does not affect within-plant virus accumulation (20). We then tested whether the complete viral genome could be reconstituted on receiver plants, either following transmission by different aphids that fed on the different sets of plants, the parallel mode, or by the same aphid fed sequentially on the different plants, the sequential mode (experiment 1). In the latter case, we also looked at how temporal spacing of the sequential acquisitions affected the probability of complete genome reconstitution (experiment 2; see Fig. 1 for a schematic of the two experiments; all experimental procedures are detailed in Materials and Methods). Our results show that it is indeed possible to reconstitute the whole virus genome, in both cases, even though this is less likely when increasing the time between sequential acquisitions. We further show that under sequential acquisition, the reconstitution occurs at the very early stage of the virus cycle within the aphid midgut cells. Our findings thus support the hypothesis that multipartite viruses may circumvent the genomic integrity maintenance cost during host-to-host transmission because concomitant transmission of all genome segments is not necessary. Their capacity to transmit genomic segments nonconcomitantly suggests that these viruses have an immense potential to exchange genetic information through reassortment. i.e., exchange of complete segments, since parental genotypes could exchange segments without ever occurring on the same host individual.
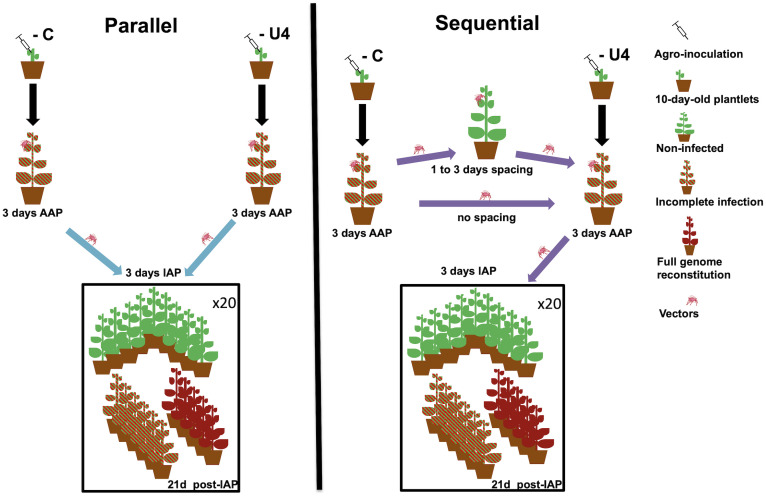
Fig. 1.
Schematic of the experimental design of parallel and sequential transmission modes. Details of the two transmission modes are schematized as well as their expected outcome using infections lacking segments C and U4 as an example. For sequential transmissions, we show only the case where aphids first fed on plants infected with FBNSVC− and then fed on plants infected with FBNSVU4−.The age of test plants, the status and age of infections, and the durations of AAPs and of IAP are indicated. The parallel and sequential modes with no time-spacing treatments were performed during experiment 1, and the sequential modes with time-spacing treatments were performed during experiment 2 (the sequential with no time-spacing treatment was independently performed in both experiments). Each experiment was replicated twice. As mentioned in Materials and Methods, the parallel transmission mode implies separate acquisition and inoculation of segments, whereas the sequential transmission mode implies temporally separated acquisition of segments but may allow concomitant inoculation.
Results
FBNSV Full-Genome Reconstitution Is Possible through Both Parallel and Sequential Transmission by Aphid Vectors.
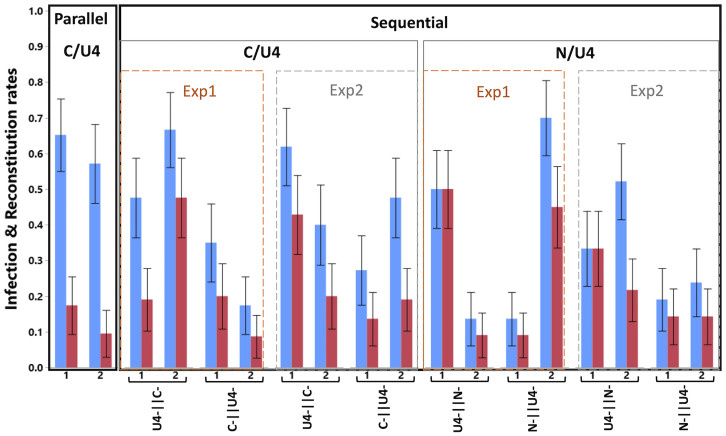
Fig. 2 presents the proportions of infected plants and of those containing all eight segments, for all conditions and replicates without any temporal spacing between sequential acquisitions. Though with variable proportions, all conditions generated full FBNSV genome reconstitutions, demonstrating that such reconstitution is possible from infections with incomplete genomes, whether transmitted separately by different aphids (parallel transmission) or acquired sequentially by the same aphid (sequential transmission).
Fig. 2.
Infection and full genome reconstitution as a function of the transmission mode. The graph shows the proportion of infected plants (blue) and plants where all three investigated segments are found (reconstitution, red) as a function of the transmission mode (parallel vs. sequential) and the segments involved (parallel: only U4 and C; sequential: either U4 and C or N and U4; the order of the segments in the labels indicates the order of infected plants on which the aphids fed). For sequential transmission and for a given pair of segments and order, we provide the results from the two different experiments, namely, parallel vs. sequential (Exp 1) and sequential with spacing in time (Exp 2), using for the latter only the 0 spacing time treatments. The two experimental replicates (1, 2) are shown for each case, and the error bars are ±1 SE of the mean. Sample size varies from 20 to 23 (Table 1).
In all replicates/conditions but two, some infections did not result in the reconstitution of the complete genome; the proportion of reconstitutions (red bars) was lower than that of infections (blue bars). Some segments are missing in these cases, and this is discussed in more length in a separate section.
Sequential transmissions performed by single aphids led, over all conditions and replicates, to ∼40% infection, which is compatible with what we previously found when transmitting the full genome with one aphid (5). Parallel transmissions performed by two aphids led to ∼60% infection. This transmission rate is very close to 64%, the proportion expected if each aphid had a 40% chance to transmit the infection and there was no interaction between them. Sequential transmission, however, led to roughly twice as many reconstitutions of the complete genome than parallel transmissions. Over all test plants, the grand mean of those containing all eight FBNSV segments after parallel transmission is 13.6% while it is of the order of 25% after sequential transmission. This observation holds whether we calculate the mean over both experiments or only the parallel vs sequential experiment, over both pairs of segments or only over C/U4 (which are the only segments involved in parallel transmissions). It is worth noting, however, that the differences in the reconstitution rate between parallel and sequential transmission modes are due only to the U4-ǁC- treatment (with the data of only the first experiment: z = 2.011, P = 0.044; with the data of both experiments in Fig. 2: z = 2.240, P = 0.025). The reconstitution rates of the C-ǁU4- reverse acquisition order do not differ statistically from those of the parallel transmission treatment (with the data of only the first experiment: z = 0.046, P = 0.963; with the data of both experiments in Fig. 2: z = 0.27, P = 0.821).
We further investigated whether the missing segments and their order of acquisition in sequential transmission affected the rates of infection and reconstitution. As already suggested by the above comparison looking at the situations involving segments C and U4, despite variation among replicates, there were more infections and reconstitutions when segment C was acquired first. Across both experiments and replicates, U4-ǁC- led to ∼54% infections and ∼32% reconstitutions vs. ∼31% infections and ∼15% reconstitutions when C was acquired second (C-ǁU4-). These differences were statistically significant (z = 2.496, P = 0.013; the experiment effect was not statistically significant: z = 0.061, P = 0.951) and are further discussed later.
For N and U4 combinations, it is highly remarkable that transmission was efficient even when segment N was not acquired first (i.e., the N-ǁU4- conditions). Indeed, it has been repeatedly shown that no transmission occurs from plants lacking segment N (14, 15), which encodes for the helper factor of FBNSV (14). We comment on this surprising result more extensively in the Discussion. For the moment, we note that, even though more variable between replicates, transmissions involving segments N and U4 led to similar proportions of infection and reconstitution as transmissions involving segments C and U4. However, in contrast to segment C and U4, the reconstitution rates resulting from the two N and U4 acquisition orders did not differ significantly (z = 0.181, P = 0.856; experiment: z = −0.908, P = 0.364; experiment by order of acquisition interaction: z = 0.633, P = 0.526).
FBNSV Infection and Reconstitution when Sequential Acquisition by Aphids Is Spaced in Time.
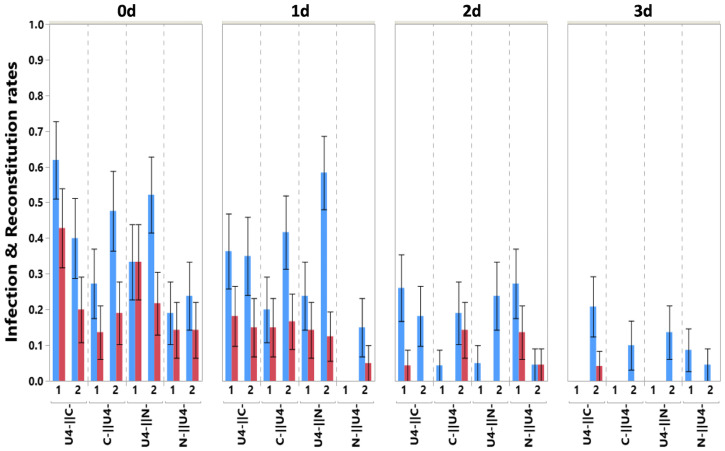
Fig. 3 presents the proportion of infected plants (blue) and of plants with the full genome reconstituted (red) when segments where acquired sequentially but with variable spacing in time of 0, 1, 2, or 3 d. The figure clearly shows that increasing spacing in time rapidly leads to a decrease both in infection and reconstitution rates (infection: z = −3.310, P < 0.001; reconstitution: z = −2.567, P = 0.010). Indeed, with no spacing (0 d), across all segment combinations and replicates, 65 plants were infected and 38 had the full genome reconstituted out of a total of 170 receiver plants, while after 3 d of spacing, only 13 plants were infected and only 1had a reconstituted genome over a total of 174. Most infections and reconstitutions occurred with no or just 1 d spacing, while very little of either occurred with more than 1 d.
Fig. 3.
Infection and full genome reconstitution as a function of time spacing between sequential acquisitions. Shows the proportion of infected plants (blue) and plants in which all three investigated segments are found (reconstitution, red) as a function of time spacing (top x-axis, expressed in days) for the different segments and order combinations. The two experimental replicates are shown for each case, and the error bars are ±1 SE of the mean. Sample size varies from 17 to 24 (Table 1).
None of the order of acquisition comparisons revealed a statistically significant effect except for the N-ǁU4- condition that resulted in fewer infections (z = −2.148, P = 0.032).
Incomplete Genome Reconstitution and Loss of Segments.
As mentioned earlier, many infections did not result in the reconstitution of the complete FBNSV genome, implying that at least one segment was missing. Here, we summarize the broad patterns, first on the replicates without time spacing and then on the replicates of time spacing under sequential transmission, and relegate the detailed account to SI Appendix, Tables S1 and S2.
Among the 385 receiver plants used in all segment combinations and transmission mode (without time spacing), 158 were infected. Only three of those did not carry segment N, indicating that this segment is rarely involved in incomplete or failed reconstitutions. Segment C was absent from 14.5% (23/158) of infected plants, and there was no clear pattern in the distribution of these cases, either between transmission modes or across segment combinations involved. Segment U4 was absent from 33% (52/158) infected plants. It was particularly absent from recipient plants of the parallel transmission mode (63%) but much less so in all sequential transmission mode treatments (27%), with no strong pattern across segment combinations involved.
Time spacing had no effect on segment N that was present in all infected plants except for the plants mentioned above plus two more plants. Segment C was present in ∼80% of infected plants for all time-spacing intervals except for 3 d where it was present in only 6/13 of infected plants. Segment U4’s presence gradually declined with the time-spacing interval, being present in 75% infected plants at 0-d spacing and down to 38% at 3 d. No clear pattern could be observed across segment combinations involved (SI Appendix, Table S2).
Reconstitution May Occur Already in Aphids upon Sequential Acquisition.
Using segment-specific fluorescence in situ hybridization (FISH) by specifically labeling the pairs of sequentially acquired segments (segment C then U4 or N then U4), we investigated whether the FBNSV genome could reconstitute in aphid vectors, when subsets of segments were acquired sequentially. Aphids were placed on plants during 7 d for each acquisition step, with 1 d on healthy plants between the two acquisition steps and 1 d of feeding on water through a Parafilm membrane after the second acquisition to eliminate the virus present in the lumen of the aphid midgut.
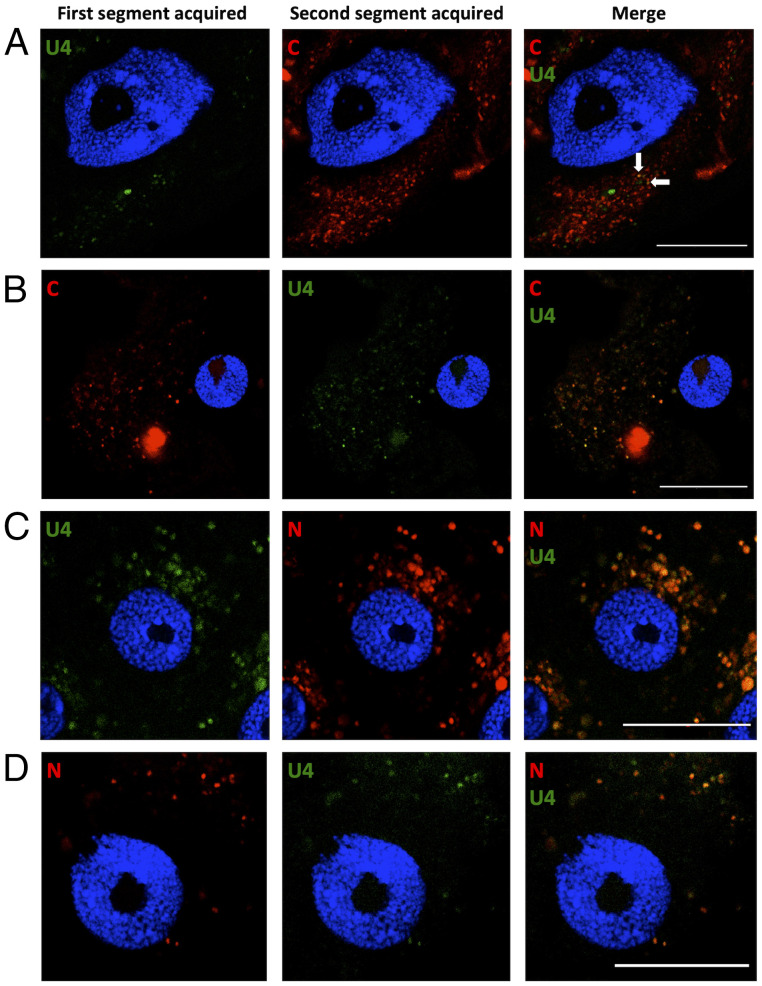
For segments C and U4, when U4 was acquired first (i.e., C-ǁU4- treatment), a few intracellular fluorescent foci showed colocalization of the two segments (Fig. 4A, white arrows). However, the majority contained only one of the two, rare foci containing only U4 (green), and numerous others containing only C (red). In contrast, when C was acquired first (U4-ǁC-), a large proportion of the foci contained both segments (Fig. 4B). Even more remarkable, for segments N and U4, we observed a near-perfect colocalization in all foci (Fig. 4 C and D) for both acquisition orders (U4-ǁN- or N-ǁU4-). These observations were very consistent across biological replicates; a total of 16, 10, 6, and 14 individual aphids were examined for treatments C-ǁU4-, U4- ǁC-, U4-ǁN-, and N-ǁU4-, respectively. At least 10 cells of the anterior midgut (AMG) were zoomed for each individual aphid. For each treatment, all zoomed cells revealed the same pattern, illustrated in Fig. 4. All controls for these experiments, particularly those confirming the segment specificity of the probes, are shown in SI Appendix, Fig. S1 (see the without C and without U4 panels).
Fig. 4.
Localization of sequentially acquired DNA segments of FBNSV in aphid AMG cells. Viral DNA is labeled by FISH in AMG cells of viruliferous aphids and observed by confocal microscopy. The green probe targets segment U4, and the red probes target either C or N in the corresponding panels. The accumulation of FBNSV DNA was similarly revealed in all observed cells (>10 cells per midgut) from 16, 10, 6, and 14 viruliferous aphids for A, B, C, and D, respectively. A representative image of each case is shown to illustrate the results. A and B represent the sequential acquisition of segments C/U4, and C and D represent that of segments N/U4, in the indicated sequential order. The white arrows show two foci containing both C and U4 segments. All images correspond to single optical sections. Cell nuclei are stained with DAPI (blue). Scale bar, 25 µm.
Altogether, our results demonstrate that full-genome reconstitution can occur at very early stages of the virus cycle within the aphid vectors, soon after entry of the segments into midgut cells.
The Reconstituted Viral Genome Can Be Transmitted as an Integral FBNSV Genome.
We finally verified that the FBNSV could be aphid transmitted from plants containing a reconstituted genome in a way similar to that observed from plants initially agro- or aphid inoculated with the complete genome, as detailed in Materials and Methods. Over the four transmission assays (one donor plant with a reconstituted genome from each of the sequential transmission treatments C-ǁU4-, U4-ǁC-, N-ǁU4-, and U4-ǁN-), we obtained 36.36% (28/77) successful transmissions. This transmission rate is close to that we typically observe for transmissions by a single aphid under our experimental conditions, from plants initially infected with the full FBNSV genome. There was variability among treatments, at least partially reflecting the relatively small sample sizes (U4-ǁC-: 45.45% (10/22); C-ǁU4-: 31.25% (5/16); U4-ǁN-: 50% (10/20); N-ǁU4-: 15.79% (3/19)).
In the 28 infected plants, we observed the absence of 1 or 2 segments in only 3 plants, as follows: segment U4 was absent from 1 plant of the U4-ǁC- treatment, segments C and N were both absent from 1 plant of the C-ǁU4- treatment, and segments C and U4 were both absent from another plant of the C-ǁU4- treatment. Finally, one plant of the U4-ǁC- treatment tested negative for segments N and U4 and had one qPCR replicate slightly above (cycle threshold [Ct] = 30.85) and one slightly below (Ct = 30.28) the threshold value of segment C (Ct = 30.43). Repeating the qPCRs for this individual plant yielded almost identical values. We decided to consider this plant as not infected, as indicated by most of the evidence.
These results thus confirm that the reconstituted FBNSV can successfully be transmitted by aphids to uninfected plants as the integral viral genome.
Discussion
Our results clearly show that the reconstitution of the complete FBNSV genome through aphid transmission from distinct plants infected with incomplete genomes is possible. This result demonstrates that all FBNSV segments do not need to be acquired and/or inoculated concomitantly and thus has important implications for our understanding of how multipartite viruses may be coping with the so far assumed cost of a multipartite way of life related to the maintenance of their genomic integrity during host-to-host transmission (1, 3–5, 14, 21).
The nonconcomitant transmission of genomic segments could potentially occur in all vector-borne multipartite viruses, even those not following the circulative transmission mode. There is no reason why the parallel mode would be restricted to such viruses. The relevance of the sequential mode would depend on the opportunity of viral particles from different acquisitions to co-occur in the vector. This should not be a problem for semipersistent viruses that may be retained in their vectors from hours to days (e.g., the bipartite criniviruses transmitted by whiteflies (22)) to weeks/months/years (e.g., the bipartite nepoviruses and tobraviruses transmitted by nematodes (23)). Even nonpersistent viruses could potentially use the sequential mode because they may cause alterations of their vectors’ behavior that make the vectors visit more plants more rapidly (e.g., the tripartite cucumber mosaic virus (CMV) (24)).
We found that genome reconstitution could occur when incomplete infections are transmitted either by distinct aphids, a mode we termed parallel transmission, or by the same aphid sequentially acquiring viral segment sets from different plants to transmit them together to a receiver plant. These modes represent the two extremes of a continuum; reconstitutions could potentially, and probably most likely, occur in nature through any mixture of these modes.
When transmission is parallel, reconstitution can only occur in the receiver plant. Our results show that the overall infection levels we observed, ∼60%, are compatible with the independent transmission of the two incomplete infections by the two aphids that potentially carry them. However, only ∼22% of these infected plants ended up with a reconstituted complete genome, with the remaining plants lacking either segment C or, for unknown reasons, more frequently U4. These segments may be missing either because they were not transmitted or because they were lost in the receiver plant after transmission. The former could occur because the aphids that were supposed to transmit them did not acquire or acquired but did not inoculate them to the receiver plant in sufficient amounts. Consistent with this is our previous demonstration that very few viral particles are transmitted from single aphids (5), likely resulting in frequent segment loss sometimes resulting in infection failure. Unfortunately, this previous study quantifying the bottleneck that aphid transmission imposes to FBNSV populations did not specifically monitor the transmission of segments C and U4, and thus, we have no data allowing a direct comparison. If missing segments were lost after transmission, we see no obvious reason explaining why segment U4 should be lost more often than segment C, as it is more frequent than segment C both in plants (8) and aphids (25). Our results on the transmission from plants with the reconstituted genomes indicate that segment loss is much rarer when donor plants have all eight segments, suggesting that transmission from plants with incomplete infections may be particular in this respect. This matter thus deserves further investigation. Incomplete infections are rarely reported on field isolates of nanoviruses, which could be due to several compatible explanations, as follows: a reporting bias, a lower performance relative to full infections, or complementation of initially incomplete infections through the processes investigated here.
Sequential transmission without time spacing led to infection levels in agreement with previous single-aphid transmission results (5). Reconstitution of the complete genome was much more frequent under this transmission mode, as ∼50% of infected plants carried all genome segments, as opposed to only 22% through parallel transmission. It is remarkable that the segments acquired sequentially colocalized within intracellular granules (fluorescent foci) in aphid gut epithelial cells, indicating that under this transmission mode, reconstitution can occur at very early stages of the virus cycle within the body of its vector. This observation suggests similar accumulation sites for the eight FBNSV segments in aphid midgut cells, whether acquired concomitantly (26) or sequentially (this study), which may explain the higher reconstitution rate, when compared to parallel transmission.
We found an unexpected effect due to the order of acquisition; more infections and reconstitutions occurred when segment C was acquired first (U4-ǁC- treatment) than second (C-ǁU4 treatment). The fact that we only rarely manage to obtain infections lacking segment C through agroinoculation may suggest that, at least under our conditions, this segment plays an important role that was not revealed in other experimental conditions (20). Alternatively, we cannot exclude that segment C plays a yet unanticipated role in FBNSV transmission in the aphids, resulting into higher transmission if it is acquired early.
A much more remarkable result, however, concerns segment N. As previously stated, it is established that the presence of this segment is mandatory for aphid transmission (20, 26). It is thus considered as the segment producing the helper component required for FBNSV transmission (27). So far, all described helper components in the vector transmission of plant viruses must be acquired together with or before the viral particles (28–31). Our sequential experiment results, however, clearly demonstrate that this is not the case for the N segment of FBNSV; it is possible to have transmission, successful infection, and even full-genome reconstitution even if segment N (and/or its expression products) is acquired after the other segments (i.e., our N-ǁU4- treatment). This suggests that the segments acquired by the aphid from plants that do not contain the segment N can survive or wait in the aphid’s gut until segment N (and/or its expression products) is acquired during the second acquisition step. The single full-genome reconstitution (out of a total of 13 infected plants) we observed with a 3-d spacing shows that this complementation can occur with at least this delay between the 2 acquisition steps. We have previously shown that when the virus is acquired from plants lacking segment N, no viral segment is visible in aphid midgut cells, while when N is present in the infected plant, they all penetrate and accumulate together into cytoplasmic granules (26). Our confocal microscopy results show that the same process occurs during the sequential acquisition, whether N is acquired prior to or after the other segments. The specific mechanisms through which the helper of FBNSV acts need further studies. We can note, however, more to the point of this study, that by being able to potentiate the transmission of viral segments previously or subsequently acquired by aphids, this helper allows for a much larger spectrum of possible reconstitutions of the virus’ full genome. Consequently, genome reconstitution through sequential acquisition and transmission may help FBNSV, and potentially other nanoviruses, alleviate the cost of the maintenance of genome integrity upon host-to-host transmission.
Our time-spacing results were very surprising. We observed that infection and reconstitution declined as time spacing increased. FBNSV, as all nanoviruses, is supposed to be transmitted according to the circulative nonpropagative mode (32) and thus travels through the aphid’s body, gut, and salivary gland cells without replicating. Depletion of the first acquired infection (drastic reduction of transmission rate) during the short time intervals we used is thus surprising (33). But what is even more surprising is that the second acquired infection should lead to successful infections on its own, even if the first infection was depleted, at a rate that in principle should not depend on the time since the first acquisition. This is clearly not what happened in our experiments. One potential explanation could be that the ability of aphids to successfully transmit viral infections decreases with time. As already noted above, this would be unlikely within the short time intervals we used (33). An alternative explanation could be that the first acquisition somehow triggers a mechanism that inhibits transmission after the second acquisition, and the strength of this inhibitory mechanism would increase with spacing time between the two acquisitions. Because it is beyond the scope of this work, we leave this very interesting phenomenon to the future investigation it deserves We note for the moment that this unanticipated negative effect of time spacing between sequential acquisitions may limit the potential of reconstitutions to help multipartite viruses overcome the cost linked to genomic integrity upon host-to-host transmission.
It is currently impossible to quantitatively evaluate the potential of complete genome reconstitution through nonconcomitant transmission of different segments to alleviate the host-to-host transmission cost of multipartitism. Such evaluation would require the precise knowledge of ecological and behavioral variables, such as the prevalence and density of infected plants, the density of vectors, how often vectors move from plant to plant, and what governs their individual host plant choice, all of which is currently unknown. Moreover, in the introduction, we stated that our study partly addresses this issue. We used this term because we acknowledge that here we demonstrate that reconstitution is possible between incomplete infections that are viable (even though they do not necessarily perform as well as complete genome infections). The potential of genome reconstitution to alleviate the cost of maintenance of genomic integrity would be much larger if it could be shown that such reconstitution occurs also from uptake within aphids or release within plants of segments sets that are not viable on their own. We relegate this to future investigation. This study nevertheless demonstrates that the concomitant transmission of the different genome components is not mandatory. Thus, after showing that multipartite viruses may solve the within-host cost of genomic integrity maintenance through a supracellular, distributed functioning (12), the present study shows that they may solve the between-host cost of genomic integrity maintenance through a supraindividual host, distributed transmission. Furthermore, this functioning suggests that these viruses have an immense potential for reassortment, compared to, e.g., segmented viruses that in order to reassort need to coinfect the same host cell since parental genotypes could exchange segments without ever having to meet each other, a feature that may greatly influence their evolution.
Materials and Methods
Virus Isolate, Clones, and Plant Agroinoculation.
The first FBNSV isolate, discovered on faba beans (Vicia faba) in Ethiopia (15), was characterized and cloned by Grigoras and colleagues (34). Each genome segment was inserted as a head-to-tail dimer into the binary plasmid pBin19. Eight plasmids, each containing a dimer of a segment, together constitute the FBNSV infectious clone. The impact on plant infection and plant-to-plant transmission of the absence of any of the segments was investigated (20). The authors showed that the presence of segments R, S, and M was mandatory for infection, but it was possible to infect plants when one of segments C, N, U1, U2, or U4 was missing. They further found that the absence of segments C or U4 did not affect virus accumulation and transmission, while the absence of U1 or U2 decreased symptom severity and virus titer. They also established that the absence of the N segment impedes plant-to-plant aphid transmission, even though it does not seem to affect within-plant viral growth.
In this study, we inoculated 10-d-old faba bean plantlets (V. faba cv. Sevilla; Vilmorin) with FBNSVcomplete, FBNSVC−, FBNSVN−, or FBNSVU4− agrobacteria solutions (strain COR308), as earlier described (34). While FNSVcomplete corresponds to a mixture at equal ratio of eight agrobacterium cultures, with each containing one of the eight plasmids described above, the indicated culture and thus the indicated segment was omitted in FBNSVC−, FBNSVN−, or FBNSVU4−. Plants were maintained as previously described in ref. 26. Plant infection was controlled by total DNA extraction and qPCR (see below) for 21 d after agroinoculation. Even though we were able to obtain relatively many infected plants through agroinoculation of FBNSVN− and FBNSVU4− at 51.06% (=96/188) and 52.81% (=94/178), respectively, we could only obtain 13.08% (=56/428) infected plants with FBNSVC−.
Aphid Transmission.
For all experiments, we used aphid colonies of Acyrthosiphon pisum (clone 210) maintained in a controlled chamber as previously described in ref. 26. Transmission tests were made under similar conditions using faba beans as host plants.
We tested whether the complete virus genome could be reconstituted if 1) different aphids fed on different plants carrying incomplete infections missing different segments came together on the same recipient plant, a mode we hereafter call parallel transmission, and if the same aphid sequentially fed on different plants carrying incomplete infections missing different segments was then transferred on one recipient plant, a mode we hereafter call sequential transmission. We use these terms for ease of language, although the parallel mode implies separate acquisition as well as separate inoculation of the distinct sets of segments, while the sequential mode implies a sequential acquisition of the distinct sets of segments and may allow their concomitant inoculation. Subsequently 2), we asked whether sequential transmission could successfully reconstitute the complete viral genome if the sequential feedings of the aphid were separated in time (from 0 up to 3 d). Finally, 3) we tested that the reconstituted genomes were successfully transmitted as the genuine parental genotype. Fig. 1 provides a schematic representation of the experimental design.
Parallel vs. sequential transmission.
In a first experiment, we tested whether the complete genome could be reconstituted through parallel or sequential transmission and at which rate. For the parallel transmission test (Fig. 1, blue arrows), 2 FBNSVC− and 2 FBNSVU4− plants, 1 of each per replicate, were used as donors and 40 aphids were placed on each plant. Aphids fed on plants during an acquisition access period (AAP) of 3 d to acquire the respective set of segments. Then, we took one aphid from each donor plant and placed them on two different leaflets of the same recipient plant in a clip-cage (cage allowing the maintenance the aphid on one leaflet) during an inoculation access period (IAP) of 3 d.
For sequential transmission, we used one FBNSVC− and two FBNSVU4− donor plants and further tested whether the order of acquisition of the different incomplete infections affected the probability of complete genome reconstitution. Thus, for each condition, 40 aphids were placed on each donor plant (either FBNSVC− or FBNSVU4−) for the first acquisition step. Then, we directly transferred aphids on the plants for the second acquisition step with the complementary set of segments. Each AAP step lasted 3 d. Finally, aphids were individually transferred into a clip-cage on a leaflet of a receiver plant (one aphid/receiver) during an IAP of 3 d.
Transmission does not occur in the absence of segment N (20, 26), which is thus considered to encode for a transmission helper component (27, 35). For this reason, we used only incomplete infections lacking either segments C or U4 for the parallel transmission mode. For the sequential transmission mode, we used the following combinations: 1) FBNSVC− then FBNSVU4− (C-ǁU4-), 2) FBNSVU4− then FBNSVC− (U4-ǁC-), 3) FBNSVN− then FBNSVU4− (N-ǁU4-), and 4) FBNSVU4− then FBNSVN− (U4-ǁN-). The third combination allowed us to test whether the successful transmission of segments can occur even if the N segment, which is necessary for aphid transmission, cannot be acquired during the first acquisition but is acquired during the second acquisition.
The number of receiver plants per transmission mode and replicates are given in Table 1. All replicates for both transmission modes were conducted at the same time.
Table 1.
Number of inoculated plants for each experiment, transmission mode, segment combination, and replicate
| Experiment | Transmission mode | Combinations* | Replicate | Number of inoculated plants† by spacing time (d) | |||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||||
| 1 | Parallel | a | 20 | ||||
| b | 23 | ||||||
| Sequential | 1 | a | 20 | ||||
| b | 23 | ||||||
| 2 | a | 21 | |||||
| b | 21 | ||||||
| 3 | a | 22 | |||||
| b | 20 | ||||||
| 4 | a | 22 | |||||
| b | 22 | ||||||
| 2 | Sequential | 1 | a | 22 | 20 | 23 | 17 |
| b | 21 | 24 | 21 | 20 | |||
| 2 | a | 21 | 22 | 23 | 21 | ||
| b | 20 | 20 | 22 | 24 | |||
| 3 | a | 21 | 21 | 22 | 23 | ||
| b | 21 | 20 | 22 | 22 | |||
| 4 | a | 21 | 21 | 20 | 23 | ||
| b | 23 | 24 | 21 | 22 | |||
*Combinations are as follows: 1, aphids fed on FBNSVC− (plants without segment C)-infected plants and then on FBNSVU4− (plants without segment U4)-infected plants; 2, aphids fed on FBNSVU4−-infected plants then on FBNSVC−-infected plants; 3, aphids fed on FBNSVN− (plants without segment N)-infected plants then on FBNSVU4−-infected plants; 4, aphids fed on FBNSVU4−-infected plants and then on FBNSVN−-infected plants.
†Median/mean sample size = 21/21.5.
Sequential transmission spaced in time.
We subsequently tested whether the time separating the two sequential acquisitions had an effect on the reconstitution of the complete viral genome.
For this type of experiment (Fig. 1, purple arrows), 200 to 250 aphids were placed on a first donor plant infected with 1 set of segments. Then, 40 aphids were transferred on a second donor plant infected with the complementary set of segments. When the two sequential acquisitions were not spaced in time (0 d), the transfer occurred immediately after the end of the first AAP. The remaining aphids were placed onto a healthy plant. Every 24 h, up to a maximum spacing of 3 d, 40 aphids were taken from the healthy plant and placed on a second donor plant infected with the complementary set of segments. Each AAP lasted 3 d. Finally, at the end of the second AAP, aphids were individually placed in a clip-cage on a leaflet of a receiver plantlet during an IAP of 3 d.
Two replicates were carried out for each combination and spacing time (Table 1). However, because the number of receiver plants involved was very large, we first conducted one replicate with all segment combinations and spacing times, and 2 wk later, conducted the second replicate.
All IAPs were stopped with a spray of insecticide (Pirimor; Certis) except where otherwise mentioned. For all experiments, 21 d after the end of the IAP, the total DNA of symptomatic plants was extracted, and qPCR was performed to test for the presence of segments C, N, and U4 (see qPCR details below).
Transmission of the reconstituted complete FBNSV genome.
To verify if the reconstituted complete FBNSV infections can be transmitted to healthy plants, just as the genuine parental genotype, we used the infected faba beans from the sequential transmission experiments with 1 d of time spacing as source plants. In this case, at the end of the IAP of the second replicate, plants were not treated with the insecticide solution. Instead, all aphids were individually removed from the plants and a spray of Marseille soap in water (∼1 wt/vol concentration) was applied onto the plants to eliminate potentially remaining aphids. We allowed for 21 d of disease progression, during which plants were isolated to avoid any aphid contamination. After this period, one plant from each of the C-ǁU4-, U4-ǁC-, N-ǁU4-, and U4-ǁN- sequential transmission experiments with the reconstituted complete FBNSV genome (checked by qPCR) was used as a donor for the transmission of the reconstituted virus. Forty aphids were placed onto one donor plant of each combination. After an AAP of 3 d, 1 aphid was placed onto each of 20 to 24 receiver plantlets for an IAP of 3 d. Finally, 21 d after the end of the IAP, the total DNA of symptomatic receiver plants was extracted and the segments C, N, and U4 were quantified by qPCR.
Viral DNA Extraction and qPCR Detection.
Total DNA extraction from plants was performed as described in ref. 36. FBNSV infection causes faba bean leaf rolling and plant stunting in the early stage of infection and chlorosis and necrosis in the late stage. We extracted DNA from all plants showing some signs of leaf rolling and stunting and quantified segments C, N, or U4 in all these plants. On the two upper leaf levels of symptomatic faba beans, three circular areas of a 0.6-cm diameter were squashed onto a Whatman paper disk. Then, we deposited each disk onto the filter of a 200-µL micropipette tip and added 100 µL of modified Edwards buffer (200 mM Tris‐HCl [pH 7.5], 25 mM EDTA, 250 mM NaCl, 0.5% SDS, 1% PVP40, and 0.2% ascorbic acid in water). Samples were centrifuged, at 5,000 g for 15 s, into a PCR plate placed underneath. Finally, we precipitated the DNA with 50% isopropanol (final concentration) and washed the sample once with 70% EtOH before resuspending it in 50 µL of distilled water.
For the qPCR quantification of the C, N, and U4 segments, each sample was diluted 10 times in distilled water. We used the LightCycler 480 thermocycler (Roche) using 2 µL of the total DNA 1/10th-diluted extract. We used the LightCycler FastStart DNA Master Plus SYBR Green I kit (Roche) according to the manufacturer’s instruction. For each sample, we added 5 µL of the 2× qPCR Mastermix and 0.3 or 0.5 µM primers (0.3 µM final for segment C and 0.5 µM for segments N and U4; specific primers for FBNSV segments were described in ref. 6), complemented at 8 µL with H2O. A total of 2 µL of the DNA sample was added to a final mix of 10 µL. All samples were analyzed with two technical replicates. Forty qPCR cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s were applied to the samples. Post-PCR data analyses were carried as described elsewhere (37).
Because the goal of this study was to investigate whether complete infections can be reconstituted from incomplete infections, we needed to define values of Ct, i.e., the number of cycles needed to reach the fluorescence threshold, beyond which a segment can be considered as absent. Indeed, it is possible in principle to obtain some amplification with the primers of a segment even when it is absent if the primers can hybridize at a low rate with other segments or plant DNA sequences. In order to establish the threshold Ct beyond which each of segments C, N, or U4 could be considered with confidence as absent, we performed qPCRs on samples of plants that were agroinoculated with all FBNSV segments but the focal one. To this end, we agroinoculated plants with FBNSVN−, FBNSVU4−, or FBNSVC−, following the procedures described earlier. These plants were grown for 21 d in growth chambers under a 13/11-h day/night photoperiod at a temperature of 26/20 °C. The total DNA of plants was then extracted, and the presence of segments C, N, and U4 was tested in each plant through qPCR. We could thus obtain the amplification level produced by the primers of a segment from plants in which the segment was absent. We then calculated for each segment the lower tolerance threshold such that 95% of potential future samples would have higher values with a 95% probability (38, 39). These thresholds were 30.37, 31.09, and 28.27 for segments C, N, and U4, respectively (SI Appendix for details on these calculations).
Preparation of Aphid Midguts and FISH.
We previously showed that, when acquired from a plant infected with FBNSVcomplete, the eight segments of FBNSV colocalized in perinuclear cytoplasmic granules in cells of the AMG of aphid vector A. pisum (26). Here, we specifically labeled the pairs of sequentially acquired segments (segment C then U4 or N then U4) to determine whether they colocalize in the AMG cells of their vector. Such eventual colocalization of the two sequentially acquired segments would indicate that the FBNSV genome reconstitution can occur at early stages of accumulation in aphid midguts.
Organ preparation and labeling were performed as detailed in ref. 26. Because 3 d of AAP do not allow sufficient accumulation of viruses into the vector AMG for visualization, aphids were placed on plants during 7 d for the first and the second acquisition steps. After the first AAP, aphids were placed on healthy plants in order to purge viral particles from their gut; in order to limit potential depletion of the first set of segments, this purge period was limited to a single day. The second AAP was followed by a 24-h purging period by feeding aphids on water through a Parafilm membrane to eliminate the virus present in the lumen of the AMG. Then, purged aphids were dissected. AMGs were soaked in 1× phosphate-buffered saline (PBS; pH 7.4) and fixed for 20 min in 4% paraformaldehyde prepared in 1× PBS. To stop the fixation reaction, AMGs were incubated in 0.1 M glycine (pH 7.4) for 15 min. Organs were washed twice (10 min each) in 1× PBS, and an additional incubation in 30% H2O2 solution for 20 min was performed to discolor AMGs and improve the posterior FISH treatment. Finally, AMGs were kept in 1× PBS at 4 °C until used for a maximum storage time of 3 wk.
FISH was performed using fluorescent probes specific to segments C, N, or U4 prepared as described in ref. 40. Amplified coding sequences of segments C and N were labeled with Alexa fluor 468 (red) and those for segment U4 were labeled with Alexa fluor 488 (green). Dissected AMGs kept in 1× PBS were washed three times (5 min each) in hybridization buffer (20 mM Tris-HCl [pH 8], 0.9 M NaCl, 0.01% SDS, and 2.7 M of urea that replaced the 30% deionized formamide used in ref. 40), before an overnight incubation at 37 °C with the segment-specific probes (diluted 1/30 in hybridization buffer). Labeling was stopped by three washes (5 min each) in the hybridization buffer and then AMGs were placed in a 1× PBS solution. Samples were mounted on microscope slides in a Vectashield antifade-mounting medium (Vector Laboratories) that contains DAPI for nuclei staining, and AMGs were observed using the Zeiss LSM700 confocal microscope equipped with a 63× objective.
Data Analysis.
Based on the Ct threshold values defined above, we determined whether each segment was present or absent from each recipient plant. In all cases but one, both technical replicates of each sample were either above or below the threshold. The only exception concerned segment C of one sample that we conservatively considered as absent.
All asymptomatic plants were considered noninfected. All plants showing symptoms were qPCR processed, and if any of the C, N, or U4 segments was present in the sample, we considered the plant as infected. When all three segments were present in recipient plants, we considered that a complete infection was reconstituted.
To investigate whether transmission modalities (parallel or sequential) affected the probability of reconstitution (P(R)) or infection (P(I)), we ran Generalized Linear Mixed Models considering that P(R) and P(I) were binomially distributed with logit-distributed errors, transmission modality and segments involved as fixed factors, and experimental replicate as a random factor. Spacing time was added as a covariate when relevant. We used the lme4 package in R to run these analyses. Figures were drawn using JMP 13.2.1. The data and R scripts for these analyses can be found at https://entrepot.recherche.data.gouv.fr/citation?persistentId=doi:10.57745/EH9KRW.
Supplementary Material
Acknowledgments
This work was funded by the Agence nationale de la recherche grants Reassort ANR-20-CE02-0016 and Nanovirus ANR-18-CE92-0028. M.Y. and S.B. acknowledge support from INRAE Département Santé des Plantes et Environnement, Y.M. from CNRS and IRD, and J.L.-Z. from IRD. The authors acknowledge valuable technical help provided by Ms Sophie Le Blaye. Many thanks to Bernard Francq and Thomas Mathew for their insights on tolerance intervals.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201453119/-/DCSupplemental.
Data Availability
Tables with results of qPCR reactions allowing us to infer the presence of viral segments have been deposited in https://entrepot.recherche.data.gouv.fr/dataverse/inrae (https://entrepot.recherche.data.gouv.fr/citation?persistentId=doi:10.57745/EH9KRW).
References
- 1.Lucía-Sanz A., Manrubia S., Multipartite viruses: Adaptive trick or evolutionary treat? NPJ Syst. Biol. Appl. 3, 34. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojosnegros S., et al. , Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genet. 7, e1001344 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sicard A., Michalakis Y., Gutiérrez S., Blanc S., The strange lifestyle of multipartite viruses. PLoS Pathog. 12, e1005819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iranzo J., Manrubia S. C., Evolutionary dynamics of genome segmentation in multipartite viruses. Proc. Biol. Sci. 279, 3812–3819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallet R., et al. , Small bottleneck size in a highly multipartite virus during a complete infection cycle. J. Virol. 92, e00139-e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gronenborn B., Nanoviruses: Genome organisation and protein function. Vet. Microbiol. 98, 103–109 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Lal A., et al. , Nanovirus disease complexes: An emerging threat in the modern era. Front Plant Sci 11, 558403 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sicard A., et al. , Gene copy number is differentially regulated in a multipartite virus. Nat. Commun. 4, 2248 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Mansourpour M., et al. , Effects of an alphasatellite on life cycle of the nanovirus Faba bean necrotic yellows virus. J. Virol. 96, e0138821 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z., et al. , Genome segments accumulate with different frequencies in Bombyx mori bidensovirus. J. Basic Microbiol. 56, 1338–1343 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wu B., Zwart M. P., Sánchez-Navarro J. A., Elena S. F., Within-host evolution of segments ratio for the tripartite genome of Alfalfa mosaic virus. Sci. Rep. 7, 5004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sicard A., et al. , A multicellular way of life for a multipartite virus. eLife 8, e43599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betancourt M., Fereres A., Fraile A., García-Arenal F., Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J. Virol. 82, 12416–12421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michalakis Y., Blanc S., The curious strategy of multipartite viruses. Annu. Rev. Virol. 7, 203–218 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Franz A., Makkouk K. M., Vetten H. J., Host range of faba bean necrotic yellows virus and potential yield loss in infected faba bean. Phytopathol. Mediterr. 36, 94–103 (1997). [Google Scholar]
- 16.Grigoras I., et al. , Genome diversity and evidence of recombination and reassortment in nanoviruses from Europe. J. Gen. Virol. 95, 1178–1191 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Abraham A. D., et al. , Two distinct nanovirus species infecting faba bean in Morocco. Arch. Virol. 155, 37–46 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotfipour M., Behjatnia S. A. A., Dall’Ara M., Ratti C., The full-length genome characterization and diversity of faba bean necrotic stunt virus in Iran. Eur. J. Plant Pathol. 157, 239–250 (2020). [Google Scholar]
- 19.Timchenko T., et al. , Infectivity of nanovirus DNAs: Induction of disease by cloned genome components of Faba bean necrotic yellows virus. J. Gen. Virol. 87, 1735–1743 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Grigoras I., et al. , Nanovirus DNA-N encodes a protein mandatory for aphid transmission. Virology 522, 281–291 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Michalakis Y., Blanc S., Editorial overview: Multicomponent viral systems. Curr. Opin. Virol. 33, vi–ix (2018). [DOI] [PubMed] [Google Scholar]
- 22.Chen A. Y. S., Walker G. P., Carter D., Ng J. C. K., A virus capsid component mediates virion retention and transmission by its insect vector. Proc. Natl. Acad. Sci. U.S.A. 108, 16777–16782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown D. J. F., Robertson W. M., Trudgill D. L., Transmission of viruses by plant nematodes. Annu. Rev. Phytopathol. 33, 223–249 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Mauck K. E., De Moraes C. M., Mescher M. C., Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. U.S.A. 107, 3600–3605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sicard A., et al. , Circulative nonpropagative aphid transmission of nanoviruses: An oversimplified view. J. Virol. 89, 9719–9726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Mattia J., et al. , Route of a multipartite Nanovirus across the body of its aphid vector. J. Virol. 94, e01998-e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franz A. W. E., van der Wilk F., Verbeek M., Dullemans A. M., van den Heuvel J. F. J. M., Faba bean necrotic yellows virus (genus Nanovirus) requires a helper factor for its aphid transmission. Virology 262, 210–219 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Kassanis B., Govier D. A., New evidence on the mechanism of aphid transmission of potato C and potato aucuba mosaic viruses. J. Gen. Virol. 10, 99–101 (1971). [DOI] [PubMed] [Google Scholar]
- 29.Kassanis B., Govier D. A., The role of the helper virus in aphid transmission of potato aucuba mosaic virus and potato virus C. J. Gen. Virol. 13, 221–228 (1971). [DOI] [PubMed] [Google Scholar]
- 30.Lung M. C. Y., Pirone T. P., Studies on the reason for differential transmissibility of cauliflower mosaic virus isolates by aphids. Phytopathology 63, 910 (1973). [Google Scholar]
- 31.Lu G., et al. , Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog. 15, e1007655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaafar Y. Z. A., Ziebell H., Aphid transmission of nanoviruses. Arch. Insect Biochem. Physiol. 104, e21668 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Watanabe S., Bressan A., Tropism, compartmentalization and retention of banana bunchy top virus (Nanoviridae) in the aphid vector Pentalonia nigronervosa. J. Gen. Virol. 94, 209–219 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Grigoras I., et al. , Reconstitution of authentic nanovirus from multiple cloned DNAs. J. Virol. 83, 10778–10787 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froissart R., Michalakis Y., Blanc S., Helper component-transcomplementation in the vector transmission of plant viruse. Phytopathology 92, 576–579 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Di Mattia J., et al. , Co-acquired nanovirus and geminivirus exhibit a contrasted localization within their common aphid vector. Viruses 12, 299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallet R., Fabre F., Michalakis Y., Blanc S., The number of target molecules of the amplification step limits accuracy and sensitivity in ultradeep-sequencing viral population studies. J. Virol. 91, e00561-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma G., Mathew T., One-sided and two-sided tolerance intervals in general mixed and random effects models using small-sample asymptotics. J. Am. Stat. Assoc. 107, 258–267 (2012). [Google Scholar]
- 39.Francq B. G., Lin D., Hoyer W., Confidence, prediction, and tolerance in linear mixed models. Stat. Med. 38, 5603–5622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vernerey M.-S., Pirolles E., Blanc S., Sicard A., Localizing genome segments and protein products of a multipartite virus in host plant cells. Bio Protoc. 9, e3443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tables with results of qPCR reactions allowing us to infer the presence of viral segments have been deposited in https://entrepot.recherche.data.gouv.fr/dataverse/inrae (https://entrepot.recherche.data.gouv.fr/citation?persistentId=doi:10.57745/EH9KRW).