Significance
Certain types of lung epithelial cells are critical for lung premetastatic niche (PMN) formation. How the primary tumor educates which subpopulation of lung epithelial cells for PMN formation remains largely unknown. Here, we describe a subpopulation of interleukin (IL)-10–producing glutathione peroxidase 3 (GPX3)+ alveolar type 2 epithelial cells (GPX3+ AT2) that has an immunosuppressive function after tumor education, proposing the concept of “epithelial cell polarization,” which was correlated with impaired T cell responses. Under the tumor stressful signals, GPX3 in AT2 cells regulates oxidative stress and promotes hypoxia-inducible factor 1 (HIF-1α) expression to facilitate the production of IL-10. Targeting of GPX3+ AT2 cells and their immunosuppressive molecules may contribute to early intervention of tumor metastasis to lung.
Keywords: premetastatic niche, alveolar type 2 epithelial cells, glutathione peroxidase 3, interleukin-10, lung metastasis
Abstract
The cellular and molecular components required for the formation of premetastatic niche (PMN) to promote lung metastasis need to be further investigated. Lung epithelial cells have been reported to exhibit immunomodulatory roles in lung homeostasis and also to mediate immunosuppressive PMN formation in lung metastasis. Here, by single-cell sequencing, we identified a tumor-polarized subpopulation of alveolar type 2 (AT2) epithelial cells with increased expression of glutathione peroxidase 3 (GPX3) and high production of interleukin (IL)-10 in the PMN. IL-10-producing GPX3+ AT2 cells inhibited CD4+ T cell proliferation but enhanced regulatory T cell generation. Mechanistically, tumor exosome-inducing GPX3 expression is required for GPX3+ AT2 cells to preferentially produce IL-10 by stabilizing hypoxia-inducible factor 1 (HIF-1α) and promoting HIF-1α-induced IL-10 production. Accordingly, conditional knockout of GPX3 in AT2 cells suppressed lung metastasis in spontaneous metastatic models. Together, our findings reveal a role of tumor-polarized GPX3+ AT2 cells in promoting lung PMN formation, adding insights into immune evasion in lung metastasis and providing potential targets for the intervention of tumor metastasis.
The immunological attributes of stromal cells modulate the development and function of immune cells to determine the types and intensity of the immune and inflammatory responses (1, 2). Under tumor conditions, tumor cells co-opt stromal cells to shape the local immune microenvironment, thus promoting tumor progression and metastasis (3, 4). The concept of premetastatic niche (PMN) has been proposed to better describe the crosstalk between disseminated tumor cells and their microenvironment, especially the stromal cells in the metastatic organ (5, 6). Tumor-derived secreted factors shed by the tumor and cellular compound in the target organ work together to prepare the “soil” for tumor transmission to distant metastatic sites (7, 8). Immunosuppression is one of the key hallmarks of PMN, which prevents the disseminated tumor cells from immune attack (9). However, the roles of stromal cells and their related molecules in immunosuppression and PMN formation in the metastatic organ need to be fully studied.
In many cancers, lung metastasis is common and is a lethal determinant (10, 11). An interconnected network of lung resident and recruited cells, including epithelial cells, lymphocytes, and macrophages, is critical for lung homeostasis in physiological conditions, but also for lung metastasis in tumor-bearing hosts (9, 12). Lung stromal cells (including fibroblasts and epithelial cells) can recruit bone marrow-derived cells by secreting chemokines, cytokines, and vesicular substances, thereby creating a supportive and immunosuppressive microenvironment for the colonization of tumor cells (13, 14). For instance, in response to sustained tumor and stromal education, accumulated neutrophils, decreased effector and cytotoxicity T cells, and polarized macrophages are important mediators for immunosuppression in lung PMN (15, 16).
Particularly in lung, lung epithelial cells such as squamous alveolar type (AT) 1 epithelial cells and surfactant-secreting AT2 cells are also important for lung inflammation and metastasis in addition to gas exchanges (17, 18). Moreover, interactions of lung Clara cells, macrophages, and AT2 cells recruit CD11b+ TLR4+ cells to the lung, promoting the formation of PMN (19). Using spontaneous metastatic models of Lewis lung carcinoma (LLC) and melanoma B16/F10, our previous studies showed that lung epithelial cells were responsible for initiating neutrophil recruitment and lung PMN formation (15). Mechanistically, tumor-derived exosomes can be taken up by AT2 cells and stimulate their TLR3 activation by exosomal RNAs, consequently inducing neutrophil-related chemokine secretion in AT2 cells and promoting PMN formation. In-depth studies on lung epithelial cells in tumor metastasis will provide new insights into immunosuppression in PMN and organotropic metastasis, suggesting a potential target to prevent lung metastasis.
Here, using single-cell sequencing, we identified a subpopulation of interleukin (IL)-10-producing GPX3+ AT2 cells in the lung of tumor-bearing mice that inhibited CD4+ T cell proliferation but enhanced regulatory T cell generation, thus promoting PMN formation and lung metastasis.
Results
Identification of GPX3+ Type 2 Lung Epithelial Cells in Tumor-Bearing Mice.
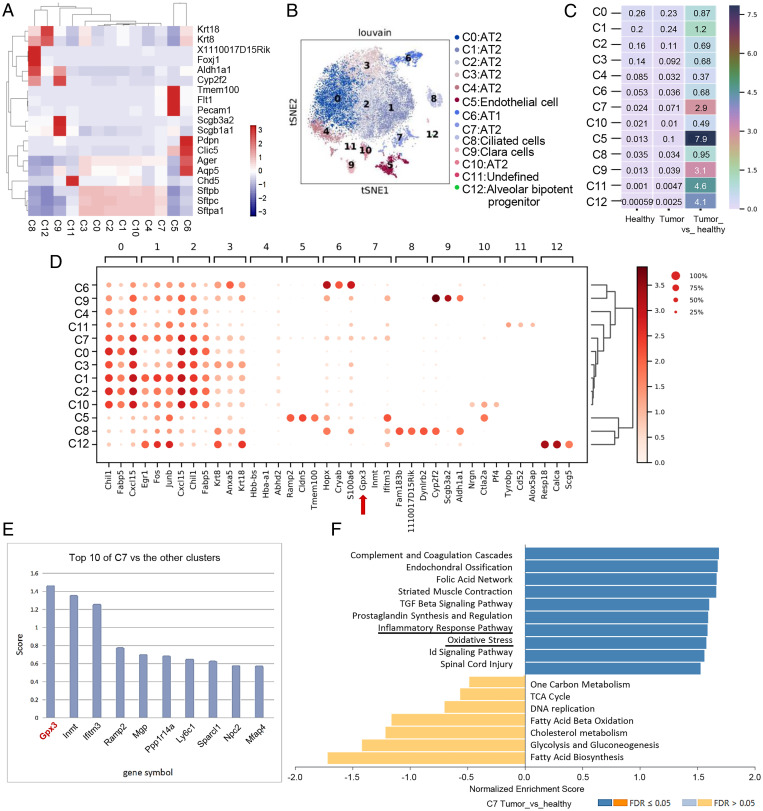
By single-cell RNA sequencing (scRNA-seq) of lung epithelial cells from healthy and LLC-bearing mice, a total of 20,354 epithelial cells were clustered into 12 cell subpopulations according to annotated marker genes (Fig. 1A and SI Appendix, Fig. S1A). They mainly contained AT1 cells, AT2 cells, and Clara cells, of which AT2 cells took up the majority of cell subsets. Dimensional reduction analysis of t-distributed stochastic neighbor embedding (t-SNE) was performed to exhibit distinct distributions of clusters (Fig. 1B). Cluster 7 ranked first in the ratio of different clusters of AT2 cells in a tumor-bearing mouse compared with a healthy one (Fig. 1C and SI Appendix, Fig. S1A). Furthermore, pseudotime analysis by Monocle 3 indicated that cluster 7, as the end of the pseudo timeline, was not in the same branch as other clusters in AT2 cells (SI Appendix, Fig. S1B). To exclude the possibility of proliferation or expansion of a normal population for cluster 7 under tumor inoculation, an in vivo proliferation assay using bromodeoxyuridine was used, indicating a minimal proliferation rate of Sftpc+ AT2 cells in the lung of tumor-bearing mice in vivo (SI Appendix, Fig. S1C). The data showed the presence of this distinct cluster in tumor-bearing mice.
Fig. 1.
Identification of GPX3+ AT2 cells in the lung of tumor-bearing mice by single-cell sequencing. (A) Visualization of clusters with annotated marker genes by heatmap. (B) t-SNE plots show 12 cell clusters of epithelial cells identified by 10× Genomics scRNA-seq. (C) The ratio of each cell type in epithelial cells of LLC tumor-bearing mouse to those of the healthy one by heatmap. (D) Expression of top three genes among each cell type in 12 cell clusters of epithelial cells shown by dot plot with algorithm of Wilcoxon. Red arrow: GPX3. (E) Scores of marker genes by comparing cluster 7 with the rest of the clusters. (F) GSEA of pathway enriched in cluster 7 in lungs of LLC-bearing mouse versus healthy one by WebGestalt (www.webgestalt.org/#) using the database of PANTHER pathway. Id, inhibitor of DNA binding; TCA, tricarboxylic acid cycle; FDR, false discovery rate.
To further determine the distinct patterns of cluster 7, we analyzed top genes highly expressed in each cell subset, of which GPX3 (glutathione peroxidase 3) ranked first among the top-three marker genes of cluster 7 compared to others (Fig. 1 D and E). GPX3, as an important metabolic enzyme for cellular oxidative metabolism (20), has been reported to be both a tumor suppressor and a promoter during tumor progression (21). However, the functions of GPX3+ AT2 cells in lung metastasis and the role of GPX3 in lung epithelial cells in PMN formation remain unknown.
Gene set enrichment analysis (GSEA) comparing cluster 7 with other clusters in AT2 cells indicated that GPX3+ AT2 cells were enriched in pathways in cancer and several inflammatory pathways (SI Appendix, Fig. S1D). Also, comparison of cluster 7 from tumor bearing and healthy mice revealed that GPX3+ AT2 cells were enriched in inflammatory-related response pathways and oxidation pressure (Fig. 1F). In particular, the function of GPX3+ AT2 cells in lungs of tumor-bearing mice was associated with alternated T cell response (SI Appendix, Fig. S1E). Thus, we identified GPX3+ AT2 cells as a tumor-induced subpopulation of AT2 cells in the lung of tumor-bearing mice, and GPX3+ AT2 cells may be related to immunoregulation.
GPX3+ AT2 Cells Promote Lung PMN Formation and Metastasis.
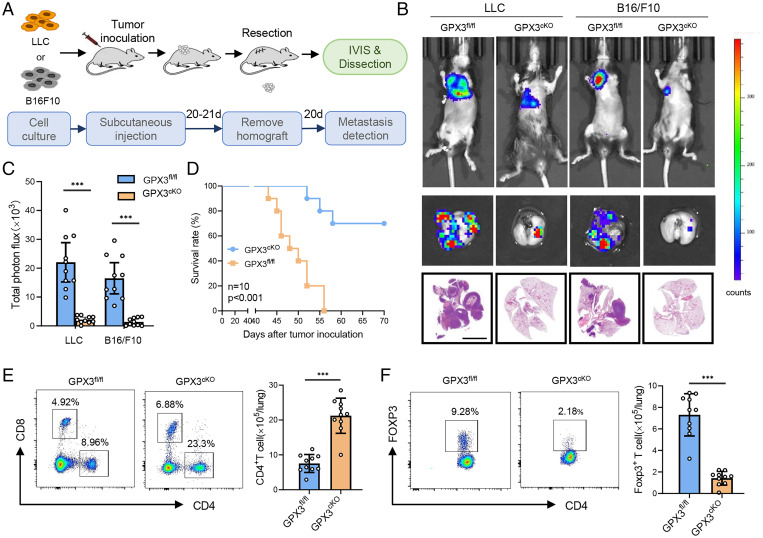
In order to investigate the function of GPX3+ AT2 cells in lung PMN formation and metastasis, we constructed mice with conditional knockout (CKO) of GPX3 in AT2 cells by cross-breeding GPX3fl/fl and AT2 cell-specific Sftpccre mice (SI Appendix, Fig. S2A). The deficiency of GPX3 in AT2 cells was further confirmed by immunoblot and in situ immunofluorescence (SI Appendix, Fig. S2 B and C). The mouse models of spontaneous lung metastasis were prepared by subcutaneously inoculating LLC or B16/F10 cells into GPX3cKO mice or GPX3fl/fl littermates, and the tumor was surgically removed ∼21 d after inoculation (Fig. 2A). Interestingly, GPX3cKO mice exhibited a remarkable reduction both in size and number of lung metastasis in two tumor models compared with GPX3fl/fl littermates (Fig. 2 B and C and SI Appendix, Fig. S2D). Moreover, GPX3cKO mice survived much longer than GPX3fl/fl littermates after tumor removal (Fig. 2D). The results suggest that CKO of GPX3 in AT2 cells inhibited tumor metastasis, thus proposing a critical role of GPX3+ AT2 cells in promoting lung PMN formation and metastasis.
Fig. 2.
CKO of GPX3 in AT2 cells inhibits tumor metastasis. (A) Diagram for the mouse model of spontaneous lung metastasis of LLC and B16/F10. IVIS, in vivo imaging system. (B and C) Representative images (B) and quantitative analysis (C) of lung metastasis of GPX3fl/fl or GPX3cKO mice detected by luciferase-based bioluminescence imaging or by H&E-stained lung sections and quantification of lung metastatic foci of GPX3fl/fl or GPX3cKO mice (n = 10) 40 d after LLC or B16/F10 inoculation. Scale bar, 5 mm. (D) Survival of GPX3fl/fl or GPX3cKO mice (n = 10 each) after LLC inoculation. Kaplan-Meier test. (E and F) Flow cytometry analysis of the proportions and absolute numbers of CD4+ T cells, CD8+ T cells (E), and CD4+ Foxp3+ Treg cells (F) in the lungs of GPX3fl/fl or GPX3cKO mice 14 d after LLC inoculation. Data are mean ± SD of one representative experiment. Similar results were seen in three independent experiments. Unpaired Student’s t tests unless noted. ***P < 0.001.
GPX3 Is Required for Tumor-Polarized Immunosuppressive Function of AT2 Cells.
GPX3+ AT2 cells were enriched in inflammatory responses and impaired T cell response (Fig. 1F and SI Appendix, Fig. S1E). First, we analyzed the immune cell subpopulations in the lungs of GPX3cKO mice and GPX3fl/fl littermates before and after tumor inoculation. There were no obvious differences in immune cell subpopulations between the lungs of GPX3cKO mice and GPX3fl/fl littermates prior to tumor inoculation (SI Appendix, Fig. S2E). After tumor inoculation, however, GPX3cKO mice showed markedly increased CD4+ T cells and decreased CD4+ FOXP3+ regulatory T (Treg) cells (Fig. 2 E and F), as well as deceased CD11b+ Ly6G+ Ly6Cint neutrophils and Ly6G− Ly6C+ monocytes in the lung (SI Appendix, Fig. S2F), which correlated with the single-cell sequencing analysis. The data indicated that tumor-polarized GPX3+ AT2 cells may exhibit immunosuppressive functions at least partially through modification of T cell responses.
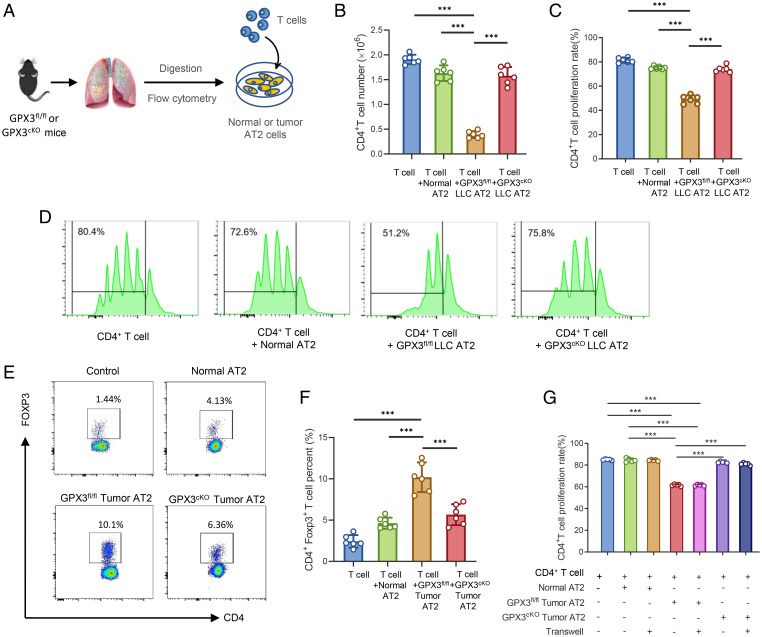
Through an in vitro coculture system of CD4+ T cells with AT2 cells sorted from healthy, tumor-bearing GPX3fl/fl, or GPX3cKO mice (Fig. 3A), we found a reduction in CD4+ T cell proliferation when cocultured with GPX3fl/fl AT2 cells from both LLC- and B16/F10-bearing mice, but not tumor GPX3cKO AT2 cells or normal AT2 cells (Fig. 3 B–D and SI Appendix, Fig. S3A). In addition, CD4+ Foxp3+ T cells notably expanded in the presence of tumor GPX3fl/fl AT2 cells, but not tumor GPX3cKO AT2 cells (Fig. 3 E and F). When cocultured with tumor GPX3fl/fl AT2 cells under transwells, the proliferation of CD4+ T cells was still inhibited (SI Appendix, Fig. S3G), indicating that soluble factors released by tumor AT2 cells were responsible for the inhibition of T cell proliferation. Importantly, in vivo depletion of CD4+ T cells using anti-CD4 antibody reversed the reduction of metastasis in tumor-bearing GPX3cKO mice (SI Appendix, Fig. S3 B and C), further confirming the essential role of CD4+ T cells for the function of GPX3+ AT2 cells in tumor-bearing mice. Therefore, primary tumors can polarize and induce the generation of immunosuppressive AT2 cells in a GPX3-dependent way.
Fig. 3.
Tumor polarizes GPX3+ AT2 cells to suppress T cell responses in a GPX3-dependent way. (A) Model diagram of coculture system for AT2 cells and T cells. (B–D) Flow cytometry analysis of CD4+ T cell number (B), proliferation rate (C), and representative images of CFSE-based proliferation assay (D) of CD4+ T cells cocultured with AT2 cells from healthy, LLC-bearing GPX3fl/fl, or GPX3cKO mice respectively. (E and F) The proportions (E) and absolute numbers (F) of Treg cells were detected by flow cytometry after CD4+ T cells cocultured with AT2 cells as in B. (G) The flow cytometry analysis of CD4+ T cell proliferation after cocultured with different AT2 cells with or without transwells (two-way ANOVA). Data are mean ± SD of one representative experiment. Similar results were seen in three independent experiments. Unpaired Student’s t tests unless noted. ***P < 0.001.
GPX3+ AT2 Cells Preferentially Secrete IL-10 to Suppress T cell Function.
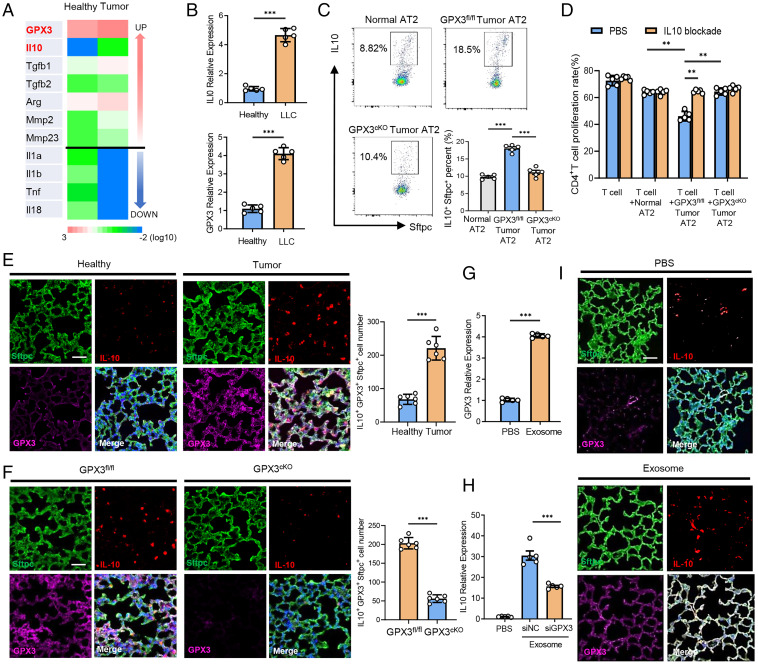
We went further to investigate how GPX3 can polarize the function of GPX3+ AT2 cells in tumor-bearing mice. AT2 cells from tumor-bearing mice and healthy ones were purified for RNA-seq. Importantly, the anti-inflammatory factors (e.g., IL-10, transforming growth factor [TGF]-β1, and TGF-β2) in AT2 cells from tumor-bearing mice were up-regulated, while their expression of proinflammatory factors (such as IL-1α, IL-1β, and tumor necrosis factor [TNF] α) were remarkably down-regulated (Fig. 4A), being charactered as the “polarization” of AT2 cells. In accordance with the RNA-seq data, GSEA analysis of scRNA-seq data for the involved inflammatory pathways was performed and showed that the genes in the C7 cluster in tumor mouse were enriched in anti-inflammatory TGF-β and IL-10 pathways, but not proinflammatory ones (i.e., IL-1, TNF, interferon [IFN]) (SI Appendix, Fig. S4 A and B).
Fig. 4.
GPX3-induced IL-10 expression is required for tumor-polarized function of GPX3+ AT2 cells. (A) Heat map of differently expressed cytokine genes of purified AT2 cells in lungs from LLC-bearing (tumor) or healthy mice as determined by RNA-seq. (B) qPCR analysis of IL-10 and GPX3 messenger RNA expression in AT2 cells purified from tumor mice 14 d after LLC inoculation or from healthy ones. (C) Flow cytometry analysis of IL-10 expression in AT2 cells of GPX3fl/fl and GPX3cKO mice after LLC inoculation. (D) Flow cytometry analysis of CD4+ T cell proliferation cocultured with AT2 cells from healthy, GPX3fl/fl, or GPX3cKO mice after LLC inoculation with or without anti-IL-10 antibody treatment. (E) Immunofluorescent analysis of Sftpc, IL-10, and GPX3 expression and absolute numbers of Sftpc+ IL-10+ GPX3+ cells in the lungs of healthy or LLC-bearing mice. Scale bar, 100 μm. (F) Immunofluorescent analysis of Sftpc, IL-10, and GPX3 expression and absolute numbers of Sftpc+ IL-10+ GPX3+ cells in the lungs of GPX3fl/fl or GPX3cKO mice after LLC inoculation. Scale bar, 100 μm. (G) qPCR analysis of GPX3 expression in MLE-12 cells treated with or without tumor exosomes for 2 h. (H) qPCR analysis of IL-10 expression in control or GPX3-silenced MLE-12 cells treated with or without tumor exosomes for 2 h. siNC, silence of negative control; siGPX3, silence of GPX3. (I) Representative immunofluorescent analysis of Sftpc, IL-10, and GPX3 expression in lungs from healthy mice after LLC inoculation with or without exosome treatment. Scale bar, 100 μm. Data are mean ± SD of one representative experiment. Similar results were seen in three independent experiments. Unpaired Student’s t tests unless noted. **P < 0.01; ***P < 0.001.
As IL-10 is reported to be critical for the inhibition of effector T cells and also a driving force in Treg cell induction (22, 23), we detected GPX3 and IL-10 expression in tumor-polarized AT2 cells. Similar to sequencing data, the expression of GPX3 and IL-10 was much higher in AT2 cells from LLC or B16/F10 tumor-bearing mice than that from healthy ones (Fig. 4B and SI Appendix, Fig. S4C). Indeed, an increase in IL-10+ AT2 cells was observed in tumor-polarized GPX3fl/fl AT2 cells, but not GPX3cKO AT2 cells, suggesting that GPX3 was required for IL-10 production by tumor-polarized AT2 cells (Fig. 4C). Accordingly, blockade of IL-10 restored the proliferation of T cells when cocultured with tumor-polarized GPX3fl/fl AT2 cells but had no influence on the effects of GPX3cKO AT2 cells (Fig. 4D). Furthermore, the proliferation of CD4+ T cells when cocultured with purified tumor AT2 cells from IL-10 knockout mice increased significantly compared to AT2 cells from littermate controls (SI Appendix, Fig. S4D). Immunofluorescence assay indicated that IL-10 was mainly expressed in AT2 cells and showed good colocalization with GPX3 in Sftpc+ AT2 cells. In addition, IL-10-producing GPX3+ AT2 cells expanded more significantly in lungs of tumor-bearing mice than in lungs of healthy ones (Fig. 4E). However, we found a decrease in the number of IL-10-producing GPX3+ AT2 cells in GPX3cKO mice compared to GPX3fl/fl mice with tumor inoculation (Fig. 4F).
We and others reported that tumor-secreted extracellular vesicles, especially exosomes, can be absorbed by lung epithelial cells to promote PMN formation (7, 15). We used MLE-12 cells (a mouse lung epithelial cell line) that were treated with tumor exosomes in vitro as the cellular model for tumor-polarized GPX3+AT2 cells and found higher expression of both IL-10 and GPX3 in MLE-12 cells after exosome treatment (Fig. 4 G and H). However, an obvious decrease in IL-10 expression was observed in GPX3-silenced MLE-12 cells treated with tumor exosomes (Fig. 4H). Tail vein injection of tumor exosomes was conducted to simulate PMN formation in vivo as described previously (15). In accordance with the above results, we found good colocalization of IL-10 with GPX3 and also increased IL-10 and GPX3 expression in the lungs of mice with tumor exosome injection (Fig. 4I). These in vitro and in vivo data demonstrated that GPX3 was required for IL-10 expression in tumor-polarized GPX3+AT2 cells, which was critical for the immunosuppressive function of AT2 cells in lung PMN formation.
GPX3 Promotes HIF-1α to Induce IL-10 Expression by Inhibiting HIF-1α Degradation.
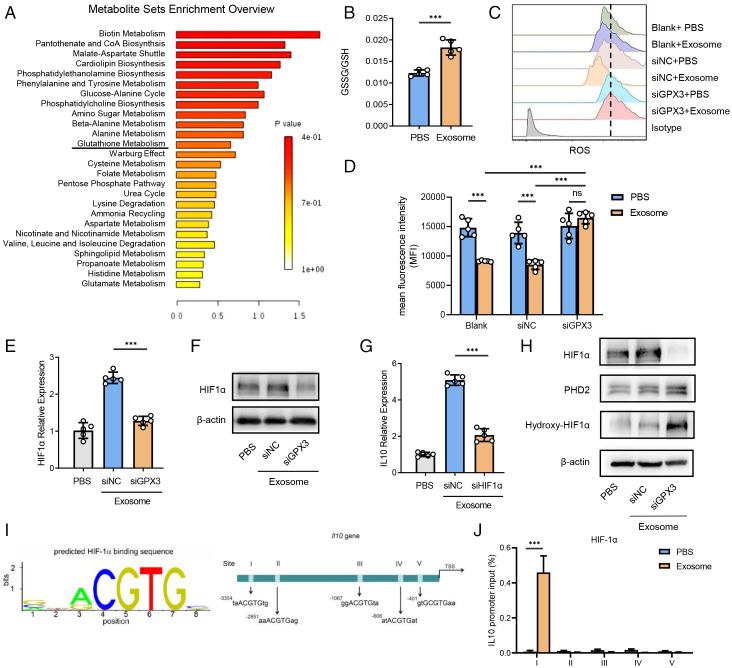
GPX3 is a tripeptide containing a sulfhydryl group bound by glutamic acid, cysteine, and glycine that plays an important role in glutathione metabolism (24). Metabolome analysis of premetastatic lung tissues from tumor-bearing mice showed enrichment of glutathione metabolism compared with tissues from healthy ones (Fig. 5A). Given that GPX3 can catalyze reductive glutathione (GSH) to oxidized glutathione (GSSG)as an important antioxidant way to scavenge reactive oxygen species (ROS) and protect cells from oxidative damage (25, 26), we detected the ratio of GSSG/GSH and found a higher ratio in MLE-12 cells after tumor exosome treatment (Fig. 5B). Indeed, an increased intracytoplasmic level of ROS was observed in GPX3-silenced MLE-12 cells after tumor exosome treatment (Fig. 5 C and D), indicating the clearance of ROS by GPX3.
Fig. 5.
GPX3 stabilizes HIF-1α and promotes HIF-1α-dependent IL-10 induction in AT2 cells. (A) Metabolite sets enrichment of metabolome by comparing the whole lungs of LLC-bearing mice with those of healthy ones. CoA, coenzyme A. (B) The proportion of GSSG/GSH of MLE-12 cells stimulated with tumor exosomes as detected by glutathione assay kit. (C and D) Representative histograms (C) and mean fluorescent intensity (MFI) (D) showing ROS levels of control or GPX3-silenced MLE-12 cells treated with or without tumor exosomes as detected by flow cytometry. The dashed line refers to the highest MFI. (E and F) Messenger RNA (mRNA) expression (E) and protein level (F) of HIF-1α expression in control or GPX3-silenced MLE-12 cells treated with or without tumor exosomes for 2 h. β-actin was used as a control. siNC, silence of negative control; siGPX3, silence of GPX3; siHIF1α, silence of HIF1α. (G) mRNA expression of IL-10 in control or HIF1α-silenced MLE-12 cells treated with or without tumor exosomes for 2 h. (H) The protein level of HIF-1α, PHD2, and hydroxy-HIF-1α in normal or GPX3-silenced MLE-12 cells treated with or without tumor exosomes. (I) Five possible HIF-1α-binding sites containing motif (ACGTG) in IL-10 promoter region. TSS, transcription start site. (J) ChIP analysis of HIF-1α-binding sites in IL-10 promoter region of MLE-12 cells treated with tumor exosomes. Immunoglobulin G was used as a control. Data are mean ± SD of one representative experiment. Similar results were seen in three independent experiments. Unpaired Student’s t tests unless noted. ***P < 0.001. ns, not significant.
To uncover how ROS can affect IL-10 expression, we searched for crucial involved transcription factors. It has been shown that ROS can inhibit hypoxic and nonhypoxic induction of hypoxia-inducible factors (HIFs), among which HIF-1α is best characterized and induces the transcription of target genes (27, 28). We found that tumor exosomes could promote HIF-1α expression in MLE-12 cells, which was greatly down-regulated by the silence of GPX3 on both transcriptional and protein levels (Fig. 5 E and F). Accordingly, immunofluorescence assay indicated that the expression of IL-10 and HIF-1α was much lower in GPX3cKO mice than in littermate controls (SI Appendix, Fig. S5A). Furthermore, HIF-1α silence inhibited IL-10 production induced by tumor exosomes in MLE-12 cells (Fig. 5G). These data suggest that GPX3 induces IL-10 expression via promotion of HIF-1α by clearing of ROS.
It has been reported that HIF-1α can be up-regulated via mammalian target of rapamycin (mTOR), extracellular regulated protein kinases (ERK), and nuclear factor kappa-B (NF-κB) pathways under various stimulus signals (29, 30). However, there were no significant changes in the activation of PI3K/Akt/mTOR, NF-κB, JNK-p38, and ERK signaling pathways in MLE-12 cells with or without GPX3 silence (SI Appendix, Fig. S5B). Furthermore, in well-oxygenated cells, HIF-1α subunits are hydroxylated on proline residues by the oxygen-dependent prolyl-4-hydroxylases (PHDs) (31), resulting in the proteasomal degradation of HIF-1α (32). Indeed, the silence of GPX3 led to increased PHD2 expression and HIF-1α hydroxylation while HIF-1α expression decreased after exosome treatment (Fig. 5H).
As HIF-1α is a transcription factor, we predicted HIF-1α-binding sites in the IL-10 promoter region (33) and found five repeats in total (Fig. 5I). In addition, chromatin immunoprecipitation (ChIP) confirmed the binding of HIF-1α with the region of the first repeat in the IL-10 promoter site in MLE-12 with exosome treatment (Fig. 5J and SI Appendix, Fig. S5C). These data indicate that GPX3 can scavenge ROS and inhibit hydroxylation degradation of HIF-1α, thus promoting IL-10 production.
Discussion
Improved understanding of the polarization of immune cells has provided insights into the complexity of the immune system and tumor microenvironment (TME) (34, 35). Here, we propose the concept of tumor-induced polarization of AT2 cells with high expression of GPX3 and production of IL-10, but not a normal expanded population. First, minimal in vivo proliferation of Sftpc+ AT2 cells were discovered in the lung of tumor-bearing mice. Second, pseudotime analysis of single-cell data indicated that GPX3+ AT2 cells were not in the same branch as other clusters in AT2 cells, indicating the presence of this distinct cluster in tumor-bearing mice. Finally, the function of GPX3+ AT2 cells was totally different after tumor education. The identification of GPX3+ AT2 cells warrants more intensive study in the future.
Both scRNA-seq and RNA-seq data showed that genes in GPX3+ AT2 cells from tumor-bearing mice were enriched in anti-inflammatory pathways (i.e., IL-10 and TGF-β) but not proinflammatory ones (i.e., IL-1, TNF, IFN). Meanwhile, increased proliferation of CD4+ T cells was observed in GPX3cKO mice under LLC or B16/F10 tumor inoculation, along with impaired induction of CD4+ Foxp3+ Treg cells, suggesting that GPX3+ AT2 cells played a crucial role in tumor metastasis, at least partially by inhibiting T cell response. Furthermore, CD4+ T cell depletion in vivo can reverse the reduction of metastasis in GPX3cKO mice, confirming a CD4+ T cell-dependent manner for the prometastatic function of GPX3+ AT2 cells. More detailed information about the mechanisms of polarized AT2 and its role in regulating the function of CD4+ T cells or other immune cells (i.e., neutrophils) is needed.
GPX3 is critical for IL-10 production in tumor-polarized AT2 cells, while IL-10 is required for the tumor-polarized immunosuppressive function of GPX3+ AT2 cells. As a glutathione peroxidase, GPX3 catalyzes GSH to GSSG to scavenge surplus ROS (36). We found that GPX3-mediated ROS consumption led to HIF-1α accumulation by inhibiting the degradation of HIF-1α. Given that HIF-1α can transcriptionally regulate IL-10 expression in B cells, possibly through two hypoxia-responsive elements (HREs) (33), the binding of HIF-1α to HRE I, including consensus core (A/GCGTG) on the IL-10 promotor, was then verified by ChIP after exosome administration. The axis of GPX3/HIF-1α/IL-10 elucidates a mechanism for the inhibition of CD4+ T cell proliferation and the induction of regulatory T cells of AT2 cells, which may be a promising target for potentiating anti-tumor immunity in PMN. More studies with IL-10 CKO mice are needed to validate its function in lung epithelial cells and the formation of lung PMN.
With accumulated evidence demonstrating that GPX3 acts as both a tumor suppressor and a promoter during tumor progression (21), mice with CKO of GPX3 in AT2 cells were therefore studied to elucidate its role in stromal cells and lung PMN formation. We found that GPX3 knockout greatly reduced lung metastasis and prolonged survival of mice with inoculation of LLC or B16/F10 tumor cells. As no other specific markers, except GPX3 and IL-10, were identified as the markers of the population, it is now difficult to confirm whether this functional population exists in GPX3cKO mice. However, it could be more reasonable to be the loss of function of this subpopulation in GPX3cKO mice because there is no evidence to show that GPX3 can control its development. Next, we will search for more markers to help identify tumor-polarized GPX3+ AT2 cells and specific transcription factors for the development of GPX3+ AT2 cells. The current data, which were presented using a spontaneous in vivo lung metastasis model, indicate that GPX3 could be a potential biomarker and therapeutic target for lung metastasis.
Taken together, our findings illustrate that IL-10-producing GPX3+ AT2, a tumor-polarized subpopulation of lung epithelial cells, can inhibit T cell responses and promote PMN formation. Thus, our study may add insights into the formation of a favorable immunosuppressive microenvironment that facilitates lung metastasis, providing potential targets for cancer intervention.
Materials and Methods
Mice and Cells.
GPX3 Cas9-CKO mice and Sftpc-IRES-iCre Cas9-KI mice with a C57BL/6J background were obtained from GemPharmatech Co., Ltd. GPX3fl/fl mice were crossed with mice expressing Sftpc-IRES-iCre recombinase, leading to the loss of function of GPX3 in AT2 cells of the mice. C57BL/6J mice were from Joint Ventures Sipper BK Experimental Animal Company. All mice were maintained in specific pathogen-free conditions. All mouse experiments were performed under the supervision of the Scientific Investigation Board of Naval Medical University, Shanghai. All mice used in experiments were of matched sex and age, ranging from 6 to 10 wk. MLE-12 cells, LLC cells, and B16/F10 cells were obtained from American Type Culture Collection.
Preparation of Tumor-Bearing Mice.
In spontaneous lung metastatic models,1*106 luciferase-labeled LLC or B16/F10 cells were subcutaneously injected into the dorsum of mice. The tumors were removed surgically after diametering approximately up to 10 mm (range between 20 and 21 d). Lung metastasis was confirmed by ex vivo bioluminescence imaging by IVIS Lumina II (Perkin-Elmer) and detected by hematoxylin/eosin (H&E) staining with paraffin sections for 40 d.
Lung Tissue Dissociation and Cell Isolation.
Lung tissues were collected from healthy mice or mice with tumor inoculation for 14 d and dissociated with 0.5 mg/mL Liberase (Sigma-Aldrich) and 25 mg/mL deoxyribonuclease (Sigma-Aldrich). The solution was pipetted every 10 min during the incubation, and the suspension was dispersed through a 70-mm cell strainer. To purify cells from lung tissues, single-cell suspensions from lungs were stained with antibodies, and then lung epithelial cells (CD45− Epcam+) or AT2 cells (CD45− Sftpc+) were sorted using a flow cytometer (SH800, Sony) with purities of ≥98%.
scRNA-seq.
A female C57BL/6J mouse at an age of 8 wk was subcutaneously inoculated with 1*106 LLC. Lung tissues were collected from this mouse 14 d after tumor inoculation or from a healthy mouse and then dissociated into single-cell suspensions. Lung epithelial cells were purified for 10× library preparation and sequencing. A total of 20,354 cells were sequenced after cell filtration, including 8,550 cells from the tumor-bearing mouse and 11,804 cells from the healthy one. All procedures were performed according to standard manufacturer’s protocol (CG000206 RevD) by Shanghai Biotechnology Corporation. Sequencing libraries were sequenced on NovaSeq6000 (Illumina) using 2 × 150 chemistry and quantified using a High-Sensitivity DNA Chip (Agilent) on a Bioanalyzer 2100 and the Qubit High Sensitivity DNA Assay (Thermo Fisher Scientific). scRNA-seq data processing reads were processed using the Cell Ranger 2.1.0 pipeline with default and recommended parameters. FASTQs generated from Illumina sequencing output were aligned to the mouse genome with the STAR algorithm (37), version GRCm38. The filtered output of gene-barcode matrix was then imported into the Seurat (v3.0.2) R toolkit (38). We excluded cells with fewer than 200 or more than 6,000 detected genes and cells with more than 10% expression of mitochondria genes using the PercentageFeatureSet function of the Seurat package (38). Data were normalized by NormalizeData of Seurat and integrated after “anchors” were identified between datasets using FindIntegrationAnchors and IntegrateData in the Seurat package (38, 39). Principal component analysis was then performed and reduced to the top 30 components. We visualized the clusters on a two-dimensional map, and cells were clustered using graph-based clustering with the Louvain Method after computing a shared nearest neighbor graph (38). For subclustering, we applied the same procedure of scaled dimensionality reduction and clustering to the specific set of data. For each cluster, we used the Wilcoxon rank sum test to find significant deferentially expressed genes compared to the remaining clusters. SingleR (40) and known marker genes were used to identify cell types. Gene Ontology, Kyoto encyclopedia of genes and genomes (KEGG), and GSEA pathway enrichment analyses of differentially expressed genes (DEGs) were performed by clusterProfiler (v 3.14.0) package.
Flow Cytometry.
For flow cytometry staining, single-cell suspensions from lungs were stained with the indicated antibodies at room temperature (RT) for 20 min, including CD4+ T cells (CD45+ CD4+), CD8+ T cells (CD45+ CD8+), B cells (CD45+ CD19+), macrophages (CD45+ CD11b+ F4/80+), monocyte-like myeloid-derived suppressor cells (MDSCs) (Ly6C+ Ly6G−), granulocyte-like cells (Ly6C− Ly6G+), natural killer (NK) cells (CD45+ NK1.1+), and epithelial cells (CD45− Epcam+). The cells were washed twice and analyzed by flow cytometry on both BD LSR Fortessa (BD Biosciences) and SONY ID7000 with FlowJo software. For intracellular staining, cells were washed, fixed, and permeabilized using the FOXP3 Fix/Perm Buffer Set (Biolegend, 421403) according to manufacturer’s protocol. Fluorescence intensity was analyzed on the fluorescence-activated cell sorter LSR Fortessa with FlowJo software (BD Biosciences).
Cell Proliferation Assay.
Splenic CD4+ T cells were purified by CD4+ T cell isolation magnetic beads (Miltenni), and 5*104 cells labeled with carboxyfluorescein succinimidyl ester (CFSE) (MCE, HY-D0938) were cocultured with 1*105 purified AT2 cells indicated above in 96-well plates. Anti-CD3/CD28 antibody (1 μg/mL) and IL-2 were added to stimulate T cell proliferation. The cells were cocultured in an incubator at 37 °C for 72 h with constant temperature and humidity. Proliferation was monitored by flow cytometry on BD LSR Fortessa (BD Biosciences).
Immunofluorescence Analysis.
In vivo lung tissues of tumor-bearing mice and wild-type (WT) littermates were fixed in formalin, embedded with paraffin, and cut into 4-μm-thick sections from paraffin-embedded samples. The formalin-fixed and parrffin-embedded (FFPE) tissue sections were dewaxed and rehydrated through an ethanol gradient. Antigen retrieval was obtained by a pressure cooker in citrate buffer (pH 6.0) for 3-min heating, and endogenous peroxidase was blocked by 3% H2O2 for 10 min at RT. The sections were then incubated with goat serum for 30 min and antibodies for 1 h at RT, after which the slides were rinsed with phosphate-buffered saline (PBS) three times and incubated with secondary antibodies for 10 min at RT. Fluorescent amplification signal solution (1:200) was then used, and DAPI was applied to label nuclear cells. Whole-tissue slides were scanned at 4× magnification performed with Vectra automatic quantitative pathological imaging analysis system by Phenochart panoramic analysis software and inForm Analysis Software (PerkinElmer).
Tumor Exosome Isolation.
Tumor-derived exosomes were separated as previously described (41). Briefly, tumor tissues were collected, cut into small pieces, added to fresh culture medium containing no serum, and incubated for 12 h, after which exosomes were purified by three successive centrifugations at 300 × g (5 min), 1,200 × g (20 min), and 10,000 × g (30 min) to remove cellular debris. The supernatant was then filtered by strainers of 1.2 μm, 0.8 μm, and 0.2 μm consecutively, followed by ultracentrifugation at 110,000 × g for 90 min at 4 °C and resuspension of sediments in PBS. For in vivo exosome treatment, the intravenous injection via tail vein of 5 mg exosomes (50 μg/μL) was conducted as described previously every 3 d (15), and tumor cells were subcutaneously injected into mice 14 d later.
LS-MS.
Lung tissues of mice burdened with LLC tumor for 14 d and WT littermates were extracted with internal standard and extraction solvent, ground at 60 Hz for 2 min, ultrasonicated at RT for 10 min, and centrifuged at 13,000 rpm, 4 °C for 15 min. The supernatants were collected, filtered with 0.22-μm strainers, and quality controlled. The metabolic profiling of both electrosprayionization (ESI)-positive and -negative ion modes was analyzed with ACQUITY UHPLC system from Waters Corporation and AB SCIEX Triple TOF 5600 System. Water and acetonitrile, both containing 0.1% formic acid, were used as mobile phases A and B, respectively. Full-scan mode (m/z ranges from 70 to 1,000) coupled with information dependent acquisition (IDA) mode were applied to acquire data, and repeats were achieved by injecting quality controls (QCs) at regular intervals of every 10 samples throughout the process. The analysis of liquid chromatography-mass spectrometry (LC-MS) was performed by Progenesis QI software from Waters Corporation based on public databases, including www.hmdb.ca/, https://www.lipidmaps.org/, and self-built databases of Shanghai Lu-Ming Biotech Co., Ltd., Shanghai, China.
Statistical Analysis.
Comparisons of the mean between groups were performed using unpaired Student’s t test and two-way ANOVA. The survival rate was determined by Kaplan-Meier test. Data analysis was performed using Statistical Package for the Social Sciences software for Windows and Prism 8 (version 8.0.2): *P < 0.05; **P < 0.01; ***P < 0.001 unless otherwise stated; ns, not significant.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (32170918, 81788101, and 81972683) and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-017).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201899119/-/DCSupplemental.
Data Availability
The single-cell sequencing and RNA-seq data from this study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under Accession Nos. GSE195754 (42) and GSE195755 (43), respectively. All other study data are included in the article and/or SI Appendix.
References
- 1.Hanahan D., Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Zhang M., et al. , Splenic stroma drives mature dendritic cells to differentiate into regulatory dendritic cells. Nat. Immunol. 5, 1124–1133 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Peinado H., et al. , Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Shen M., et al. , Tinagl1 suppresses triple-negative breast cancer progression and metastasis by simultaneously inhibiting integrin/FAK and EGFR signaling. Cancer Cell 35, 64–80.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Gu Y., et al. , Tumor-educated B cells selectively promote breast cancer lymph node metastasis by HSPA4-targeting IgG. Nat. Med. 25, 312–322 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Kaplan R. N., et al. , VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoshino A., et al. , Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong J., et al. , Extracellular vesicles of carcinoma-associated fibroblasts creates a pre-metastatic niche in the lung through activating fibroblasts. Mol. Cancer 18, 175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Cao X., Characteristics and significance of the pre-metastatic niche. Cancer Cell 30, 668–681 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Barlow J. L., McKenzie A. N. J., Innate lymphoid cells of the lung. Annu. Rev. Physiol. 81, 429–452 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Headley M. B., et al. , Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 531, 513–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian B.-Z., et al. , CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., et al. , Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 18, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T., et al. , Loss of SMAD4 promotes lung metastasis of colorectal cancer by accumulation of CCR1+ tumor-associated neutrophils through CCL15-CCR1 axis. Clin. Cancer Res. 23, 833–844 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., et al. , Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Liang W., Li Q., Ferrara N., Metastatic growth instructed by neutrophil-derived transferrin. Proc. Natl. Acad. Sci. U.S.A. 115, 11060–11065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkauskas C. E., et al. , Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtzman M. J., Byers D. E., Alexander-Brett J., Wang X., The role of airway epithelial cells and innate immune cells in chronic respiratory disease. Nat. Rev. Immunol. 14, 686–698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomita T., Sakurai Y., Ishibashi S., Maru Y., Imbalance of Clara cell-mediated homeostatic inflammation is involved in lung metastasis. Oncogene 30, 3429–3439 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Barrett C. W., et al. , Tumor suppressor function of the plasma glutathione peroxidase gpx3 in colitis-associated carcinoma. Cancer Res. 73, 1245–1255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C., Worley B. L., Phaëton R., Hempel N., Extracellular glutathione peroxidase GPx3 and its role in cancer. Cancers (Basel) 12, 2197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaguchi S., et al. , Regulatory T cells and human disease. Annu. Rev. Immunol. 38, 541–566 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez P. C., et al. , Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 64, 5839–5849 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Weinberg J. M., Davis J. A., Abarzua M., Rajan T., Cytoprotective effects of glycine and glutathione against hypoxic injury to renal tubules. J. Clin. Invest. 80, 1446–1454 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mak T. W., et al. , Glutathione primes T cell metabolism for inflammation. Immunity 46, 675–689 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Peng D. F., et al. , DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut 58, 5–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flügel D., Görlach A., Kietzmann T., GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 119, 1292–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao D., Corle C., Seagroves T. N., Johnson R. S., Hypoxia-inducible factor-1α is a key regulator of metastasis in a transgenic model of cancer initiation and progression. Cancer Res. 67, 563–572 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Karar J., Maity A., PI3K/AKT/mTOR pathway in angiogenesis. Front. Mol. Neurosci. 4, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee K. E., Simon M. C., SnapShot: Hypoxia-inducible factors. Cell 163, 1288–1288.e1 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Leite de Oliveira R., et al. , Gene-targeting of Phd2 improves tumor response to chemotherapy and prevents side-toxicity. Cancer Cell 22, 263–277 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Gilkes D. M., Semenza G. L., Wirtz D., Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat. Rev. Cancer 14, 430–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng X., et al. , Hypoxia-inducible factor-1α is a critical transcription factor for IL-10-producing B cells in autoimmune disease. Nat. Commun. 9, 251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivashkiv L. B., Nγ I. F., IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 18, 545–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponzetta A., et al. , Neutrophils driving unconventional T cells mediate resistance against murine sarcomas and selected human tumors. Cell 178, 346–360.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X., Kang S., Tanaka S., Park S., Monitoring the glutathione redox reaction in living human cells by combining metabolic labeling with heteronuclear NMR. Angew. Chem. Int. Ed. Engl. 55, 7939–7942 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Le D. T., et al. , Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aran D., et al. , Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen T., Guo J., Yang M., Zhu X., Cao X., Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J. Immunol. 186, 2219–2228 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Y. Gu, Z. Wang, Single-cell transcriptome of tumor-educated lung epithelial cells. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195754. Deposited 21 July 2022. [Google Scholar]
- 43.Y. Gu, Z. Wang, Transcriptome sequencing of tumor-educated lung epithelial cells in the pre-metastatic niche. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195755. Deposited 21 July 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell sequencing and RNA-seq data from this study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under Accession Nos. GSE195754 (42) and GSE195755 (43), respectively. All other study data are included in the article and/or SI Appendix.