Significance
Xylomyrocins are a unique family of nonribosomal peptides (NRPs) from a limited cohort of phytopathogenic fungi. Xylomyrocin biosynthesis utilizes a sugar acid, derived from carbohydrate metabolism, to initiate the assembly of depsipeptides. This unprecedented metabolic interchange extends the precursor structural space available for NRP synthetase–based natural product assembly. The xylomyrocin NRP synthetase further increases the structural diversity of xylomyrocins by leveraging the multiple alcohol functionalities of the polyol to generate macrocyclic ester regioisomers. Some xylomyrocins display promiscuous peptide backbone N-methylation, despite an otherwise collinearly programmed NRP synthetase. Understanding such rare, flexible biosynthetic programs will improve genome mining and sequence-based NRP structure prediction and open new avenues to generate “unnatural” NRPs for crop protection or pharmaceutical drug discovery.
Keywords: fungal nonribosomal peptides, xylonic acid, cyclodepsipeptides, molecular networking, natural product dereplication
Abstract
Xylomyrocins, a unique group of nonribosomal peptide secondary metabolites, were discovered in Paramyrothecium and Colletotrichum spp. fungi by employing a combination of high-resolution tandem mass spectrometry (HRMS/MS)–based chemometrics, comparative genome mining, gene disruption, stable isotope feeding, and chemical complementation techniques. These polyol cyclodepsipeptides all feature an unprecedented d-xylonic acid moiety as part of their macrocyclic scaffold. This biosynthon is derived from d-xylose supplied by xylooligosaccharide catabolic enzymes encoded in the xylomyrocin biosynthetic gene cluster, revealing a novel link between carbohydrate catabolism and nonribosomal peptide biosynthesis. Xylomyrocins from different fungal isolates differ in the number and nature of their amino acid building blocks that are nevertheless incorporated by orthologous nonribosomal peptide synthetase (NRPS) enzymes. Another source of structural diversity is the variable choice of the nucleophile for intramolecular macrocyclic ester formation during xylomyrocin chain termination. This nucleophile is selected from the multiple available alcohol functionalities of the polyol moiety, revealing a surprising polyspecificity for the NRPS terminal condensation domain. Some xylomyrocin congeners also feature N-methylated amino acid residues in positions where the corresponding NRPS modules lack N-methyltransferase (M) domains, providing a rare example of promiscuous methylation in the context of an NRPS with an otherwise canonical, collinear biosynthetic program.
Small-molecule nonribosomal (depsi)peptide secondary metabolites (SMs) of bacteria and fungi are important allelochemicals that contribute to pathogenesis as virulence factors and toxins, mediate intra- and interspecies communication, and increase the fitness of their producers as siderophores and chemoprotectors against environmental stressors (1, 2). Many of these compounds have also been developed as marketed drugs or pesticides or serve as probes for chemical biology or lead compounds for drug discovery.
Nonribosomal (depsi)peptide scaffolds are assembled by nonribosomal peptide synthetase (NRPS) enzymes by recursive condensation of a large variety of aminoacyl, ketoacyl, or hydroxyacyl monomers. NRPSs are complex multidomain megasynthases that contain three types of highly conserved core domains: 1) adenylation (A) domains that are responsible for the activation and loading of the monomer units onto the NRPS; 2) peptidyl carrier domains (also known as thiolation domains) that covalently tether the precursors and the intermediates; and 3) condensation (C) domains that catalyze the sequential condensation of the monomers with the growing peptide chain (1–3). NRPSs may feature a chain assembly initiation domain for N-terminal acylation and contain a peptide chain assembly-terminating domain such as a thioesterase, a terminal C domain (CT), or a reductase domain. In addition, NRPSs may also incorporate a variety of additional domains such as N-methyltransferase (M) domains that modify the monomers or the intermediates. Fungal NRPSs may contain one or more modules (assemblies of single copies of core, modification, initiation, and termination domains). These modules often act in a sequential, noniterative manner with a single monomer incorporated by each module (Type A, collinear NRPSs). In other cases, certain modules (Type B, iterative NRPSs) or specific domains (Type C, nonlinear NRPSs) are reused during biosynthesis, making a priori structure predictions precarious from sequence information alone (1, 4). Structural diversity may be further increased by modifications of the nonribosomal peptide (NRP) scaffold, conducted by additional enzymes whose encoding genes are typically colocated in the genome with the NRPS gene, forming a biosynthetic gene cluster (BGC).
With 11,039 fungal genome assemblies in the National Center for Biotechnology Information (NCBI) GenBank and 2,271 in the Joint Genome Institute (JGI) MycoCosm (May 23, 2022), genome mining was hoped to provide a treasure throve of structurally novel small-molecule SM candidates whose deduced structures could then be verified by targeted metabolomics (4–8). However, fungal genomic information is a surprisingly inadequate basis for targeting unprecedented SM scaffolds. Thus, iterative Type B and Type C fungal NRPSs are governed by an idiosyncratic and largely cryptic program. Worse, as opposed to bacterial NRPSs where the specificity signatures of the A domains (9, 10) are good predictors for the amino acid being incorporated, fungal A domain specificity remains remarkably unpredictable by bioinformatic means. This is further exacerbated when non–amino acid building blocks are incorporated, whose nature, and even presence, remains largely opaque to bioinformatic predictions. Together with the unpredictability of the regiospecificity (and often the exact nature) of post-NRPS tailoring steps and the fact that fungal BGCs frequently remain silent under routine laboratory growth conditions (11), genome mining is often constrained to the prediction of the theoretical capacity of a fungal strain to produce members of an already known SM family (12).
The recent development of untargeted SM metabolomics, together with chemometric, molecular networking-based dereplication (5, 13, 14) and prioritization (15) techniques, provides practical means to identify potentially novel SMs in prokaryotes (4, 8, 13, 16) and increasingly in fungi (17). Shortlisting candidate BGCs from the genome sequence with gene content and NRPS characteristics congruent with the SM structure empowers the validation of these BGCs by functional assays (18–21). Next, comparative genomic analyses may reveal additional congeners of the founding SM or highlight compounds with shared, distinguishing structural motifs installed by characteristic biosynthetic enzymes (17). In this work, we adapted such a workflow to discover an unprecedented family of fungal polyol cyclodepsipeptides by using rapid matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) to scan a large collection of fungal isolates, followed by liquid chromatography–high-resolution tandem mass spectrometry (LC-HRMS/MS)–based molecular networking. Identification of the BGC in the newly sequenced genome of the producer fungus allowed us to expand the family by comparative genome mining and targeted metabolomics in taxonomically related isolates. The resulting polyol cyclodepsipeptides showed moderate antimalarial activity, demonstrated unusual NRPS programming, and most importantly, highlighted unprecedented connections between NRP biosynthesis and carbohydrate catabolism.
Results
Discovery of Polyol Cyclodepsipeptides by HRMS-Guided Chemometrics.
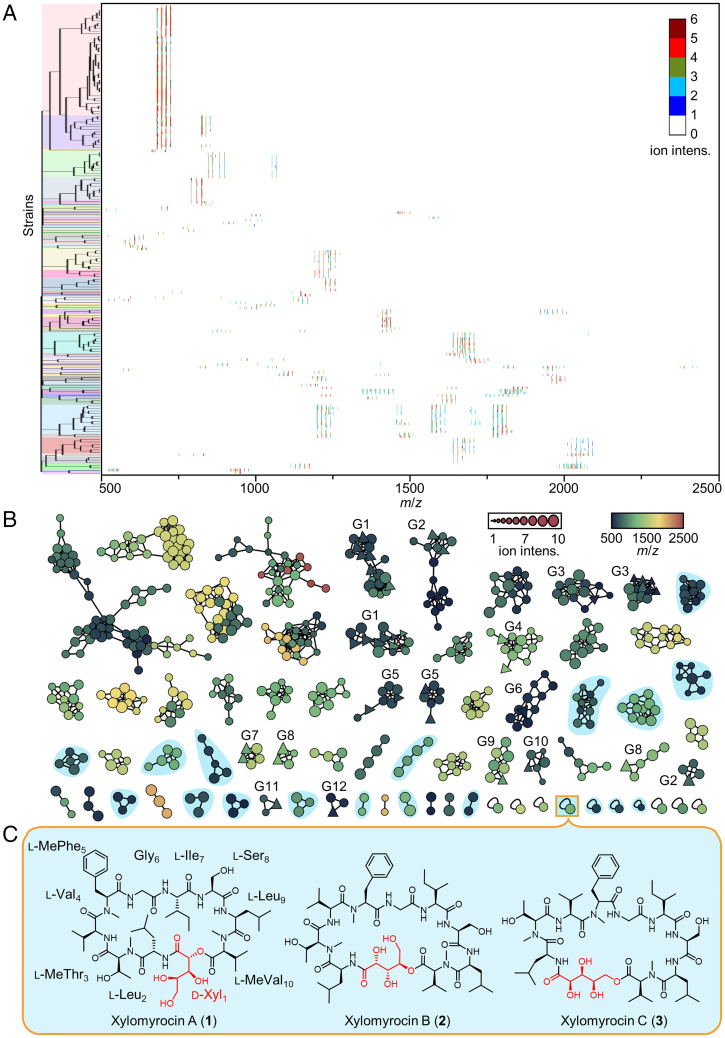
We surveyed a library of 1,748 fungal strains for the production of peptide SMs. We utilized a rapid and cost-effective miniaturized assay system involving the in situ extraction of a loopful (<5 mg) of mycelia from agar media, followed by direct MALDI-TOF MS analysis. Of the 1,748 strains (SI Appendix, Table S1), 182 were found to produce at least one set of peptide-like SM each. These peptides differed in their mass to charge (m/z) ratios and ion intensities. Next, we used a hierarchical clustering approach to deconvolute the 182 peptide-containing SM extracts, revealing 61 clades where members of a given clade share a characteristic peptide fingerprint in their MALDI-TOF MS profiles (Fig. 1A). We selected one representative strain for each of the 61 clades to conduct solid-state fermentations and monitored SM production by LC-HRMS/MS. The resulting data were used to generate a molecular network using the Global Natural Products Social (GNPS) platform (14). This molecular network not only established structural relationships among the peptides but also dereplicated congeners of known SMs (Fig. 1B) by cross-referencing the public MS/MS database of the GNPS platform. Two criteria were then formulated to further prioritize candidate strains for large-scale fermentation and detailed chemical analyses. First, those peptides that cluster with known compounds were excluded, since they likely represent analogs of the known peptides with only minimal structural variations. Contrarily, members of the clades without any GNPS library hit (clades with round nodes only on Fig. 1B), as well as the peptides corresponding to the unconnected nodes (stand-alone round nodes at the bottom of Fig. 1B), were considered to have a better chance of delivering novel structures. Second, we ruled out those candidates that presented a clear and evident MS/MS fragmentation pattern (a characteristic of linear peptides) and concentrated on those with complex MS/MS profiles (an indicator of cyclic peptides). By applying these filters, we were able to pinpoint 10 promising candidate strains that may have produced multiple novel cyclic peptides (nodes shaded in blue, Fig. 1B).
Fig. 1.
MS-guided chemometric discovery of polyol cyclodepsipeptides. (A) Hierarchical clustering of the MALDI-TOF MS spectra of SM extracts from 182 fungal isolates (SI Appendix, Table S1). The resulting 61 clades are highlighted by different colors on the tree. The heat map shows the MS fingerprints of the extracts. The gradient color indicates the intensity (intens.) of each ion signal. (B) Molecular network analysis of the peptide SMs, based on the LC-HRMS/MS fingerprints of metabolite extracts from fermentations with 61 fungal strains, exemplifying the 61 clades identified in A. Each node represents the parent ion of one single peptide. The shape of the node indicates whether the MS/MS spectrum of the peptide has a GNPS database hit (triangle, known peptide; circle, unknown peptide). The size and color of the node indicates the MS intensity and the m/z of the parent ion, respectively. Peptides with similar MS/MS spectra are connected by edges and form a clade. The length of the edge indicates the structural similarity between two peptides. Clades labeled as G1–G12 contain putative analogs of SMs with known structures (G1: enniatins and bassianolides; G2: beauvericins; G3: hormonemates; G4: acremostatins; G5: arenamide A and emericellamide A; G6: communesin B; G7: GNPS spectrum ID CCMSLIB00000001578 and CCMSLIB00000001580; G8: cyclosporins; G9: trikoningin-KB-I; G10: ferrichrome; G11: GNPS spectrum ID CCMSLIB00000853575; G12: 27-epi-tryptoquivaline); those highlighted in cyan indicate putative novel cyclic peptides, collectively produced by 10 representative fungal strains. [M+H]+ and [M+NH4]+ adducts of the same peptide tend to group together to form one cluster, while [M+Na]+ adducts of the same peptide routinely form a separate cluster. (C) The chemical structures of xylomyrocins A–C (1–3), polyol cyclodepsipeptides isolated from Paramyrothecium sp. XJ0827. d-Xyl, d-xylonic acid; l-Leu, l-leucine; l-MeThr, N-methyl- l-threonine; l-Val, l-valine; l-MePhe, N-methyl- l-phenylalanine; Gly, glycine; l-Ile, l-isoleucine; l-Ser, l-serine; l-MeVal, N-methyl- l-valine.
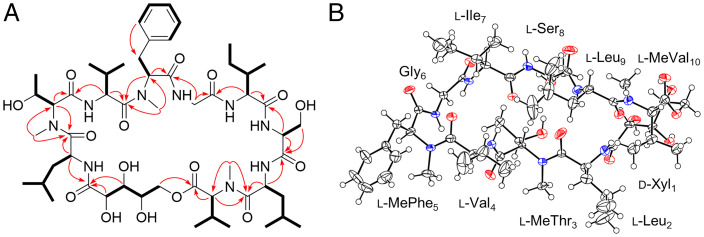
Among these prioritized strains, Paramyrothecium sp. XJ0827 was found to biosynthesize xylomyrocins A–C (1–3), three SMs of the same molecular mass (1,119.64 Da; Fig. 1C). The 13C NMR spectrum of 3 displayed multiple carbonyl carbon signals at δC 171.2 to 176.1 and heteroatom-bound carbon signals at δC 54.8 to 74.8 (SI Appendix, Table S2.1), strongly indicating that it is a peptide natural product. A comprehensive analysis of the two-dimensional (2D) NMR spectra (Fig. 2A) revealed that 3 consists of a 2,3,4,5-tetrahydroxypentanoic acid (THP) moiety and nine amino acid residues, including glycine (Gly), isoleucine (Ile), leucine (Leu, 2×), N-methylated phenylalanine (MePhe), N-methylated threonine (MeThr), N-methylated valine (MeVal), serine (Ser), and valine (Val). Detailed examination of the correlations from the α-proton(s)/N-methyl protons to the amide carbonyl carbons in heteronuclear multiple bond correlation (HMBC) spectra (Fig. 2A) established a primary sequence of THP1–Leu2–MeThr3–Val4–MePhe5–Gly6–Ile7–Ser8–Leu9–MeVal10 for 3, while the HMBC correlation between the δ-protons of THP1 (δH 4.41, 4.09) and the carbonyl carbon (δC 171.87) of MeVal10 confirmed the connecting site between THP1 and MeVal10, thus fully establishing the planar structure of 3 (Fig. 2A). De novo sequencing of 3 by analyzing the tandem mass spectrum was inconclusive due to the complex fragmentation pattern resulting from the random ring opening of the cyclodepsipeptide backbone during collision-induced dissociation. To avoid such complexity, 3 was first linearized by methanolysis upon treatment with CH3ONa/CH3OH, and the resulting methoxy-substituted derivative (19) was subjected to tandem mass analysis (SI Appendix, Fig. S1.1). The peptide sequence of 3 deduced from the MS/MS fragmentation data of 19 was in complete agreement with that elucidated by NMR spectrometry, thus further supporting the established planar structure of 3. Similarly, the planar structures of 1 and 2 were elucidated by interpreting the NMR and HRMS/MS data (SI Appendix, Table S2.1 and Figs. S1.2, S1.3, and S2). Although 1 and 2 share the same peptide sequence with 3, they differ in the macrocycle connectivity, utilizing the α or the γ hydroxyl group of the THP unit to form an ester bond with the carbonyl group of MeVal10, respectively (Fig. 1C). Incubation of xylomyrocins 1–3 in mildly acidic solvents or buffers failed to lead to the facile interconversion of these regioisomers (SI Appendix, Fig. S3). This indicates that these metabolites are genuine biosynthetic congeners and not products of spontaneous intramolecular transesterification or artifacts of isolation.
Fig. 2.
Structure elucidation of xylomyrocin C (3). (A) Key 1H-1H TOCSY/COSY (bold lines) and HMBC (plain arrows) correlations of 3. (B) Oak Ridge Thermal-Ellipsoid Plot Program (ORTEP) representation of 3 obtained from X-ray crystallographic analysis. Red and blue circles represent O and N atoms, respectively.
All the amino acid components of 1–3 have l-configuration, as determined by the advanced Marfey’s method (22). The overall spatial structure of xylomyrocin C (3) was established using X-ray single crystal diffraction, also verifying the identity of the THP moiety as d-xylonic acid (Fig. 2B). Incorporation of a d-xylonic acid building block into an NRP and the amino acid sequences of 1–3 are both unprecedented for peptide SMs (based on search results in SciFinder, the Dictionary of Natural Products, and the NORINE (23), COCONUT (24), and NP Atlas (25) online databases), thereby establishing xylomyrocins A–C as the founding members of polyol cyclodepsipeptides, a novel family of NRPs.
Identification of the Xylomyrocin BGC of Paramyrothecium sp. XJ0827.
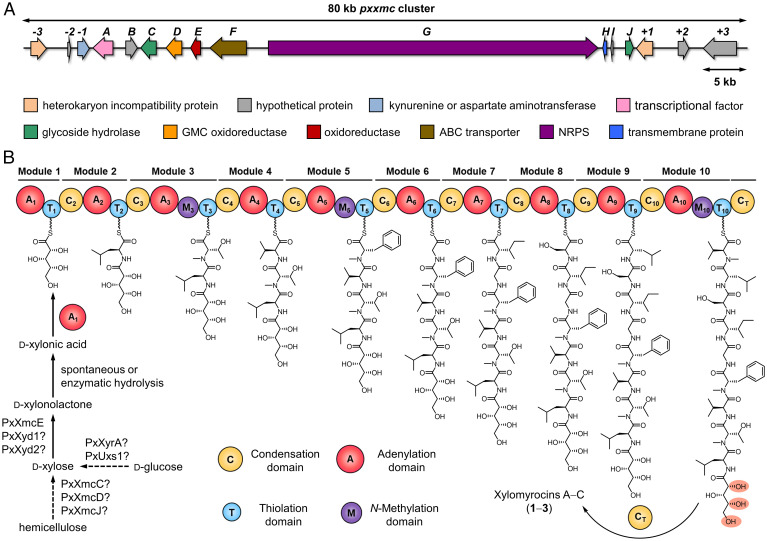
To gain insight into the biosynthesis of xylomyrocins A–C (1–3), we determined the genome sequence of Paramyrothecium sp. XJ0827 using Illumina and Nanopore sequencing platforms (SI Appendix, Table S3; NCBI BioProject: PRJNA770833, Accession No.: JAJFPB000000000). Bioinformatic analysis of the genome assembly using antiSMASH 5.0 (26) revealed that Paramyrothecium sp. XJ0827 has a rich repertoire of SM BGCs (SI Appendix, Table S4). Twenty-six out of the 98 predicted BGCs encode an NRPS or an NRPS-like enzyme as the core biosynthetic enzyme (SI Appendix, Table S5). Among the NRPSs, PxNRPS6.4, PxNRPS6.5, and PxNRPS11.3 each contain enough modules to produce the decapeptide scaffolds of compounds 1–3 using a sequential (noniterative) assembly process. Iterative or nonlinear assembly modes were deemed unlikely since xylomyrocins A–C do not contain successive, repeated amino acid blocks. A closer look at the domain organizations of these NRPSs revealed that PxNRPS11.3 would likely synthesize a tetradecapeptide with N-methylated residues at the 3rd, 5th, 7th, 10th, and 14th positions, while PxNRPS6.5 may assemble a peptaibiotic-like unmethylated tetracosapeptide. In contrast, PxNRPS6.4 not only has the exact number of modules necessary for the collinear assembly of 1–3 but also harbors an N-methylation (M) domain each in the 3rd, 5th, and 10th modules, matching the N-methylation pattern of 1–3 perfectly (Fig. 3B). PxNRPS6.4 also features a C domain at its C-terminus (CT), which is a strong indicator for macrocyclic peptide products in fungal NRPSs (27, 28). The predicted substrate specificity signatures (10, 29) of the A domains in all modules of PxNRPS6.4 were in broad agreement with the amino acids found in xylomyrocins A–C (SI Appendix, Table S6). The first A domain (A1) of PxNRPS6.4 shares high sequence identities with α-hydroxy acid–activating A domains of well-characterized fungal cyclodepsipeptide synthetases (SI Appendix, Table S7) (30, 31). The replacement of a highly conserved aspartic acid with a glycine in the A1 domain (SI Appendix, Table S6) also predicts activation of a non–amino acid substrate (30–32), in agreement with the presence of d-xylonic acid in 1–3 as the first building block. Taken together, bioinformatic analyses strongly suggest that PxNRPS6.4 (heretofore, PxXmcG) is responsible for the biosynthesis of the cylodepsi-decapeptide core of xylomyrocins in Paramyrothecium sp. XJ0827.
Fig. 3.
Biosynthesis of xylomyrocins A–C (1–3). (A) Schematic representation of the pxxmc biosynthetic locus of Paramyrothecium sp. XJ0827. ABC, ATP-binding cassette; NAD, nicotinamide adenine dinucleotide. (B) Module and domain organization of the Paramyrothecium sp. XJ0827 NRPS PxXmcG (12,303 amino acids; 1,364.6 kDa) and the proposed biosynthesis of 1–3.
Detailed analysis of the genomic locus containing pxxmcG revealed a plausible BGC for xylomyrocin production (Fig. 3A). This BGC contains four genes, pxxmcC, pxxmcD, pxxmcE, and pxxmcJ, whose encoded enzymes are likely involved in the biosynthesis of the unprecedented d-xylonic acid building block (SI Appendix, Table S8). Thus, PxXmcD is a putative glucose-methanol-choline (GMC) superfamily oxidoreductase. Together with fungal or plant-derived phenols, GMC oxidoreductases are part of an electron transfer system that reduces the active site copper of lytic polysaccharide monooxygenases to initiate hemicellulose degradation and subsequent xylooligosaccharide formation in phytopathogenic or saprobic fungi (Fig. 3B) (33, 34). Such xylooligosaccharides may be further broken down by glycoside hydrolases, such as the secreted β-1,4-xylosidase PxXmcC and the glycoside hydrolase PxXmcJ, to yield d-xylose monomers. Finally, PxXmcE is an oxidized nicotinamide-adenine dinucleotide (NAD+)–dependent oxidoreductase with a WcaG (COG0451) conserved domain. In analogy with uronate dehydrogenases (35) that also feature such a domain, PxXmcE may produce d-xylonic acid from d-xylose via the oxidation of the anomeric carbon.
The pxxmc BGC also features genes encoding a fungal transcription factor with a GAL4-like C6 zinc binuclear cluster DNA-binding domain (pxxmcA); an adenosine 5′-triphosphate (ATP)–binding cassette transporter (pxxmcF); a transmembrane protein of unknown function (pxxmcH); and two hypothetical proteins (pxxmcB and pxxmcI) without identifiable conserved domains (Fig. 3A and SI Appendix, Table S8). Genes flanking the pxxmc BGC encode proteins with no identifiable function in xylomyrocin biosynthesis (SI Appendix, Table S8). The genes encoding the PxXmcG NRPS, the PxXmcE dehydrogenase, the PxXmcF exporter, and the PxXmcA regulator were found to be expressed under cultivation conditions conducive to xylomyrocin biosynthesis. Since glucose was the sole carbon source in this medium, putative hemicellulose catabolic enzymes PxXmC, D, and I were only weakly expressed (SI Appendix, Fig. S4).
Functional Analysis of the pxxmc Biosynthetic Genes.
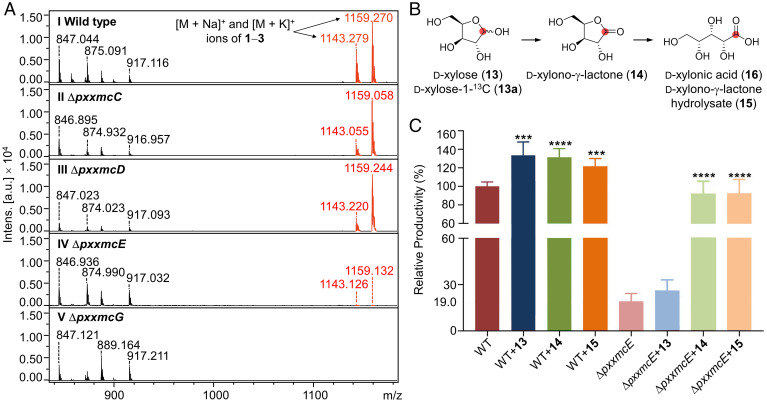
To verify the involvement of the pxxmc BGC in the biosynthesis of compounds 1–3, we carried out targeted gene disruptions in Paramyrothecium sp. XJ0827. Gene disruption cassettes were constructed by inserting the hph hygromycin resistance gene between targeting arms for the pxxmcC, pxxmcD, pxxmcE, or pxxmcG genes, respectively (SI Appendix, Fig. S5), and then introduced separately into Paramyrothecium sp. XJ0827 protoplasts. The knockouts of the targeted genes in the resulting hygromycin B–resistant transformants were validated by PCR analysis (SI Appendix, Fig. S5), and the strains where the desired double homologous recombination events had taken place were evaluated for the production of 1–3 using MALDI-TOF MS. No alterations in the morphologies or growth rates were observed for any of the knockout strains. However, as anticipated, the production of 1–3 was specifically and completely abolished in all ΔpxxmcG isolates (Fig. 4A), confirming PxXmcG as the xylomyrocin NRPS.
Fig. 4.
Functional analysis of pxxmc biosynthetic genes. (A) MALDI-TOF MS traces of extracts from (I) Paramyrothecium sp. XJ0827 wild-type or (II to V) mutant strains. All strains were cultivated for 10 d on PDA media. Ions with m/z of 847, 875, 889, and 917 derive from unrelated metabolites. They show little variation in their intensities (Intens.) among the various strains, illustrating that the ΔpxxmcE and ΔpxxmcG mutations specifically affect the productivities of xylomyrocins 1–3. (B) Proposed conversion of d-xylose (13) or d-xylose-1-13C (13a) to d-xylonic acid (16). The position of the label in 13a is indicated with a red dot. (C) Relative xylomyrocin productivities of the wild-type and ΔpxxmcE strains upon supplementation with d-xylose (13), d-xylono-γ-lactone (14), and d-xylono-γ-lactone hydrolysate (15). Hydrolysate 15 was prepared by treating 14 with 1 M aqueous NaOH solution. Both strains were cultivated for 6 d in liquid M-100 media before supplementation of 13, 14, or 15 at 3 mM (final concentration). Fermentation was continued for an additional 3 d to facilitate complete utilization of the supplemented substrate. Data were collected from three independent experiments of three replicates each (n = 9). Columns represent the mean ± SD. ***P < 0.001 and ****P < 0.0001 as determined with unpaired Student two-tailed t tests comparing the yields of the substrate-supplemented fermentations with that of the nonsupplemented fermentation. a.u., arbitrary units.
The biosynthesis of xylomyrocins was severely impaired upon disruption of the NAD+-dependent oxidoreductase-encoding gene pxxmcE, consistent with the proposed role of its encoded enzyme in the generation of the d-xylonic acid building block. However, trace amounts of 1–3 were still produced by the ΔpxxmcE strain (Fig. 4A). Since we did not detect d-xylonic acid or d-xylono-γ-lactone in xylomyrocin production media using LC-MS, we speculated that the ΔpxxmcE mutant may harbor PxXmcE homologs or nonhomologous isofunctional enzymes (36) that could still supply some d-xylonic acid for the low-level production of 1–3. To our surprise, no homologs of PxXmcE or the bacterial NAD+-dependent d-xylose dehydrogenase XylB (GenBank: ACL94329) (37) were found in the deduced proteome of Paramyrothecium sp. XJ0827 upon comprehensive BLASTp searches. However, two genes not clustered with the pxxmc BGC encode the putative proteins PxXyd1 and PxXyd2 (SI Appendix, Table S9), each similar to the oxidized nicotinamide-adenine dinucleotide phosphate–dependent d-xylose dehydrogenase Xyd1 of Trichoderma reesei (GenBank: A0A024SMV2; 72% and 52% identity, respectively) (38). Both of these genes were expressed under conditions conducive to xylomyrocin production (SI Appendix, Fig. S4). Recombinant T. reesei Xyd1 converts d-xylose to d-xylono-γ-lactone both in vitro and in vivo upon expression in Saccharomyces cerevisiae, yet with a lower efficiency than the NAD+-dependent XylB (39, 40). d-xylono-γ-lactone will then undergo spontaneous or lactonase-catalyzed hydrolysis to yield d-xylonic acid (40).
Supplementation of d-xylose-1-13C (13a) to both the wild-type and ΔpxxmcE strains significantly increased the abundancy of the +1-Da isotopic peak (m/z 1,121.8) of the [M+H]+ ion of 1 (m/z 1,120.8) in both strains (SI Appendix, Fig. S6). MS/MS analysis of the 13C-labeled 1 after methanolysis-mediated linearization revealed that it was the Xyl1 unit that was isotopically labeled, probably at the carbonyl carbon (SI Appendix, Fig. S7), indicating that the d-xylonic acid unit of 1 is indeed derived from d-xylose in both strains. Incremental increases of the +2-, +3-, and +4-Da peaks were also detected, likely due to incorporation of the label from 13a into amino acids during fungal metabolism (SI Appendix, Fig. S6).
Next, we supplemented cultures of the wild-type and ΔpxxmcE strains with d-xylose (13), d-xylono-γ-lactone (14), and d-xylono-γ-lactone hydrolysate (15, consisting predominantly of d-xylonic acid 16 [SI Appendix, Fig. S8]: d-xylonic acid is cost prohibitive for feeding experiments). LC-MS analysis showed that supplementation of 13, 14, and 15 each improved the xylomyrocin productivity of the wild-type strain (Fig. 4 B and C). However, feeding 13 did not increase the production of 1–3 in the ΔpxxmcE strain, indicating that the d-xylose pool is abundant even in the mutant and that the rate-limiting step of xylomyrocin production in this mutant is a reaction downstream of d-xylose during d-xylonic acid production. Correspondingly, supplementation with either 14 or 15 was able to restore the production of xylomyrocins in the ΔpxxmcE mutant strain to the wild-type level (Fig. 4C), indicating that neither the hydrolysis of d-xylono-γ-lactone (14) in the culture nor the utilization of d-xylonic acid is abrogated upon knockout of pxxmcE. Together with the bioinformatic analyses, these experiments show that the oxidase PxXmcE has a major role in the provisioning of d-xylono-γ-lactone (14) and d-xylonic acid (16) from d-xylose (13) during the biosynthesis of xylomyrocins A–C. However, basal levels of 16 may still be provided by PxXyd1, PxXyd2, or other carbohydrate catabolic enzymes of the cell.
Strains with the pxxmcC or the pxxmcD knockouts were still able to produce 1–3 without an observable decrease in productivity (Fig. 4A). Thus, the PxXmcC glycoside hydrolase and the PxXmcD GMC oxidoreductase are not necessary for xylomyrocin biosynthesis under our fermentation conditions. Plant pathogenic fungi like Paramyrothecium sp. are known to employ a wide variety of lignocellulolytic enzymes that release d-xylose monomers from hemicellulosic polysaccharides (41–43). Accordingly, the Paramyrothecium sp. XJ0827 genome encodes a large panel of xylan esterases, xylanases, and xylosidases (SI Appendix, Table S10). d-xylose may also be produced in strain XJ0827 by pentose interconversions (e.g., from xylitol using a d-xylose reductase such as the predicted PxXyrA) or by uridine 5'-diphosphoglucuronate decarboxylation with the putative PxUxs1, both of which were expressed in xylomyrocin production media with glucose as the sole carbon source (SI Appendix, Fig. S4 and Table S9). Indeed, fermentations with both the wild-type and ΔpxxmcE strains on media with d-glucose-13C6 as the sole carbon source showed that the [M+H]+ ion of 1 exhibits a +54-Da mass shift (m/z 1,175.2, compared to unlabeled 1, m/z 1,120.8), confirming that all carbons in 1 (C54H89N9O16), including those of the d-xylonic acid unit, may be derived from glucose (SI Appendix, Fig. S6). Biosynthetic contingencies are commonly observed in secondary metabolism, enlisting alternative precursor supply pathways (44, 45), and with homologous or nonhomologous isofunctional counterparts of primary metabolic enzymes integrated into BGCs (46–48), as may be the case with PxXmcE, PxXmcC, PxXmcD, and PxXmcJ.
Genome Mining for Members of the Xylomyrocin Family.
To discover additional members of the xylomyrocin family by genome mining, we identified BGCs that encode close homologs of both PxXmcE and PxXmcG in the NCBI GenBank and the JGI MycoCosm (SI Appendix, Fig. S9 and Table S11). While PxXmcE homologs are widespread in fungi, they are rarely clustered with NRPSs that are homologs of PxXmcG. Such clusters exist only in a few selected Sordariomycetes genomes, such as in those of several (but not all) strains of Colletotrichum spp. (Glomerellales, Glomerellaceae); two strains of Dactylonectria and one of Ilyonectria spp. (Hypocreales, Nectriaceae); and one strain each of Paramyrothecium (Hypocreales, Stachybotryaceae) and Phaeoacremonium spp. (Togniniales, Togniniaceae). These strains with published genome sequences are prospective producers of xylomyrocin-type polyol cyclodepsipeptides that utilize a d-xylonic acid starter unit. Satisfyingly, the 10- and 34-aa specificity signatures (10) of the A1 domains of these XmcG orthologs are nearly identical (SI Appendix, Fig. S10), defining a clear diagnostic motif for d-xylonic acid activation in fungal NRPSs. Indeed, a position-specific iterative (PSI)-BLASTp with the 34-aa d-xylonic acid A domain signature was sufficient to retrieve the same set of fungal NRPSs from GenBank that had previously been obtained with the combined BLASTP searches with the complete PxXmcE and PxXmcG proteins as baits.
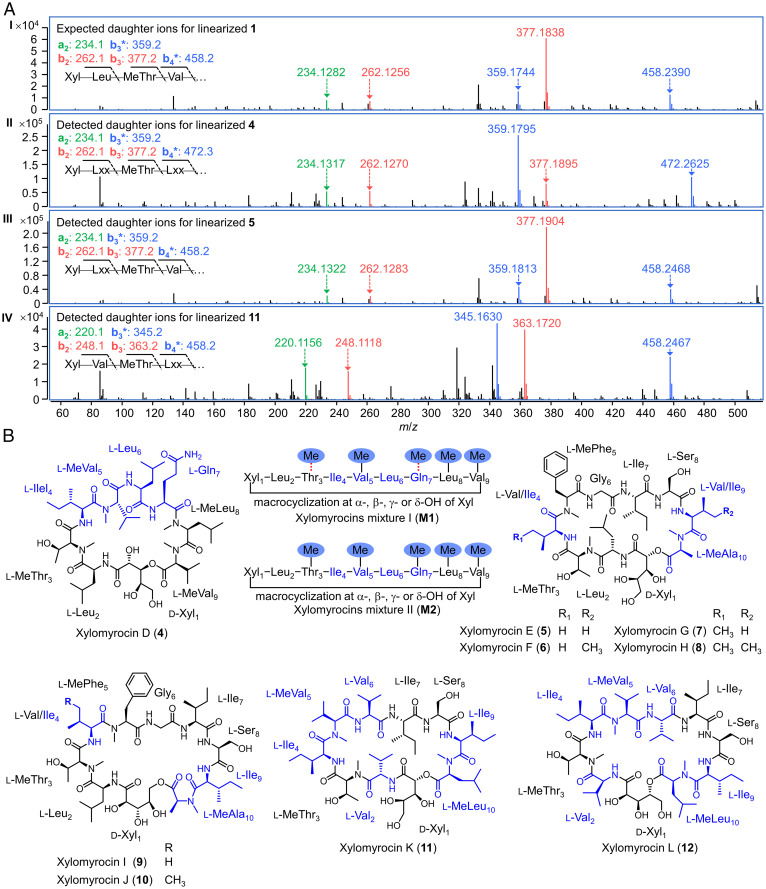
Next, we explored the production of xylomyrocin analogs by selected strains from the genome survey (if easily accessible) and included additional Paramyrothecium and Colletotrichum spp. from commercial or in-house strain collections (SI Appendix, Table S12). We surveyed SM extracts for 50 of these strains cultivated on seven different media each (SI Appendix, SI Materials and Methods) using our established MALDI-TOF MS screen. After dereplication by hierarchical clustering of SM profiles, we identified seven representative strains that produced 10 groups of putative peptides under different culture conditions (SI Appendix, Table S13). Direct tandem HRMS analysis of cyclic peptides often results in complex MS/MS spectra with numerous ion series due to multiple sites for ring opening (49). Thus, we opted to linearize any potential cyclodepsipeptides by treating the fungal crude extracts with MeONa/MeOH solution before LC-MS injection. The resulting methanolysis specifically breaks the lactone bond between the N-terminal d-xylonic acid and the C-terminal amino acid residues in xylomyrocin-type cyclic peptides while leaving all amide bonds intact, generating linear, full-length xylomyrocin derivatives with O-methylated C termini. On the contrary, peptides that lack ester bonds remain unaffected. This method revealed three groups of peptides that yielded derivatives with a +32-Da mass shift after the MeONa/MeOH treatment, corresponding to the cleavage of the lactone ring and the addition of the methoxy functionality. Detailed comparisons of the MS/MS spectra of the linearized peptides with those of compounds 1–3 uncovered similar fragmentation patterns at their N termini (Fig. 5A) and thus strongly indicated that these peptides are putative xylomyrocins. Finally, large-scale fermentations and structure elucidation of the isolated target peptides confirmed that Paramyrothecium roridum NRRL 2183, P. roridum MN113194, and Colletotrichum sp. XJ1040 indeed produce new members of the xylomyrocin family of polyol cyclodepsipeptides (Fig. 5B).
Fig. 5.
Discovery of additional xylomyrocin-type cyclodepsipeptides. (A) MS/MS spectra (m/z range, 0 to 500) of linearized xylomyrocins from (I) Paramyrothecium sp. XJ0827; (II) P. roridum NRRL 2183; (III) P. roridum MN131194; and (IV) Colletotrichum sp. XJ1040. Daughter ions of the a-, b-, and b−H2O (b*)-type, resulting from collision-induced dissociation are shown in green, red, and blue, respectively. Xyl, d-xylonic acid; Leu, l-leucine; MeThr, N-methyl-l-threonine; Val, l-valine; Lxx, l-leucine, l-isoleucine, or N-methyl-l-valine. (B) Structures of xylomyrocins isolated from P. roridum NRRL 2183 (4, M1, and M2), P. roridum MN131194 (5–10), and Colletotrichum sp. XJ1040 (11 and 12). Red dashed lines connecting the methyl functionality and amino acid residues in M1 indicate that N-methylation occurs at either amino acid Thr3 or Gln7, but not both. Amino acid residues that are different from those of compounds 1–3 are colored in navy.
P. roridum NRRL 2183, selected by the comparative genomics survey, produces an assortment of xylomyrocin congeners that all feature a Xyl1–Leu2–(Me)Thr3–Ile4–MeVal5–Leu6–(Me)Gln7–MeLeu8–MeVal9 nonadepsipeptide backbone. They share the Xyl-Leu-(Me)Thr N-terminus and the (Me)Leu-MeVal C terminus with xylomyrocins A–C (1–3) from strain XJ0827, but the identities of the remaining constituent amino acids, the overall sizes of the macrocyclic rings, and the N-methylation patterns are quite different (Fig. 5B and SI Appendix, Fig. S11). Structural diversity among the xylomyrocin congeners produced by P. roridum NRRL 2183 arises from variable N-methylation at the third and seventh residues and the existence of macrocycle regioisomers due to the utilization of any of the four d-xylonic acid hydroxyl groups as the nucleophile during peptide chain termination (Fig. 5B). Unlike 1–3, the xylomyrocin macrocycle regioisomer mixtures M1 and M2 of P. roridum NRRL 2183 could not be separated, despite testing a variety of LC columns and solvent systems. However, xylomyrocin D (4) was obtained as a pure compound that allowed the unambiguous characterization of its chemical structure (Fig. 5B).
Analysis of the predicted xylomyrocin synthetase of P. roridum NRRL 2183 (PrXmcG) revealed that the number of NRPS modules (SI Appendix, Fig. S12) and the specificity signatures of the A domains (SI Appendix, Table S14) matched perfectly the structures of 4, M1, and M2. In some xylomyrocin M1 congeners, methylation at the third residue (Thr3) was skipped by the M domain of module 3, although this domain is evidently active as shown by the presence of MeThr3 in xylomyrocin D (4) and the M2 congeners. Surprisingly, N-methylation was also observed at the seventh and eighth residues (MeGln7 and MeLeu8, respectively) despite the fact that PrXmcG has N-methylation domains only in modules 3, 5, and 9 and not in modules 7 and 8 (SI Appendix, Figs. S11 and S12).
The prxmc BGC is divided into two subclusters that sit at the ends of separate contigs in the published genome sequence of strain NRRL 2183 (GenBank: GCA_003012165, prxmc-5 to prxmcE: PXOD01000192; prxmcH to prxmc+5: PXOD01000490; SI Appendix, Fig. S12), likely due to assembly artifacts. However, the cluster is isogenic with the pxxmc BGC of strain XJ0827, except for the xmcI gene for a hypothetical protein that is completely missing from the genome assembly of P. roridum NRRL 2183 (SI Appendix, Table S15). Compared to the xylomyrocin cluster in strain XJ0827, the prxmcE and prxmcF genes underwent a translocation with reversion to the upstream side of prxmcG in NRRL 2183, and the XJ0827 and NRRL 2183 BGCs are bracketed by nonsyntenic regions (SI Appendix, Fig. S12 and Tables S8 and S15).
Xylomyrocins E–J (5–10) isolated from the other selected P. roridum stain, MN131194, and xylomyrocins K–L (11–12) from Colletotrichum sp. XJ1040 share the same ring size as xylomyrocins A–C (1–3) from Paramyrothecium sp. XJ0827. Xylomyrocins 5–8 from strain MN131194 also share high sequence identities with 1–3 from XJ0824, but they are distinguished by the C-terminal residue (MeAla vs. MeVal, respectively) and are further differentiated by variable amino acids at positions 4 and 9 (Fig. 5B and SI Appendix, Fig. S11). Xylomyrocins K and L (11 and 12) from strain XJ1040 differ from 1–3 at residues 2, 4 to 6, and 9 and 10 (Fig. 5B and SI Appendix, Fig. S11). N-methylation was not variable in the xylomyrocin series of P. roridum MN131194 or Colletotrichum sp. XJ1040, but additional macrocycle regioisomers were also detected by MS/MS in the corresponding extracts, in amounts insufficient for isolation and structure elucidation.
Xylomyrocins Display Antimalarial Activity.
The isolated xylomyrocin congeners 1–3 were evaluated for anti-infective activity in antimicrobial assays. No antibiotic or antifungal activities were detected against Staphylococcus aureus American Type Culture Collection (ATCC) 6538, methicillin-resistant S. aureus (MRSA) 11646, Escherichia coli ATCC 25922, Cryptococcus neoformans H99, and Candida albicans SC5314 with these xylomyrocins in concentrations up to 20 µg/mL (SI Appendix, Table S16). However, most xylomyrocin congeners, except for xylomyrocin G (7) and xylomyrocin I (9), displayed moderate antimalarial activity against the human protozoal parasite Plasmodium falciparum 3D7. The half-maximal inhibition concentrations (IC50) ranged from 10 to 43 μM, with no hemolytic activity observed (Table 1).
Table 1.
Antimalarial activity of xylomyrocins
| Compound | Antiprotozoal activity (IC50, μM)* against P. falciparum 3D7 |
|---|---|
| 1 | 13.5 ± 0.8 |
| 2 | 20.9 ± 0.2 |
| 3 | 23.7 ± 2.4 |
| 4 | 43.3 ± 5.1 |
| M1 | 31.0 ± 0.9 |
| M2 | 36.7 ± 0.9 |
| 6 | 40.2 ± 1.1 |
| 7 | >50 |
| 8 | 25.5 ± 0.3 |
| 9 | >50 |
| 11 | 10.0 ± 0.4 |
| Artemisinin† | 0.008 ± 0.0002 |
*All values represent the means ± SD based on three independent experiments of three replicates each (n = 9).
†Artemisinin was used as the positive control.
Discussion
We used MALDI-TOF and LC-HRMS/MS–based chemometrics and comparative genome mining to discover a group of NRP SMs produced by Paramyrothecium and Colletotrichum spp. fungi. Xylomyrocin-type BGCs were also detected in members of a few additional Sordariomycetes families (SI Appendix, Table S11). These verified and putative xylomyrocin producer fungi are endophytic, predominantly destructive hemibiotrophic phytopathogens (50) that must successfully compete with fungivorous protozoal grazers (51), a need perhaps matched by the antiprotozoal activity of xylomyrocins.
Xylomyrocins are polyol cyclodepsipeptides that feature an unprecedented aldonic acid unit (d-xylonic acid) in their backbones, derived from hemicellulose catabolism and monosaccharide interconversions. Fatty acid, polyketide, aryl acid, α-keto acid, and α-hydroxy acid building blocks are often incorporated into peptide scaffolds by NRPSs (52), demonstrating intersections of NRP biosynthesis with fatty acid or polyketide biosynthesis, shikimic acid metabolism, and amino acid catabolism. Glycosylation of NRPS-assembled peptide backbones by post-NRPS tailoring enzymes is also ubiquitous. However, the direct utilization of a sugar acid biosynthon such as d-xylonic acid for the assembly of a depsipeptide NRP scaffold is unprecedented.
Peptidyl SMs with seemingly sugar acid–derived moieties within their backbones have been described. Thus, THP motifs were found in NRPs such as the methionine aminopeptidase inhibitor bengamides and the transcription factor Rho inhibitor bicyclomycins. However, the apparent sugar acid units originate from polyketide biosynthesis in bengamides (53) and from the stepwise oxidation of a leucine unit in the case of bicyclomycins (54, 55). In contrast, xylomyrocins incorporate a preformed d-xylonic acid building block supplied by carbohydrate metabolism, as shown by our stable isotope incorporation studies.
NRPS-independent mechanisms for amide bond formation between an aminosugar and an amino acid have been elucidated for mycothiol and puromycin biosynthesis (56, 57). Similarly, lincosamide antibiotics from Streptomyces spp. feature an aminosugar biosynthon, but the condensing enzyme complex is not NRPS related (58). Bacterial peptidyl nucleoside antibiotics such as miharamycin or amipurimycin contain a guanine-glucuronate–derived moiety that is temporarily sequestered by an A-T di-domain, but none of the following biosynthetic steps involves an NRPS-catalyzed condensation (59). In contrast, we propose that it is the XmcG NRPS that incorporates the d-xylonic acid monomer during xylomyrocin biosynthesis.
A hydroxyethyl moiety derived from ketose phosphates has been described as an NRPS extender unit for the tetrahydroisoquinoline family of alkaloids (60, 61). Instead of assembling hydroxymalonyl-acyl carrier protein (ACP) as in polyketide biosynthesis (62), this building block is produced by a transketolase enzyme system that transfers the C2 unit to a freestanding ACP without the use of an A domain (61). In contrast, we propose that the A-T di-domain in module 1 of the XmcG NRPSs activates and loads a preformed aldonic acid as the starter unit for xylomyrocin biosynthesis. This is completely distinct from the transketolase-ACP platform of tetrahydroisoquinolones and is more akin to the activation and loading of amino acid–derived α-hydroxy acids seen in fungal cyclodepsipeptides (30, 63–65). Despite our best efforts (SI Appendix, SI Materials and Methods), neither the dissected A1 domain nor the A1-T1 di-domain of PxXmcG could be expressed as a soluble recombinant protein (SI Appendix, Fig. S13).
The polyol-activating A1 domains in the XmcG NRPSs of strains XJ0827 and NRRL 2183 are highly similar to each other (82% identity) but show very low (<29%) identity to the other A domains within the same XmcG NRPSs that are themselves >50% identical across the board. Indeed, the 10-aa and 34-aa extended specificity signatures of the A1 domains of the verified and putative xylomyrocin producers with known genome sequences are highly similar to each other and define a robust d-xylonic acid signature for fungal A domains (SI Appendix, Fig. S10). This high conservation may also indicate that the A1 domains of the xylomyrocin synthetases derive from a common depsipeptide-producing NRPS ancestor and are more recent additions to the rest of the XmcG proteins.
The uronate dehydrogenase-like XmcE homologs that are involved in d-xylonic acid provisioning for xylomyrocin biosynthesis in strains XJ0827 and NRRL 2183 may also be recent recruits to the BGC, considering that although such enzymes are widely distributed in fungi, their encoding genes very rarely colocate with NRPSs or other SM biosynthetic genes. Indeed, we were able to exploit the colocalization of xmcE homologs and xmcG-like NRPSs to identify candidate BGCs for xylomyrocin-type polyol cyclodepsipeptide biosynthesis (SI Appendix, Fig. S9 and Table S11).
The 10-aa and 34-aa extended specificity signatures of the amino acid–incorporating A domains of the XmcG NRPSs of strains XJ0827 and NRRL 2183 (SI Appendix, Tables S6 and S14) fail to form clear clades according to their apparent substrate specificities (SI Appendix, Fig. S14 A and B) (66, 67). Instead, these A domains clade pairwise according to their position in the two XmcG NRPSs (SI Appendix, Fig. S14C). For example, the Phe-activating A5 domain from PxXmcG of XJ0827 is most similar to domain A5 from PrXmcG of NRRL 2183, although that domain evidently activates Val. This may indicate that A domains in equivalent positions in these orthologous proteins evolve to be paralogues, with keyhole surgery-type changes that allow alternate amino acids to be recognized and incorporated into different series of xylomyrocins.
Another intriguing feature of xylomyrocin biosynthesis is the variable N-methylation seen in the xylomyrocin series isolated from NRRL 2183. Skipping the N-methylation of Thr3 in the M1 congeners may be explained by the kinetic competition between the M3 domain and chain extension by the C4 domain. However, the presence of N-methyl amino acids at positions 7 and 8 are more intriguing, given the absence of M domains in the corresponding modules of PrXmcG. Such deviations from the collinear programming of N-methylation are extremely rare in fungal NRP biosynthesis. In one such example, trans-acting N-methyltransferases encoded in the cycloaspeptides and the ditryptophenaline BGCs (PscyA and DtpB, respectively) (68, 69) catalyze the N-methylation of Phe and/or Tyr residues before these are uploaded onto their cognate NRPSs, which feature no integrated M domains. Occasional utilization of the non methylated residues by these NRPSs would then lead to variable methylation at these positions. In a somewhat similar scenario, the MtfA stand-alone N-methyltransferase of Amycolatopsis orientalis was shown to tailor the NRP after that had already been released from the synthetase (70). However, no dissociated N-methyltransferase was found to be encoded in the xmc BGCs, nor were any close homologs of PscyA, DtmB, or MtfA encoded in the genomes of strains XJ0827 and NRRL 2183.
In another example of promiscuous methylation, thalassospiramide biosynthesis in Thalassospira sp. and Tistrella sp. utilizes a cis M domain to modify residues incorporated by upstream domains of the same NRPS complex (71). However, these iterative M domains form a distant, basal clade compared to the xylomyrocin M domains, with sequence identities only in the 15 to 17% range. The thalassospiramide NRPSs also manifest other nonlinear behaviors such as intermodule substrate activation, module skipping, and pass-back chain extension, none of which are encountered with the XmcG NRPSs.
A closer look at the xylomyrocin NRPSs did not provide any sequence clues for which, if any, of the M domains would be responsible for the variable methylation observed in strain NRRL 2183. The M domains of the two XmcG proteins also clade pairwise according to their module position and not per the amino acids they are supposed to modify (SI Appendix, Fig. S14D). Thus, further studies are necessary to establish the molecular basis for the variable backbone methylation in P. roridum NRRL 2183.
A final source of xylomyrocin structural variety is the apparent polyspecificity of the chain-terminating macrocyclization. Considering that the xylomyrocin macrocycle regioisomers are remarkably stable and fail to interconvert to any appreciable degree in acidic media and buffers (SI Appendix, Fig. S3), the terminal CT domains of the xylomyrocin synthases must be able to accept any of the α, β, γ, or δ hydroxyls of d-xylonic acid as a nucleophile. This then implies that the substrate-binding pockets of these domains may accommodate the xylonate moiety in several alternative binding poses. While the biosynthetic basis is currently unknown, a similar flexibility is observed for the inositol phosphorylceramide inhibitor haplofungins produced by Lauriomyces bellulus, where either of the C-4 or the C-5 alcohol functionalities of arabinonic acid is competent to act as the acceptor to download a polyketide chain, resulting in haplofungin regioisomers (72).
Conclusions
Rapid in situ extraction and derivatization, MALDI-TOF MS, LC-HRMS/MS, chemometric dereplication, and genome mining led to the discovery of an unprecedented family of nonribosomal cyclodepsipeptides produced by a limited cohort of phytopathogenic fungi.
Xylomyrocin biosynthesis revealed an unexpected connection between carbohydrate metabolism and NRPS-based peptidyl SM biosynthesis. Enriching precursor diversity by recruiting novel building blocks is a prominent strategy of NRPS evolution in nature and may be reproduced by synthetic biology to generate unnatural products (uNPs) that augment the structure space accessible by NRP biosynthesis.
Xylomyrocin biosynthesis also features a polyspecific chain-terminating CT domain that affords xylomyrocin macrocycle regioisomers. Together with promiscuous peptide backbone N-methylation events and subtle changes in precursor-activating A domains that lead to amino acid replacements among different congeners, the producer fungi seem to engage in structure-activity relationship studies to optimize these polyol cyclodepsipep tides toward their presumed allelochemical role. A mechanistic understanding of such rare, flexible programming events in NRP biosynthesis will improve genome mining and sequence-based SM structure prediction. Such insights will also open additional avenues for NRPS synthetic biology to generate uNPs for crop protection research or pharmaceutical drug discovery.
Materials and Methods
Fungal Strains and Culture Conditions.
Paramyrothecium sp. XJ0827 and Colletotrichum sp. XJ1040 were isolated from soil samples collected from the northern region of Xinjiang province, China. P. roridum NRRL 2183 was purchased from the Agriculture Research Service Culture Collection. Other fungal strains listed in the SI Appendix, Table S12 were acquired from the Agriculture Culture Collection of China or MoonBiotech. After growing on potato dextrose agar (PDA) medium at 28 °C for 5 d, fungal strains were subcultured on PDA, MMK2, YM, YES, LSF1, MV8, or super malt agar medium at 28 °C for 10 to 14 d to monitor the production of peptide-like SMs (SI Appendix provides details).
MALDI-TOF MS Data Acquisition and Hierarchical Cluster Analysis.
trans-2-[3-(4-tert-butylphenyl)-2-methyl-2-propenylidene]malononitrile (Sigma-Aldrich) was used as the MALDI-TOF MS matrix. Details of the sample preparation are described in the SI Appendix, SI Materials and Methods. The MALDI-TOF MS spectra were recorded in reflector/positive ion mode, and each sample was ablated 1,000 times within 1 s with a laser energy output of 70%. A mass-to-charge (m/z) window of 500 to 2,500 was applied to monitor the production of peptide-like SMs. All mass data were analyzed in the FlexAnalysis software (Bruker). The mass peak list of peptide-producing fungal isolates was exported, and all peaks were aligned according to their m/z values using the Mass Profiler Professional software (Agilent). Background noise and mass peaks corresponding to nonspecific metabolites (ubiquitously present in over 50% of fungal isolates) were removed. The resulting dataset was subjected to hierarchical clustering and heatmap plotting using imageGP (73).
HRMS/MS Data Acquisition and Molecular Network Generation.
Representative peptide-producing fungal strains were cultivated on PDA medium for 10 to 14 d, and the mycelium was scraped off and extracted with 90% aqueous methanol. The resulting extract was concentrated and subjected to full-scan ultra-high performance liquid chromatography-high resolution electrospray tandem mass spectrometry (UHPLC-HRESI-MS/MS) analysis. The obtained tandem LC-MS data were used to generate the initial peptide molecular network using the GNPS server (14) following the well-established protocol (74) (SI Appendix provides details).
Isolation and Characterization of Xylomyrocins.
After growing on appropriate medium for 14 d, the medium with the fungal culture was cut into small pieces (1 cm × 1 cm) and extracted with 90% aqueous ethanol three times. The extract was concentrated under vacuum to afford a crude extract, which was fractionized by macroporous D101 resin (Macklin Biochemical Co. Ltd.) and octadecylsilyl (ODS, Fuji Silysia Chemical Ltd.) chromatography. The xylomyrocin-containing fractions were identified by MALDI-TOF MS and then purified by semipreparative high performance liquid chromatography (HPLC) using an Agilent Eclipse XDB-C18 reversed-phase column (5 μm, 9.4 mm × 250 mm) to yield the corresponding xylomyrocins. The chemical structure of each xylomyrocin was elucidated by analyzing the HRMS/MS, one-dimensional and 2D NMR spectroscopic, and X-ray crystallographic data (SI Appendix provides details).
Genome Sequencing and Knockout of the pxxmc Biosynthetic Genes.
The genome of Paramyrothecium sp. XJ0827 was sequenced using a combination of Illumina NovaSeq and Oxford Nanopore technologies (SI Appendix, Table S3), and the assembly was deposited in the NCBI GenBank as BioProject: PRJNA770833 with the accession No. JAJFPB000000000. Gene disruptions were conducted with linear gene deletion cassettes transformed into Paramyrothecium sp. XJ0827 protoplasts using polyethylene glycol-mediated transformation, as described for Aspergillus nidulans (75). Details on the gene disruption procedure can be found in the SI Appendix, SI Materials and Methods. PCR primers used for gene disruption cassette amplification and gene-knockout verification are listed in the SI Appendix, Table S17.
Reverse Transcription–Quantitative PCR (RT-qPCR) Analysis.
RT-qPCR was performed using the ABI Prism 7500 real-time PCR system (Applied Biosystems), in a final reaction volume of 20 µL containing complementary DNA, Power SYBR Green PCR master mix (Thermo Fisher Scientific), and the appropriate primers (final concentration: 200 nM each) as listed in SI Appendix, Table S17. Details of the PCR amplification procedure are described in SI Appendix, SI Materials and Methods. The tubulin-encoding gene was used as the reference gene, and the relative expression fold-change was calculated using the comparative threshold cycle 2−ΔΔCT method. Three biological replicates were analyzed, and for each sample, three technical replicates for each targeted gene were performed.
Antimicrobial Activity Evaluation.
P. falciparum strain 3D7 was used in antimalaria tests. Fresh O-type human erythrocytes were obtained from volunteers, with the samples de-identified before they were used in our study. Cultivation of P. falciparum and antiprotozoal assay were carried out as described previously (76). Two strains of the Gram-positive bacteria S. aureus: subsp. aureus Rosebach (ATCC 6538) and MRSA 11646, and one Gram-negative bacterium E. coli ATCC 25922 were used to evaluate the antibacterial activity of isolated xylomyrocins. Antibacterial assays were carried out utilizing the two-fold serial broth microdilution method with slight modifications (77). Minimum inhibitory concentration (MIC) values were defined as the lowest concentrations of the test compounds that inhibited bacterial growth. Antifungal activity was evaluated against two fungal pathogens of high medical relevance, C. neoformans H99 and C. albicans SC5314 as described previously with slight modifications (78). MIC value was defined as the lowest concentration of the test compound that resulted in a culture with unchanged density (100% inhibition compared to the growth of the untreated control). The SI Appendix provides details.
Supplementary Material
Acknowledgments
This article is dedicated to Professor Xiaoyi Wei, South China Botanical Garden, Chinese Academy of Sciences, on the occasion of his retirement. This work was supported by the National Key Research and Development Program of China (Grant No. 2018YFA0901800 to Y.X.); the National Natural Science Foundation of China (Grant Nos. 32070053 and 31870076 to Y.X. and 32170070 to C.W.); the Youth Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. Y2022QC13 to C.W.); the Central Public-Interest Scientific Institution Basal Research Fund (Grant Nos. 1610392021004 to C.W. and Y2020XK20 to L.Z.); the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (Grant No. CAAS-ASTIP to Y.X.); the US Department of Agriculture, National Institute of Food and Agriculture (Hatch project Grant No. ARZT-1361640-H12-224 to I.M.); and VTT Technical Research Centre of Finland (to I.M.). We thank Dr. Mei Zhang (Beijing Center for Physical and Chemical Analysis) for providing access to their NMR device; Dr. Lida Han (Research Facility Center of Biotechnology Research Institute) for the high resolution electrospray mass spectrometry measurements; and Dr. Kate de Mattos-Shipley (Bristol University UK) for the PscyA sequence data.
Footnotes
Competing interest statement: I.M. has disclosed financial interests in TEVA Pharmaceuticals Hungary, which are unrelated to the subject of the research presented here. All other authors declare no competing financial interests.
This article is a PNAS Direct Submission. B.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123379119/-/DCSupplemental.
Data Availability
X-raycrystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC 2119453) [79]. Some study data are available. Copies of the X-ray crystallographic data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/, or by emailing data_request@ccdc.cam.ac.uk, or by contacting the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223–336-033.
References
- 1.Süssmuth R. D., Mainz A., Nonribosomal peptide synthesis—Principles and prospects. Angew. Chem. Int. Ed. Engl. 56, 3770–3821 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Alonzo D. A., Schmeing T. M., Biosynthesis of depsipeptides, or Depsi: The peptides with varied generations. Protein Sci. 29, 2316–2347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molnár I., Gibson D. M., Krasnoff S. B., Secondary metabolites from entomopathogenic Hypocrealean fungi. Nat. Prod. Rep. 27, 1241–1275 (2010). [DOI] [PubMed] [Google Scholar]
- 4.van der Hooft J. J. J., et al. , Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 49, 3297–3314 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Sukmarini L., Recent advances in discovery of lead structures from microbial natural products: Genomics- and metabolomics-guided acceleration. Molecules 26, 2542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziemert N., Alanjary M., Weber T., The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 33, 988–1005 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Bills G. F., Gloer J. B., Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 4, 10.1128/microbiolspec.FUNK-0009-2016 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Atanasov A. G., Zotchev S. B., Dirsch V. M., Supuran C. T.; International Natural Product Sciences Taskforce, Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stachelhaus T., Mootz H. D., Marahiel M. A., The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6, 493–505 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Röttig M., et al. , NRPSpredictor2–A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutledge P. J., Challis G. L., Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Li Y. F., et al. , Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet. Biol. 89, 18–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen D. D., et al. , MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. U.S.A. 110, E2611–E2620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M., et al. , Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Elimat T., et al. , High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 76, 1709–1716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covington B. C., McLean J. A., Bachmann B. O., Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 34, 6–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hautbergue T., Jamin E. L., Debrauwer L., Puel O., Oswald I. P., From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 35, 147–173 (2018). [DOI] [PubMed] [Google Scholar]
- 18.van der Lee T. A. J., Medema M. H., Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet. Biol. 89, 29–36 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Goering A. W., et al. , Metabologenomics: Correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2, 99–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson E. I., et al. , Discovery of the tyrobetaine natural products and their biosynthetic gene cluster via metabologenomics. ACS Chem. Biol. 13, 1029–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClure R. A., et al. , Elucidating the rimosamide-detoxin natural product families and their biosynthesis using metabolite/gene cluster correlations. ACS Chem. Biol. 11, 3452–3460 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada K.-i., et al. , A method using LC/MS for determination of absolute configuration of constituent amino acids in peptide—Advanced Marfey’s method. Tetrahedron Lett. 36, 1515–1518 (1995). [Google Scholar]
- 23.Flissi A., et al. , Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 48, D465–D469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorokina M., Steinbeck C., Review on natural products databases: Where to find data in 2020. J. Cheminform. 12, 20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Santen J. A., et al. , The Natural Products Atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 5, 1824–1833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blin K., et al. , antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X., et al. , Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 8, 823–830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., et al. , Structural basis of nonribosomal peptide macrocyclization in fungi. Nat. Chem. Biol. 12, 1001–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belshaw P. J., Walsh C. T., Stachelhaus T., Aminoacyl-CoAs as probes of condensation domain selectivity in nonribosomal peptide synthesis. Science 284, 486–489 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Xu Y., et al. , Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem. Biol. 15, 898–907 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Xu Y., et al. , Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 46, 353–364 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Schwarzer D., Finking R., Marahiel M. A., Nonribosomal peptides: From genes to products. Nat. Prod. Rep. 20, 275–287 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Kracher D., et al. , Extracellular electron transfer systems fuel cellulose oxidative degradation. Science 352, 1098–1101 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Sützl L., Foley G., Gillam E. M. J., Bodén M., Haltrich D., The GMC superfamily of oxidoreductases revisited: Analysis and evolution of fungal GMC oxidoreductases. Biotechnol. Biofuels 12, 118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon S. H., Moon T. S., Iranpour P., Lanza A. M., Prather K. J., Cloning and characterization of uronate dehydrogenases from two pseudomonads and Agrobacterium tumefaciens strain C58. J. Bacteriol. 191, 1565–1573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omelchenko M. V., Galperin M. Y., Wolf Y. I., Koonin E. V., Non-homologous isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens C., et al. , Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 189, 2181–2185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toivari M. H., Ruohonen L., Richard P., Penttilä M., Wiebe M. G., Saccharomyces cerevisiae engineered to produce D-xylonate. Appl. Microbiol. Biotechnol. 88, 751–760 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Berghäll S., Hilditch S., Penttilä M., Richard P., Identification in the mould Hypocrea jecorina of a gene encoding an NADP+: d-xylose dehydrogenase. FEMS Microbiol. Lett. 277, 249–253 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toivari M., et al. , Metabolic engineering of Saccharomyces cerevisiae for bioconversion of D-xylose to D-xylonate. Metab. Eng. 14, 427–436 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Filho E. X. F., Puls J., Coughlan M. P., Physicochemical and catalytic properties of a low-molecular-weight endo-1,4-β-d-xylanase from Myrothecium verrucaria. Enzyme Microb. Technol. 15, 535–540 (1993). [Google Scholar]
- 42.Tiwari R., et al. , Biological delignification of paddy straw and Parthenium sp. using a novel micromycete Myrothecium roridum LG7 for enhanced saccharification. Bioresour. Technol. 135, 7–11 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Piyaboon O., Pawongrat R., Unartngam J., Chinawong S., Unartngam A., Pathogenicity, host range and activities of a secondary metabolite and enzyme from Myrothecium roridum on water hyacinth from Thailand. Weed Biol. Manage. 16, 132–144 (2016). [Google Scholar]
- 44.Chen L., et al. , Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis. BMC Genomics 14, 339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wohlleben W., et al. , Synthetic biology of secondary metabolite biosynthesis in actinomycetes: Engineering precursor supply as a way to optimize antibiotic production. FEBS Lett. 586, 2171–2176 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Rangaswamy V., Hernández-Guzmán G., Shufran K. A., Bender C. L., Analysis of rILERS, an isoleucyl-tRNA synthetase gene associated with mupirocin production by Pseudomonas fluorescens NCIMB 10586. DNA Seq. 13, 343–351 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Moghe G. D., Last R. L., Something old, something new: Conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 169, 1512–1523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cruz-Morales P., et al. , Phylogenomic analysis of natural products biosynthetic gene clusters allows discovery of arseno-organic metabolites in model Streptomycetes. Genome Biol. Evol. 8, 1906–1916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassman M. E., Kulagina N., Taitt C. R., Fragmentation of biotinylated cyclic peptides. Rapid Commun. Mass Spectrom. 18, 1277–1285 (2004). [DOI] [PubMed] [Google Scholar]
- 50.da Silva L. L., Moreno H. L. A., Correia H. L. N., Santana M. F., de Queiroz M. V., Colletotrichum: Species complexes, lifestyle, and peculiarities of some sources of genetic variability. Appl. Microbiol. Biotechnol. 104, 1891–1904 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Bonkowski M., Protozoa and plant growth: The microbial loop in soil revisited. New Phytol. 162, 617–631 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Walsh C. T., O’Brien R. V., Khosla C., Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew. Chem. Int. Ed. Engl. 52, 7098–7124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenzel S. C., et al. , Production of the bengamide class of marine natural products in Myxobacteria: Biosynthesis and structure-activity relationships. Angew. Chem. Int. Ed. Engl. 54, 15560–15564 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Witwinowski J., et al. , Study of bicyclomycin biosynthesis in Streptomyces cinnamoneus by genetic and biochemical approaches. Sci. Rep. 9, 20226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng S., et al. , A six-oxidase cascade for tandem C-H bond activation revealed by reconstitution of bicyclomycin biosynthesis. Angew. Chem. Int. Ed. Engl. 57, 719–723 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Jothivasan V. K., Hamilton C. J., Mycothiol: Synthesis, biosynthesis and biological functions of the major low molecular weight thiol in actinomycetes. Nat. Prod. Rep. 25, 1091–1117 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Angel Rubio M., Barrado P., Carlos Espinosa J., Jiménez A., Fernández Lobato M., The pur6 gene of the puromycin biosynthetic gene cluster from Streptomyces alboniger encodes a tyrosinyl-aminonucleoside synthetase. FEBS Lett. 577, 371–375 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Janata J., et al. , Lincosamide synthetase—A unique condensation system combining elements of nonribosomal peptide synthetase and mycothiol metabolism. PLoS One 10, e0118850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F., et al. , Characterization of miharamycin biosynthesis reveals a hybrid NRPS-PKS to synthesize high-carbon sugar from a complex nucleoside. J. Am. Chem. Soc. 142, 5996–6000 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Le V. H., Inai M., Williams R. M., Kan T., Ecteinascidins. A review of the chemistry, biology and clinical utility of potent tetrahydroisoquinoline antitumor antibiotics. Nat. Prod. Rep. 32, 328–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng C., et al. , Hijacking a hydroxyethyl unit from a central metabolic ketose into a nonribosomal peptide assembly line. Proc. Natl. Acad. Sci. U.S.A. 109, 8540–8545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan Y. A., et al. , Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl. Acad. Sci. U.S.A. 103, 14349–14354 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magarvey N. A., Ehling-Schulz M., Walsh C. T., Characterization of the cereulide NRPS α-hydroxy acid specifying modules: Activation of α-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc. 128, 10698–10699 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Feifel S. C., et al. , In vitro synthesis of new enniatins: Probing the α-D-hydroxy carboxylic acid binding pocket of the multienzyme enniatin synthetase. ChemBioChem 8, 1767–1770 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Süssmuth R., Müller J., von Döhren H., Molnár I., Fungal cyclooligomer depsipeptides: From classical biochemistry to combinatorial biosynthesis. Nat. Prod. Rep. 28, 99–124 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Boettger D., Bergmann H., Kuehn B., Shelest E., Hertweck C., Evolutionary imprint of catalytic domains in fungal PKS-NRPS hybrids. ChemBioChem 13, 2363–2373 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Xu W., Gavia D. J., Tang Y., Biosynthesis of fungal indole alkaloids. Nat. Prod. Rep. 31, 1474–1487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Mattos-Shipley K. M. J., et al. , The cycloaspeptides: Uncovering a new model for methylated nonribosomal peptide biosynthesis. Chem. Sci. (Camb.) 9, 4109–4117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saruwatari T., et al. , Cytochrome P450 as dimerization catalyst in diketopiperazine alkaloid biosynthesis. ChemBioChem 15, 656–659 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Shi R., et al. , Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics. Chem. Biol. 16, 401–410 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Zhang J. J., Tang X., Huan T., Ross A. C., Moore B. S., Pass-back chain extension expands multimodular assembly line biosynthesis. Nat. Chem. Biol. 16, 42–49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohnuki T., Yano T., Takatsu T., Haplofungins, new inositol phosphorylceramide synthase inhibitors, from Lauriomyces bellulus SANK 26899 II. Structure elucidation. J. Antibiot. (Tokyo) 62, 551–557 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Chen T., Liu Y.-X., Huang L., ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 1, e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aron A. T., et al. , Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 15, 1954–1991 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Tilburn J., et al. , Transformation by integration in Aspergillus nidulans. Gene 26, 205–221 (1983). [DOI] [PubMed] [Google Scholar]
- 76.Smilkstein M., Sriwilaijaroen N., Kelly J. X., Wilairat P., Riscoe M., Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 48, 1803–1806 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Onishi H. R., et al. , Antibacterial agents that inhibit lipid A biosynthesis. Science 274, 980–982 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Li Y., et al. , Emestrins: Anti-Cryptococcus epipolythiodioxopiperazines from Podospora australis. J. Nat. Prod. 79, 2357–2363 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Wang C., et al., Chemometrics and genome mining reveal an unprecedented family of sugar acid–containing fungal nonribosomal cyclodepsipeptides. CCDC. https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=2119453&DatabaseToSearch=Published. Deposited 1 November 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-raycrystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC 2119453) [79]. Some study data are available. Copies of the X-ray crystallographic data can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/, or by emailing data_request@ccdc.cam.ac.uk, or by contacting the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223–336-033.