Abstract
Aims: Familial hypercholesterolemia (FH) is currently a worldwide health issue. Understanding the characteristics of patients is important for proper diagnosis and treatment. This study aimed to analyze the phenotypic and genetic features, including threshold cholesterol levels, of Korean patients with FH.
Methods: A total of 296 patients enrolled in the Korean FH registry were included, according to the following criteria: low-density lipoprotein-cholesterol (LDL-C) >190 mg/dL with tendon xanthoma or family history compatible with FH, or LDL-C >225 mg/dL. DNA sequences of three FH-associated genes were obtained using whole-exome or target exome sequencing. Threshold cholesterol levels for differentiating patients with FH/pathogenic variant (PV) carriers and predictors of PVs were identified.
Results: Of the 296 patients, 104 had PVs and showed more obvious clinical findings, including higher cholesterol levels. PV rates ranged from 30% to 64% when patients were categorized by possible or definite type according to the Simon Broome criteria. Frequent PV types included missense variants and copy number variations (CNVs), while the most frequent location of PVs was p.P685L inLDLR. The threshold LDL-C levels for patient differentiation and PV prediction were 177 and 225 mg/dL, respectively. Younger age, tendon xanthoma, and higher LDL-C levels were identified as independent predictors of PVs, while traditional cardiovascular risk factors were predictors of coronary artery disease.
Conclusions: Korean patients with FH had variable PV rates depending on diagnostic criteria and distinctive PV locations. The reported threshold LDL-C levels pave the way for efficient patient care in this population.
Keywords: Diagnosis, Far East, Hyperlipoproteinemia type II, Mutation
Abbreviations: PV: pathogenic variant; CNV: copy number variation
Introduction
Familial hypercholesterolemia (FH) is currently considered a worldwide health priority 1) . Active lipid-lowering therapy for cardiovascular prevention in patients with FH cannot be overemphasized 2 , 3) ; thus, early and appropriate diagnosis is of significant importance. The Dutch Lipid Clinic Network and Simon Broome criteria are most commonly used for clinical diagnosis of FH 4) . However, the phenotypic and genetic characteristics of FH might differ between patients with different ethnicities. Hence, some countries have developed their own indices for FH, including diagnostic criteria and cholesterol levels, for patient recognition 5 , 6) .
Data from different regions in the world revealed that pathogenic variants (PVs) causing FH were most frequently found in LDLR. However, diversity in the specific locations of PVs in LDLR and other FH-associated genes was noted, with people from different ethnicities harboring unique genetic characteristics 7 - 9) . Assessment and genetic diagnosis of a PV in suspected individuals was reported to be useful for effective diagnosis, risk stratification, and initiation of proper monitoring or treatment 10) . For instance, compared to PV non-carriers, patients with FH harboring PVs showed 3- to 4-fold higher risk of developing coronary artery disease (CAD) 11) . In addition, the presence and type of PVs in patients with FH were associated with patient response to lipid-lowering therapy 12) . Given its commonness and impact, copy number variation (CNV), a PV that has not been sufficiently investigated in patients with FH, has recently come under the spotlight 13) .
Although several studies have focused on Asian patients with FH 9 , 14) , including ours 8 , 12 , 15) , it is clear that more data are needed for a better understanding of the disease and for better patient management in this region. In Korea, a registry of patients with FH started several years ago; however, the final phenotypic and genetic results only became available in 2020. Therefore, the aim of this study was to analyze the phenotypic and genetic characteristics of Korean patients with FH. In addition, we also attempted to identify the threshold cholesterol level that could help distinguish patients with FH from the general population, and PV carriers among the clinically diagnosed population. We also investigated the clinical predictors of FH-related PVs.
Patients and Methods
Study Population
Sixteen university hospitals in Korea were involved in, and the Korean Society of Lipid and Atherosclerosis supported the study. The institutional review board at each center, including Severance Hospital (4-2008-0267), Korea, approved the study protocol, and all the participants provided written informed consent. The entire study was performed in accordance with the Declaration of Helsinki.
We combined data of 97 patients, which had been published in 2015 15) , with those of 199 patients who were enrolled between August 2014 and September 2020. Unrelated men and women (age >19 years) with any of the following features were included in the study: 1) low-density lipoprotein-cholesterol (LDL-C) >190 mg/dL and tendon xanthoma, 2) LDL-C >190 mg/dL and a family history of CAD at age <60 years or LDL-C >190 mg/dL, 3) LDL-C >190 mg/dL with incomplete family history data regarding onset age of CAD or LDL-C levels, or 4) LDL-C >225 mg/dL even without family history. In Korean guidelines for dyslipidemia, it is recommended to use major international criteria such as Simon Broome to clinically diagnose FH 2) . Although the inclusion criteria used in our registry are similar to Simon Broome, they are not exactly the same but slightly wider than the criteria. For example, our criteria have the same items for number 1) and 2) in the text, like LDL-C >190 mg/dL plus tendon xanthoma or family history of coronary artery disease. However, in some cases, we also enrolled patients with eligible LDL-C levels even though detailed family history was difficult to obtain. An LDL-C level of 225 mg/dL was chosen as a cut-off value, since we had identified this level as the best for predicting PV carriers in a previous study 15) . Secondary causes of lipid disorder were excluded through clinical and biochemical assessments.
Clinical and Laboratory Data Collection
At the time of enrolment, clinical history was collected for each patient, and physical and laboratory assessments were performed. Information on demographic variables, past medical history, family history of LDL-C >190 mg/dL and premature CAD (<60 years in first-degree relatives), body mass index, tendon xanthoma, and lipid profile were collected. Tendon xanthoma was defined as hard subcutaneous nodule or thickening occurring on the Achilles tendon or extensor area of the hands, elbows, or knees. Although some guidelines 6) recommend imaging tools, such as X-ray for more precise diagnosis of xanthoma, these were not used in the present study. Patients exhibiting angiographically proven coronary artery stenosis (>50%) at more than one coronary artery, or positive results of exercise or pharmacologic stress testing were defined as having a history of CAD. Patients fasted for at least 12 h before blood sampling, and samples were analyzed within 4 h at the Korean Society of Laboratory Medicine certified laboratories.
PV Analysis
Genomic DNA was extracted using the commercially available QuickGene SP kit DNA whole blood (Fujifilm, Tokyo, Japan). For 199 patients not reported in the previous study 15) , target enrichment was performed using a Target Enrichment kit (Celemics, Seoul, Korea) with a custom panel comprising three FH-associated genes (LDLR, APOB, and PCSK9), according to the manufacturer’s instructions. Sequencing reads obtained from the NextSeq 500 platform (Illumina, San Diego, CA, USA) were further analyzed using BWA-MEM, Picard (v1.115), Samtools (v1.1), GATK (v4.0.4.0), and Varscan (v2.4.0) to call single nucleotide variants and insertions/deletions. A variant was considered as pathogenic if it fulfilled any of the following criteria: 1) protein truncated mutation, or 2) registered as pathogenic in the ClinVar (v2020.05.13) database, or meeting the criteria of ACMG guideline. For variants not previously reported, we evaluated pathogenicity considering the clear deleterious effects of amino acid changes, such as frameshift insertions/deletions. All variants listed as either known or novel pathogenic were validated using Sanger sequencing.
For CNV analysis, the Celemics ‘in-house CNV detection tool’ was used to measure copy number alterations of target genes. The program uses the normalized mean depth of target gene generated by dividing the mean depth to 1) on-target median depth of each sample, and 2) regional mean depth of several baselines. All CNVs were also validated using multiplex ligation-dependent probe amplification or Taqman.
Statistical Analysis
Continuous variables were tested for normality using the Shapiro-Wilk normality test. Variables with normal distribution are reported as means±standard deviations and analyzed using the Student’s t-test. Non-normally distributed variables are presented as median (interquartile range: IQR) and examined using the Mann-Whitney U test. Categorical variables are reported as counts and proportions and were analyzed using the Pearson’s Chi-square or Fisher’s exact test. Kernel density was used to view and compare the multi-modal distribution of total cholesterol and LDL-C of the general population and patients with FH. The proportion of values was determined by the total area contained at a specific interval. The serum cholesterol data of the general population were obtained from the Korean National Health Insurance Service (2014–2015). Receiver operating characteristic (ROC) curves were designed to facilitate the evaluation of several cut-off points for different pairs of sensitivity and specificity and were used to identify the optimum threshold value for PV prediction. To identify the independent predictors for PVs and CAD, we used multivariable logistic regression analysis. Only variables with p<0.05 in the univariable analysis were included in the multivariable analysis. Variables were checked for collinearity using the variance inflation factor (VIF); those with VIF >5 were excluded. A p<0.05 was considered significant. The Statistical Package for the Social Science v17.0 (SSPS Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Clinical Characteristics
The mean age of participants was 51.0 years. Of the total 296 participants, 143 (48.3%) were males. Fifty-seven patients (19.3%) had history of CAD, and 105 (35.5%) revealed a family history of premature CAD. Tendon xanthoma was observed in 59 patients (19.9%). The median total cholesterol and LDL-C levels were 306 and 221 mg/dL, respectively. PVs in LDLR, APOB, or PCSK9 were found in 104 patients (35.1%) ( Table 1 ) . Herein, using the Simon Broome criteria, the rate of PVs in patients with definite type FH was 64.4%, whereas that in definite or probable type FH determined using the Dutch criteria was 51.3% ( Table 2 ) . Compared to PV-negative individuals, PV-positive patients were younger and more likely to have CAD, tendon xanthoma, and family history of tendon xanthoma. PV-positive patients showed higher cholesterol and lower triglyceride levels than PV-negative ones ( Table 1 ) .
Table 1. Clinical characteristics of the study population according to the presence of pathogenic variants (PVs).
| Characteristics | Total (n = 296) | PV-positive (n = 104) | PV-negative (n = 192) | p |
|---|---|---|---|---|
| Age, years | 51.0±13.1 | 46.0±15.2 | 53.7±10.9 | <0.001 |
| Male | 143 (48.3) | 51 (49.0) | 92 (47.9) | 0.85 |
| Medical history | ||||
| Diabetes mellitus | 24 (8.1) | 10 (9.6) | 14 (7.3) | 0.48 |
| Hypertension | 102 (34.5) | 32 (30.8) | 70 (36.5) | 0.33 |
| CAD | 57 (19.3) | 27 (26.0) | 30 (15.6) | 0.03 |
| Smoking | 56 (18.9) | 22 (21.4) | 34 (17.9) | 0.74 |
| Family history | ||||
| Hypercholesterolemia | 177 (59.8) | 67 (64.4) | 110 (58.5) | 0.32 |
| Premature CAD | 105 (35.5) | 42 (40.8) | 63 (33.5) | 0.22 |
| Tendon xanthoma | 9 (3.0) | 7 (6.7) | 2 (1.0) | 0.01 |
| Body mass index | 24.4 (22.1, 26.7) | 24.5 (21.3, 26.3) | 24.4 (22.5, 26.8) | 0.20 |
| Tendon xanthomas | 59 (19.9) | 28 (26.9) | 31 (16.1) | 0.03 |
| Laboratory values, mg/dL | ||||
| Total cholesterol | 306 (281, 336) | 334 (301, 369) | 296 (277, 322) | <0.001 |
| Triglycerides | 146 (103, 201) | 132 (93, 188) | 149 (110, 209) | 0.02 |
| HDL-C | 52 (45, 62) | 49 (42, 58) | 54 (45, 62) | 0.02 |
| LDL-C | 221 (202, 250) | 246 (218, 286) | 210 (198, 234) | <0.001 |
| Conditions for inclusion | ||||
| LDL-C > 190 mg/dL and xanthoma | 59 (19.9) | 28 (26.9) | 31 (16.1) | 0.03 |
| LDL-C > 190 mg/dL and FHx | 184 (62.2) | 66 (63.5) | 118 (61.5) | 0.73 |
| LDL-C > 190 mg/dL and incomplete FHx | 26 (8.8) | 4 (3.8) | 22 (11.5) | 0.03 |
| LDL-C > 225 mg/dL without FHx | 27 (9.1) | 6 (5.8) | 21 (10.9) | 0.14 |
Abbreviations: PV, pathogenic variant; CAD, coronary artery disease; HDL-C, high-density lipoprotein-cholesterol; LDL-C, low-density lipoprotein-cholesterol; FHx, family history.
Data are presented as number (percentage), mean±SD, or median (interquartile range).
Table 2. Prediction of pathogenic variants (PVs) using three clinical diagnostic criteria for familial hypercholesterolemia.
| Criteria | Type | PV-positive (n = 104) | PV-negative (n = 192) | PV rate (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Simon Broome Register | Definite (n = 90) | 58 | 32 | 64.4 | 55.8 | 83.1 |
| Definite + possible (n = 213) | 63 | 150 | 29.6 | 60.6 | 20.6 | |
| Dutch Lipid Clinic Network | Definite (n = 74) | 37 | 37 | 50.0 | 35.6 | 80.6 |
| Definite + probable (n = 117) | 60 | 57 | 51.3 | 57.7 | 70.2 | |
| Definite + probable + possible (n = 295) | 104 | 191 | 35.4 | 100 | 0.5 | |
| MEDPED | Total cholesterol (n = 57) | 39 | 18 | 68.4 | 37.5 | 90.6 |
| LDL-C (n = 71) | 47 | 24 | 66.2 | 45.1 | 87.5 |
Abbreviations: PV, pathogenic variant; MEDPED, Make Early Diagnosis to Prevent Early Deaths; LDL-C, low-density lipoprotein-cholesterol.
Genetic Characteristics
In total, 65 PVs were identified in 104 patients. Regarding frequency, LDLR missense variants, CNVs, and frameshift variants were detected in 46%, 11%, and 9% of patients, respectively ( Fig.1A ) . Fifty-eight single nucleotide changes or small insertions/deletions were discovered in 92 patients: 54 LDLR variants in 86 patients, 2 APOB variants in 6 patients, and 2 PCSK9 variants in 4 patients ( Fig.1B and Supplementary Table 1 ) . Conversely, seven CNVs in LDLR were identified in 12 patients ( Fig.1B and Supplementary Table 2 ) .
Fig.1. (A) Frequency of pathogenic variant (PV) types in familial hypercholesterolemia (FH)-associated genes (LDLR, APOB, and PCSK9) in Korean patients with FH. (B) Location and characteristics of PVs displayed on each gene .
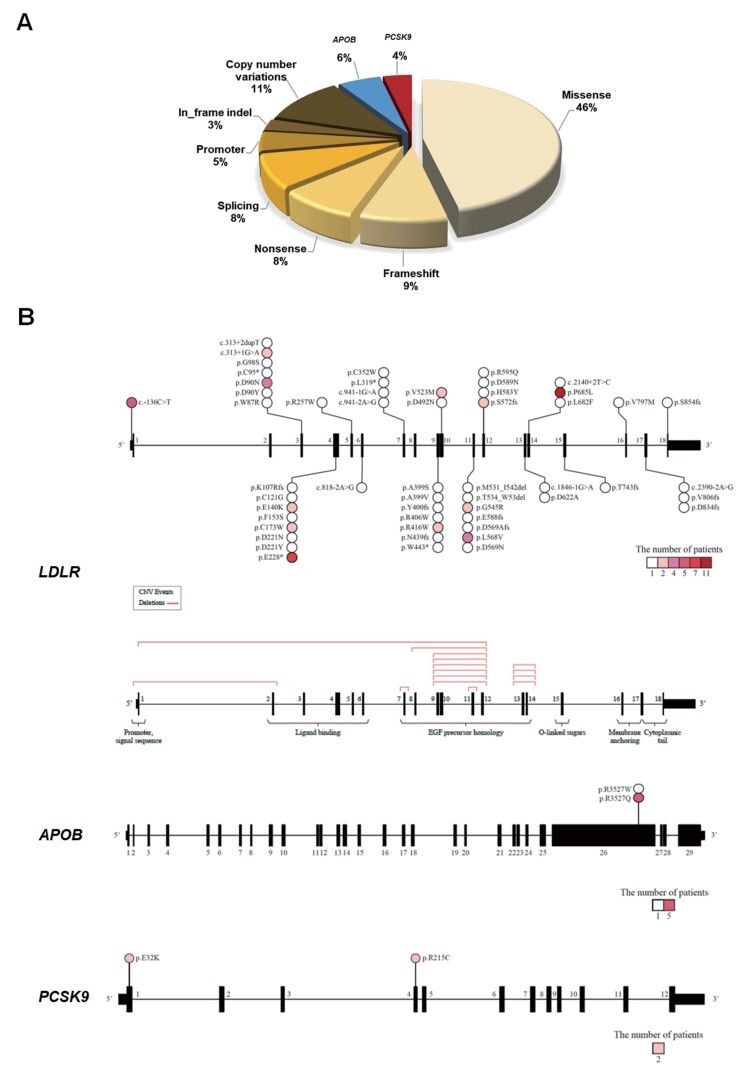
LDLR copy number variations are described on the 180 exons of the gene.
Supplementary Table 1. List of pathogenic single nucleotide changes and small insertions/deletions in familial hypercholesterolemia (FH)-associated genes.
| Gene | Nucleotide change (amino acid change)* | Accession no. | Patients (n)† |
|---|---|---|---|
| LDLR | c.-136C> T | rs879254374 | 1 |
| c.1671_1672delTG (p.E588fs) | rs879254979 | ||
| c.-136C> T | rs879254374 | 4 | |
| c.259T> C (p.W87R) | rs121908025 | 1 | |
| c.268G> T (p.D90Y) | rs749038326 | 1 | |
| c.268G> A (p.D90N) | rs749038326 | 4‡ | |
| c.285C> A (p.C95*) | rs139400379 | 1 | |
| c.292G> A (p.G98S) | rs750474121 | 1 | |
| c.313+1G> A | rs112029328 | 2 | |
| c.313+2dupT | rs875989897 | 1 | |
| c.320_332delAGACGTGCTCCCA (p.K107Rfs) | rs879254474 | 1 | |
| c.361T> G (p.C121G) | rs879254492 | 1 | |
| c.418G> A (p.E140K) | rs748944640 | 2 | |
| c.458T> C (p.F153S) | rs1568723797 | 1 | |
| c.519C> G (p.C173W) | rs769318035 | 2 | |
| c.661G> A (p.D221N) | rs875989906 | 1 | |
| c.661G> T (p.D221Y) | rs875989906 | 1 | |
| c.682G> T (p.E228*) | rs121908029 | 7 | |
| c.769C> T (p.R257W) | rs200990725 | 1 | |
| c.1765G> A (p.D589N) | rs201971888 | ||
| c.818-2A> G | rs879254687 | 1 | |
| c.941-2A> G | rs112366278 | 1 | |
| c.941-1G> A | rs879254735 | 1 | |
| c.956T> A (p.L319*) | - | 1 | |
| c.1056C> G (p.C352W) | rs13306515 | 1 | |
| c.1195G> T (p.A399S) | rs730882099 | 1 | |
| c.1196C> T (p.A399V) | - | ||
| c.1198_1199insTC (p.Y400fs) | - | ||
| c.1216C> T (p.R406W) | rs121908043 | 1 | |
| c.1246C> T (p.R416W) | rs570942190 | 2 | |
| c.1316delA (p.N439fs) | rs1057519670 | 1 | |
| c.1329G> A (p.W443*) | rs879254867 | 1 | |
| c.1474G> A (p.D492N) | rs373646964 | 1 | |
| c.1567G> A (p.V523M) | rs28942080 | 2 | |
| c.1592_1627del (p.Met531_Ile542del) | rs879254950 | 1 | |
| c.1600_1608delACTGACTGG (p.Thr534_Trp536del) | rs879254953 | 1 | |
| c.1633G> C (p.G545R) | rs879254965 | 2 | |
| c.1702_1705dupCTAG (p.D569Afs) | rs879254990 | 4 | |
| c.1702C> G (p.L568V) | rs746959386 | 1 | |
| c.1705G> A (p.D569N) | rs879254993 | 1 | |
| c.1714_1715delAG (p.S572fs) | - | 2 | |
| c.1747C> T (p.H583Y) | rs730882109 | 1 | |
| c.1784G> A (p.R595Q) | rs201102492 | 1 | |
| c.1846-1G> A | rs879255051 | 1 | |
| c.1865A> C (p.D622A) | rs879255060 | 1 | |
| c.2044C> T (p.L682F) | rs1131692220 | 1 | |
| c.2054C> T (p.P685L) | rs28942084 | 11 | |
| c.2140+2T> C | rs879255147 | 1 | |
| c.2224_2227dupACCA (p.T743fs) | - | 1 | |
| c.2389G> A (p.V797M) | rs750518671 | 1 | |
| c.2390-2A> G | rs767790696 | 1 | |
| c.2416dupG (p.V806fs) | rs773618064 | 1 | |
| c.2500_2502delinsC (p.D834fs) | rs879255219 | 1 | |
| c.2560_2563delAGTC (p.S854fs) | - | 1 | |
| APOB | c.10580G> A (p.R3527Q) | rs5742904 | 5 |
| c.10579C> T (p.R3527W) | rs144467873 | 1 | |
| PCSK9 | c.94G> A (p.E32K) | rs564427867 | 2 |
| c.643C> T (p.R215C) | rs753505066 | 2 |
*Nucleotide location number was assigned according to the LDLR (Transcript ID: NM_000527.4), APOB (Transcript ID: NM_000384.2), and PCSK9 (Transcript ID: NM_174936.3) mRNA sequences. †Patients with compound heterozygous pathogenic variants are underlined. ‡One patient had a homozygous pathogenic variant.
Supplementary Table 2. List of pathogenic whole-exon copy number variations in LDLR.
| MLPA or Taqman | NGS data | Patients (n) | |
|---|---|---|---|
| Type | Region | Detection | |
| Heterozygous deletion | Promoter–Exon 2 | 1 | 1 |
| Heterozygous deletion | Exon 7 | 1 | 1 |
| Heterozygous deletion | Exons 9–12 | 6 | 6 |
| Heterozygous deletion | Exon 11 (33 bp deletion) | 1 | 1 |
| Heterozygous deletion | Exons 13–14 | 1 | 1 |
| Heterozygous deletion | Exons 1–12 | 1 | 1 |
| Heterozygous deletion | Exons 8–12 | 1 | 1 |
Abbreviations: MLPA, multiplex ligation-dependent probe amplification; NGS, next-generation sequencing. For multi-exon copy number variants, the reported ratio values are averaged across each affected region.
Four patients were genetically diagnosed as homozygous FH: one was simple homozygous and three were compound heterozygous. One of the compound heterozygous patients showed three different PVs in LDLR ( Supplementary Table 1 ) . Frequent variant locations were c.2054C>T (p.P685L) and c.682G>T (p.E228) in LDLR and c.10580G>A (p.R3527Q) in APOB, harbored by 11, 7, and 5 patients, respectively ( Fig.1B and Supplementary Table 1 ) . Among the CNVs, deletion of exons 9–12 was the most frequent and was observed in six patients ( Fig.1B and Supplementary Table 2 ) .
The variants on ABCG5 or ABCG8, gene related to sitosterolemia 25) were examined in 68 individuals who underwent targeted exome sequencing. Among them, we found two patients without FH-associated PVs who showed two different variants in sitosterolemia genes: ABCG5 p.I609V and ABCG8 p.G110R heterozygous variants. These variants did not show clinical significance in ClinVar. Another heterozygous variant, ABCG5 p.L103P, was found in a patient with PV on LDLR. The significance of this variant is also uncertain.
Threshold Cholesterol Levels for Patient Differentiation and PV Carriers
Total cholesterol and LDL-C levels of the general population and patients with FH showed symmetric normal distribution. Multi-modal distribution suggested possible points that could help differentiate patients with FH from the general population. Based on our data, the threshold levels for total cholesterol and LDL-C were 250 and 177 mg/dL, respectively ( Fig.2A ) . According to the ROC curve used to define thresholds for PV carriers, the best cut-off value for total cholesterol was 325 mg/dL, with sensitivity, specificity, and area under the curve values of 0.784, 0.567, and 0.704, respectively. For the LDL-C level of 225 mg/dL, the sensitivity, specificity, and area under the curve values were 0.665, 0.712, and 0.745, respectively ( Fig.2B ) .
Fig.2. (A) Distribution of plasma cholesterol in individuals with familial hypercholesterolemia and general population. (B) Receiver operating characteristic curves for total and low-density lipoprotein-cholesterol and the presence of pathogenic variants.
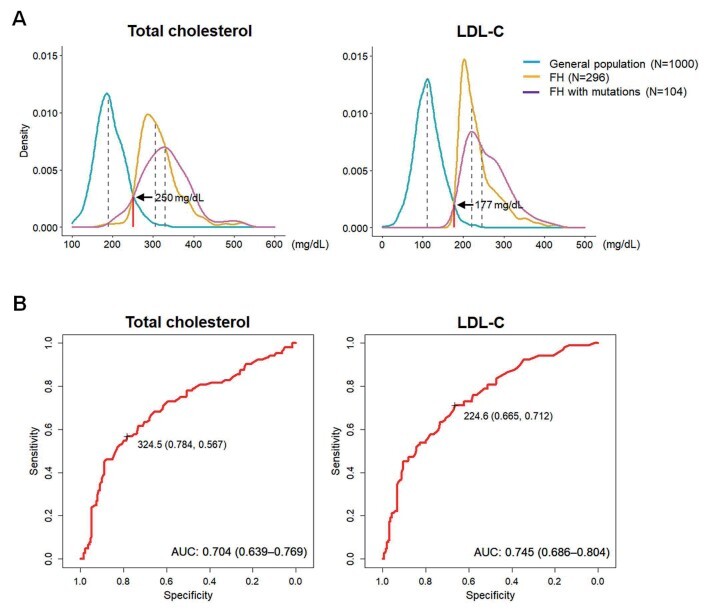
The optimal threshold levels identified based on the sensitivity and specificity are indicated.
Clinical Predictors of PVs
Univariable analysis revealed a significant association of age, CAD, tendon xanthoma, and LDL-C levels with the presence of PVs. After adjusting for potential confounders, age (hazard ratio [HR]: 0.96, p<0.001), tendon xanthoma (HR: 1.99, p=0.048), and LDL-C levels (HR: 1.02, p=0.001) were found to be independent predictors of PVs ( Table 3 ) .
Table 3. Univariable and multivariable logistic regression analysis for predictors of pathogenic variants.
| Characteristics | Univariable analysis HR (95% CI) | p | Multivariable analysis HR (95% CI) | p |
|---|---|---|---|---|
| Age, years | 0.95 (0.94–0.97) | <0.001 | 0.96 (0.94–0.98) | <0.001 |
| Male | 1.05 (0.65–1.69) | 0.85 | ||
| CAD | 1.89 (1.05–3.40) | 0.033 | 1.80 (0.92–3.53) | 0.087 |
| FHx of hypercholesterolemia | 1.28 (0.78–2.11) | 0.32 | ||
| FHx of premature CAD | 1.37 (0.83–2.24) | 0.22 | ||
| Tendon xanthomas | 1.91 (1.07–3.42) | 0.028 | 1.99 (1.01–3.92) | 0.048 |
| LDL-C, mg/dL | 1.02 (1.01–1.03) | <0.001 | 1.02 (1.01–1.03) | 0.001 |
Abbreviations: HR, hazard ratio; CI, confidence interval; CAD, coronary artery disease; FHx, family history; LDL-C, low-density lipoprotein- cholesterol.
Clinical Predictors of CAD
In univariable analysis, age, hypertension, smoking, family history of hypercholesterolemia, family history of premature of CAD, tendon xanthoma, LDL-C, HDL-C, and mutation positivity showed significant associations with CAD. In multivariable analysis adjusting confounding variables, age (HR 1.04, p=0.030), hypertension (HR 5.10, p<0.001), smoking (HR 3.58, p=0.003), LDL-C (HR 1.01, p=0.041), and HDL-C (HR 0.93, p<0.001) were identified as independent predictors of CAD ( Supplementary Table 3 ) .
Supplementary Table 3. Univariable and multivariable logistic regression analysis for predictors of CAD.
| Characteristics | Univariable analysis | p | Multivariable analysis | p |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Age, years | 1.03 (1.00–1.05) | 0.032 | 1.04 (1.00–1.07) | 0.030 |
| Male | 1.48 (0.83–2.66) | 0.19 | 1.87 (0.92-3.87) | 0.086 |
| Diabetes mellitus | 1.83 (0.68-4.49) | 0.21 | ||
| Hypertension | 5.47 (2.98-10.35) | <0.001 | 5.10 (2.46-11.05) | <0.001 |
| Smoking | 3.39 (1.76-6.49) | <0.001 | 3.58 (1.57-8.27) | 0.003 |
| FHx of hypercholesterolemia | 0.51 (0.28-0.91) | 0.024 | 0.74 (0.36-1.51) | 0.40 |
| FHx of premature CAD | 1.97 (1.09-3.55) | 0.024 | 1.91 (0.93-3.95) | 0.080 |
| Tendon xanthoma | 1.99 (1.02-3.81) | 0.040 | 1.73 (0.74-4.02) | 0.20 |
| Triglyceride, mg/dL | 1.00 (1.00-1.00) | 0.25 | ||
| HDL-C, mg/dL | 0.93 (0.91-0.96) | 0.006 | 0.93 (0.90-0.97) | <0.001 |
| LDL-C, mg/dL | 1.01 (1.00-1.01) | 0.016 | 1.01 (1.00-1.02) | 0.041 |
| Mutation positivity | 1.89 (1.05-3.41) | 0.033 | 1.36 (0.61-3.00) | 0.44 |
Abbreviations: CAD, coronary artery disease; HR, hazard ratio; CI, confidence interval; FHx, family history; HDL-C: high-density lipoprotein- cholesterol; LDL-C, low-density lipoprotein-cholesterol.
Discussion
This study comprehensively characterized Korean patients with FH, using the largest available registry data. The major findings were the following: 1) Of the 296 patients, 104 had PVs and stronger phenotype. PV rates ranged from 30% to 64% when patients were classified by possible or definite type criteria; 2) In order of frequency, missense, CNV, and frameshift variants were the most common PVs, and LDLR c.2054C>T (p.P685L) was the most frequent PV location; 3) According to multi-modal distribution, threshold levels of total cholesterol and LDL-C that helped identify patients with FH were 250 and 177 mg/dL, respectively; 4) According to ROC curves, the threshold levels of total cholesterol and LDL-C for PV carriers were 325 and 225 mg/dL, respectively; and 5) Lower age, tendon xanthoma, and higher LDL-C levels were identified as independent predictors of PVs after adjusting for confounders; 6) Older age, hypertension, smoking, low HDL-C, and high LDL-C levels were found to be independent predictors of CAD. Altogether, these results provide further genetic insights and tools for more efficient patient care in Korean populations with FH.
In the previous study published in 2015 15) , we analyzed 97 patients with FH and reported clinical features, LDL-C levels predictive of the presence of PVs, and predictors of coronary artery disease. However, the sample size was small and clinical data in that study needed to be confirmed or revised by a larger study. In the current study, we analyzed three-times larger number of individuals also including the previous 97 patients. By systemically presenting genetic and phenotypic characteristics in this larger study population, we provided better information of use in patient care than the older study. Furthermore, we compared the distribution of LDL-C in patients with FH and general population and obtained total cholesterol and LDL-C levels for patient differentiation.
PVs were reported in 35% of the study population. The relatively low rate of PV positivity could be due to the considerable portion of patients with possible type FH in our study. Compared to the incidence in the definite type, PV positivity in possible type FH has been reported to be approximately half or lower 7 , 16) . The median total cholesterol and LDL-C levels were 306 and 221 mg/dL, respectively. These values were comparable to those obtained in studies that analyzed populations of European ancestry 17 , 18) . Conversely, when patients with PVs were separately analyzed, the total cholesterol and LDL-C levels were 334 and 246 mg/dL, respectively. These levels were substantially lower than those of patients with FH in the Netherlands 19) and other Asian countries 14 , 20) . The underlying reasons for these differences, including the effects of distinct genetic backgrounds, remain to be clarified. As expected, patients with PVs were younger, more likely to have tendon xanthoma or CAD, and had higher cholesterol levels than PV-negative patients. Lower age in PV carriers suggests that the use of different cholesterol levels for screening patients with FH, such as the Make Early Diagnosis to Prevent Early Deaths criteria, may be reasonable. In addition, patients with PVs showed lower triglyceride levels and tended to have lower high-density lipoprotein-cholesterol levels than PV non-carriers, which is in agreement with the findings of a recent study from the Netherlands 21) . Among the different criteria for enrolment in our study, the PV-positive rate in patients who complied with the third and fourth criteria with uncertain family history was lower than that in the other patients. This finding indicates the potential value of family history in predicting PVs in individuals suspected to have FH.
Common types of PVs in our study were missense, CNV, and frameshift variants (in this order of frequency). On the other hand, missense, frameshift, and nonsense variants were common in a recent German study 22) . Of note, herein, CNVs were detected in a considerable proportion (11.5%) of PV carriers, similar to Japanese data 9) . Moreover, the prevalence of CNVs in PV-positive patients with FH from Canada was approximately 10% 13) . CNVs, which comprise large-scale mutations with deletion or duplication greater than 50 bp, have been overlooked for various reasons in diagnosis, including technical complexity. However, their importance is being gradually accepted, given their significant impact on clinical phenotype 23) .
The most frequent PVs in our study were c.2054C>T (p.P685L) and c.682G (p.E228*) in LDLR. A review article indicated that K811X and IVS15-3C>A are frequent variants in a group of Japanese patients with FH 9) . Furthermore, a recent study cataloging pathogenic mutations of LDLR from two major Japanese FH-care centers revealed that K811X, IVS12+2 t/c, and P685L were the three most common mutations 24) . It is interesting that P685L, the most common PV in our study, was not infrequently found in Japanese FH patients. In contrast, a recent Chinese report revealed that c.1879G>A, c.1747C>T, and c313+1G>A were highly prevalent among PV carriers 14) . These findings highlight that the characteristics of LDLR variants in each East Asian country are distinctive. The present study reported six APOB PV carriers (5.8% of total PV carriers), which indicates a relatively higher frequency of this PV in Korea than in neighboring countries. The most frequent CNV in our study was exon 9–12 deletion. Conversely, promoter–exon 1 deletion was the most common variant in a Canadian study 13) , whereas exon 2–3 deletion was the most prevalent CNV in Japan 9) . Therefore, it is highly probable that CNVs in FH also have a diverse spectrum among different ethnicities.
Multi-modal distribution data revealed that the threshold total cholesterol and LDL-C levels for differentiating patients with FH were 250 and 177 mg/dL, respectively, being higher than the 225 and 162 mg/dL reported in similar analyses in Japanese patients 20) . These values may facilitate efficient screening of individuals suspected to have FH. Furthermore, threshold total cholesterol and LDL-C levels for PV prediction using ROC curves were identified to be 325 and 225 mg/dL, respectively. Although these values are less sensitive for screening patients, they can be indicative of a stronger possibility of harboring PVs. Recently, Benn et al. reported 4.4 mmol/L (172 mg/dL) as the optimal threshold level for LDL-C to discriminate PV carriers using an ROC curve 5) . These different threshold levels may stem from differences in the study population. While the ROC curve in the study by Benn et al. was based on data from the general population, ours was from patients with FH only. Noteworthy, the threshold projected by Benn et al. (172 mg/dL) and that projected by us (177 mg/dL), obtained from the distribution curves of data from the general population and others, are similar, thereby representing a useful tool for screening patients from the general population.
Five traditional cardiovascular risk factors were found to be independent predictors of CAD in the present study. In our prior study analyzing smaller number of patients, we learned that only hypertension and low HDL-C were significant factors 15) . According to the current results, global risk factor control beyond lipid-lowering needs to be further stressed.
The current study has potential limitations. Although we tried to collect extensive data of patients with FH in Korea, the patient number was approximately 300, which is much lower than the estimated numbers of the Korean population with FH. Therefore, our results may underrepresent the phenotypic and genetic characteristics of patients with FH in this country. However, since we obtained data from 16 university hospitals from across Korea, we believe that the best available data on patients with FH to date was accessed. In addition, we established technical processes for analyzing and verifying the sequenced data published in our prior study 8) , which may have strengthened the validity of the present results.
In conclusion, the rate of PVs, including CNVs, considerably differed in patients classified by different diagnostic criteria. Frequent locations of PVs were distinctive from those observed in foreign patients. The threshold LDL-C levels for differentiation of patients with FH and PV prediction were identified to be 177 and 225 mg/dL, respectively. Altogether, these results provide new tools for more efficient patient care in the Korean population with FH.
Author Contributions
HK: Investigation, Validation, Writing-original draft; CJL: Investigation, Resources, Writing-review and editing; SHK: Resources, Writing-review and editing; JYK: Resources, Writing-review and editing; SHC: Resources, Writing-review and editing; HJK: Conceptualization, Resources, Writing-review and editing; KSP: Conceptualization, Resources, Writing-review and editing; BRC: Resources, Writing-review and editing; BJK: Resources, Writing-review and editing; KCS: Resources, Writing-review and editing; IKJ: Resources, Writing-review and editing; JOK: Resources, Writing-review and editing; JWB: Resources, Writing-review and editing; JMP: Investigation, Validation, Writing-review and editing; YL: Investigation, Writing-review and editing; IJ: Investigation, Writing-review and editing; HH: Investigation, Writing-review and editing; JHL: Investigation, Validation, Writing-original draft; SHL: Conceptualization, Funding acquisition, Supervision, Resources, Investigation, Validation, Writing-original draft; All gave final approval and agree to be accountable for all aspects of work ensuring integrity and accuracy.
KFH Registry Investigators
Hyoeun Kim, Chan Joo Lee, and Sang-Hak Lee; Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
Sang-Hyun Kim; Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea
Jang Young Kim; Yonsei University Wonju College of Medicine, Wonju, Korea
Sung Hee Choi; Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
Hyun-Jae Kang and Kyong Soo Park; Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Byung Ryul Cho; Kangwon National University, School of Medicine, Chuncheon, Korea
Byung Jin Kim and Ki Chul Sung; Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Korea
In-Kyung Jeong; Kyung Hee University Hospital at Gangdong, Kyung Hee University School of Medicine, Seoul, Korea
Jin-Ok Jeong; Chungnam National University Hospital, Chungnam National University School of Medicine, Daejeon, Korea
Jang-Whan Bae; Chungbuk National University College of Medicine, Cheongju, Korea
Doo Il Kim; Haeundae Paik Hospital, Inje University College of Medicine, Busan, Korea
Moo-Yong Rhee; Dongguk University Ilsan Hospital, Goyang, Korea
Byoung Kwon Lee; Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
Youngkeun Ahn; Chonnam National University Hospital, Gwangju, Korea
Jeong-Taek Woo; Kyung Hee University School of Medicine, Seoul, Korea
Seung-Ho Hur; Keimyung University Dongsan Medical Center, Daegu, Korea
Financial Support
This work was supported by the Korean Society of Lipid and Atherosclerosis.
Acknowledgements
We are grateful to Jiyeong Jeong, RN, and Yoo Kyung Jung, RN, for their excellent assistance in clinical data collection and patient care and to Jin Woo Im for the management of patient samples.
Conflict of Interest Statement
The authors report no conflict of interest.
References
- 1).Stock, J: Familial hypercholesterolemia: an urgent public health priority. Atherosclerosis, 2020; 308: 48-49 [DOI] [PubMed] [Google Scholar]
- 2).Rhee EJ, Kim HC, Kim JH, Lee EY, Kim BJ, Kim EM, Song Y, Lim JH, Kim HJ, Choi S, Moon MK, Na JO, Park KY, Oh MS, Han SY, Noh J, Yi KH, Lee SH, Hong SC, Jeong IK; Committee of Clinical Practice Guideline of Korean Society of Lipid and Atherosclerosis: 2018 guidelines for the management of dyslipidemia in Korea. J Lipid Atheroscler, 2019; 8: 78-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Choi JY and Na JO: Pharmacological strategies beyond statins: ezetimibe and PCSK9 inhibitors. J Lipid Atheroscler, 2019; 8: 183-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Authors/Task Force Members, ESC Committee for Practice Guidelines (CPG), ESC National Cardiac Societies: 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Atherosclerosis, 2019; 290: 140-205 [DOI] [PubMed] [Google Scholar]
- 5).Benn M, Watts GF, Tybjærg-Hansen A, and Nordestgaard BG: Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J, 2016; 37(17): 1384-1394 [DOI] [PubMed] [Google Scholar]
- 6).Harada-Shiba M, Arai H, Ishigaki Y, Ishibashi S, Okamura T, Ogura M, Dobashi K, Nohara A, Bujo H, Miyauchi K, Yamashita S, and Yokote K; Working Group by Japan Atherosclerosis Society for Making Guidance of Familial Hypercholesterolemia: Guidelines for diagnosis and treatment of familial hypercholesterolemia 2017. J Atheroscler Thromb, 2018; 25: 751-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Futema M, Whittall RA, Kiley A, Steel LK, Cooper JA, Badmus E, Leigh SE, Karpe F, Neil HA; Simon Broome Register Group, Humphries SE: Analysis of the frequency and spectrum of mutations recognised to cause familial hypercholesterolaemia in routine clinical practice in a UK specialist hospital lipid clinic. Atherosclerosis, 2013; 229: 161-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Han SM, Hwang B, Park T-G, Kim D-I, Rhee M-Y, Lee B-K, Ahn Y-K, Cho B-R, Woo J, Hur S-H, Jeong J-O, Park S, Jang Y, Lee M-G, Bang D, Lee J-H, and Lee S-H: Genetic testing of Korean familial hypercholesterolemia using whole-exome sequencing. PLoS One, 2015; 10: e0126706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Mabuchi H: Half a century tales of familial hypercholesterolemia (FH) in Japan. J Atheroscler Thromb, 2017; 24: 189-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Sturm AC, Knowles JW, Gidding SS, Ahmad ZS, Ahmed CD Ballantyne CM, Baum SJ, Bourbon M, Carrié A, Cuchel M, de Ferranti SD, Defesche JC, Freiberger T, Hershberger RE, Hovingh GK, Karayan L, Kastelein JJP, Kindt I, Lane SR, Leigh SE, Linton MF, Mata P, Neal WA, Nordestgaard BG, Santos RD, Harada-Shiba M, Sijbrands EJ, Stitziel NO, Yamashita S, Wilemon KA, Ledbetter DH, and Rader DJ; Convened by the Familial Hypercholesterolemia Foundation: Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol, 2018; 72: 662-680 [DOI] [PubMed] [Google Scholar]
- 11).Tada H, Kawashiri M-A, Nohara A, Inazu A, Mabuchi H, and Yamagishi M: Impact of clinical signs and genetic diagnosis of familial hypercholesterolaemia on the prevalence of coronary artery disease in patients with severe hypercholesterolaemia. Eur Heart J, 2017; 38: 1573-1579 [DOI] [PubMed] [Google Scholar]
- 12).Kim H, Lee CJ, Pak H, Kim D-I, Rhee M-Y, Lee B-K, Ahn Y, Cho B-R, Woo J-T, Hur S-H, Jeong J-O, Lee J-H, and Lee S-H: GENetic characteristics and REsponse to lipid-lowering therapy in familial hypercholesterolemia: GENRE-FH study. Sci Rep, 2020; 10: 19336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Iacocca MA, Wang J, Dron JS, Robinson JF, McIntyre AD, Cao H, and Hegele RA: Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia. J Lipid Res, 2017; 58: 2202-2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Wang H, Yang H, Liu Z, Cui K, Y. Zhang Y, Zhang Y, Zhao K, Yin K, Li W, and Zhou Z: Targeted genetic analysis in a Chinese cohort of 208 patients related to familial hypercholesterolemia. J Atheroscler Thromb, 2020; 27: 1288-1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Shin DG, Han SM, Kim DI, Rhee M-Y, Lee B-K, Ahn YK, Cho BR, Woo JT, Hur SH, Jeong JO, Jang Y, Lee JH, and Lee SH: Clinical features of familial hypercholesterolemia in Korea: predictors of pathogenic mutations and coronary artery disease - a study supported by the Korean Society of Lipidology and Atherosclerosis. Atherosclerosis, 2015; 243: 53-58 [DOI] [PubMed] [Google Scholar]
- 16).Taylor A, Wang D, Patel K, Whittall R, Wood G, Farrer M, Neely RD, Fairgrieve S, Nair D, Barbir M, Jones JL, Egan S, Everdale R, Lolin Y, Hughes E, Cooper JA, Hadfield SG, Norbury G, and Humphries SE: Mutation detection rate and spectrum in familial hypercholesterolaemia patients in the UK pilot cascade project. Clin Genet, 2010; 77: 572-580 [DOI] [PubMed] [Google Scholar]
- 17).Rizos CV, Elisaf MS, Skoumas I, Tziomalos K, Kotsis V, Rallidis L, Garoufi A, Athyros VG, Skalidis E, Kolovou G, Koutagiar I, Papagianni M, Antza C, Katsiki N, Ganotakis E, and Liberopoulos EN: Characteristics and management of 1093 patients with clinical diagnosis of familial hypercholesterolemia in Greece: data from the Hellenic Familial Hypercholesterolemia Registry (HELLAS-FH). Atherosclerosis, 2018; 277: 308-313 [DOI] [PubMed] [Google Scholar]
- 18).Brunham LR, Ruel I, Khoury E, Hegele RA, Couture P, Bergeron J, Baass A, Dufour R, Francis GA, Cermakova L, Mancini GBJ, Brophy JM, Brisson D, Gaudet D, and Genest J: Familial hypercholesterolemia in Canada: initial results from the FH Canada national registry. Atherosclerosis, 2018; 277: 419-424 [DOI] [PubMed] [Google Scholar]
- 19).Reeskamp LF, Tromp TR, Defesche JC, Grefhorst A, Stroes ES, Hovingh GK, and Zuurbier L: Next-generation sequencing to confirm clinical familial hypercholesterolemia. Eur J Prev Cardiol, 2020; 2047487320942996 [DOI] [PubMed] [Google Scholar]
- 20).Mabuchi H, Higashikata T, Nohara A, Lu H, Yu WX, Nozue T, Noji Y, Katsuda S, Kawashiri MA, Inazu A, Kobayashi J, and Koizumi J: Cutoff point separating affected and unaffected familial hypercholesterolemic patients validated by LDL-receptor gene mutants. J Atheroscler Thromb, 2005; 12: 35-40 [DOI] [PubMed] [Google Scholar]
- 21).Besseling J, Reitsma JB, Gaudet D, Brisson D, Kastelein JJP, Hovingh GK, and Hutten BA: Selection of individuals for genetic testing for familial hypercholesterolaemia: development and external validation of a prediction model for the presence of a mutation causing familial hypercholesterolaemia. Eur Heart J, 2017; 38: 565-573 [DOI] [PubMed] [Google Scholar]
- 22).Rieck L, Bardey F, Grenkowitz T, Bertram L, Helmuth J, Mischung C, Spranger J, Steinhagen-Thiessen E, Bobbert T, Kassner U, and Demuth I: Mutation spectrum and polygenic score in German patients with familial hypercholesterolemia. Clin Genet, 2020; 98: 457-467 [DOI] [PubMed] [Google Scholar]
- 23).M.A. Iacocca MA, Dron JS, and Hegele RA: Progress in finding pathogenic DNA copy number variations in dyslipidemia. Curr Opin Lipidol, 2019; 30: 63-70 [DOI] [PubMed] [Google Scholar]
- 24).Tada H, Hori M, Nomura A, Hosomichi K, Nohara A, Kawashiri MA, Harada-Shiba M: A catalog of the pathogenic mutations of LDL receptor gene in Japanese familial hypercholesterolemia. J Clin Lipidol, 2020; 14: 346-351 [DOI] [PubMed] [Google Scholar]
- 25).Tada H, Okada H, Nomura A, Yashiro S, Nohara A, Ishigaki Y, Takamura M, Kawsshiri MA: Rare and deleterious mutations in ABCG5/ABCG8 genes contribute to mimicking and worsening of familial hypercholesterolemia phenotype. Circ J, 2019; 83: 1917-1924 [DOI] [PubMed] [Google Scholar]


