Abstract
Penicillin-binding proteins (PBPs) are bacterial cytoplasmic membrane proteins that catalyze the final steps of the peptidoglycan synthesis. Resistance to β-lactams in Streptococcus pneumoniae is caused by low-affinity PBPs. S. pneumoniae PBP 2a belongs to the class A high-molecular-mass PBPs having both glycosyltransferase (GT) and transpeptide (TP) activities. Structural and functional studies of both domains are required to unravel the mechanisms of resistance, a prerequisite for the development of novel antibiotics. The extracellular region of S. pneumoniae PBP 2a has been expressed (PBP 2a*) in Escherichia coli as a glutathione S-transferase fusion protein. The acylation kinetic parameters of PBP 2a* for β-lactams were determined by stopped-flow fluorometry. The acylation efficiency toward benzylpenicillin was much lower than that toward cefotaxime, a result suggesting that PBP 2a participates in resistance to cefotaxime and other β-lactams, but not in resistance to benzylpenicillin. The TP domain was purified following limited proteolysis. PBP 2a* required detergents for solubility and interacted with lipid vesicles, while the TP domain was water soluble. We propose that PBP 2a* interacts with the cytoplasmic membrane in a region distinct from its transmembrane anchor region, which is located between Lys 78 and Ser 156 of the GT domain.
Penicillin-binding proteins (PBPs) are components of bacterial cytoplasmic membranes that catalyze the final steps of the peptidoglycan synthesis. These steps require three types of enzymatic activities: the glycosyltransferase (GT), which polymerizes the glycan strand by using the lipid II as the substrate; the transpeptidase (TP), which cross-links these strands via their peptide side chains; and the d-alanine carboxypeptidase, which is believed to play a regulatory role (7). Reactions catalyzed by PBPs occur either on the outer surface of the cytoplasmic membrane (GT) or further outside (TP and d-alanine carboxypeptidase); the major fraction of the proteins is therefore localized in the periplasm. PBPs can be divided into two groups by molecular sizes and amino acid sequence comparison (7): low-molecular-weight (low-Mr) PBPs that catalyze d-alanine carboxypeptidase and endopeptidase reactions, and high-molecular-weight (high-Mr) PBPs that carry GT and TP activities. Class A high-Mr PBPs are bifunctional enzymes bearing both GT and TP activities, while class B high-Mr PBPs have so far only been associated with the TP activity.
Penicillin and other β-lactam antibiotics are specific inhibitors of TP and d-alanine carboxypeptidase reactions because of their structural similarity to the natural substrates, the stem peptides (25). When β-lactams form a covalent complex with the active serine of TP domain enzymatic cavity, this event prevents the cross-linking of peptide chains of the peptidoglycan, leading to cell death.
Streptococcus pneumoniae, a major human pathogen of the upper respiratory tract possesses six PBPs, including three that are class A (PBPs 1a, 1b, and 2a), two that are class B (PBP 2x and PBP 2b), and a low-Mr PBP, PBP 3 (10). Individual deletion of either of the genes pbp2x or pbp2b is lethal for S. pneumoniae (14). However, neither the pbp1a, pbp1b, nor pbp2a gene is required for growth when deleted individually, but the presence of at least a pbp1a or pbp2a gene is essential for cell viability (12).
The increased occurrence of S. pneumoniae strains resistant to β-lactam antibiotics has become a worldwide health problem. In penicillin-resistant strains of S. pneumoniae, high-Mr PBPs have a decreased affinity for the β-lactams, driving the requirement for a higher antibiotic dosage. The low-affinity PBP variant genes conferring resistance have a mosaic structure, in which parts of genes are replaced by equivalent parts from other bacterial sources (4, 15, 17). PBP 2b and PBP 2x constitute the most common resistance determinants and confer low-level resistance to piperacillin and cefotaxime, respectively (9). A higher level of resistance to these agents (but not to benzylpenicillin) is conferred by an additional alteration of PBP 2a (11), while the highest levels of resistance require further modifications of PBP 1a (2, 11, 19, 22).
S. pneumoniae PBP 2a is an attractive target for antibacterial chemotherapy. Indeed, PBP 2a plays an important role in cell viability and in acquisition of β-lactam resistance. Furthermore, PBP 2a, like all class A PBPs, is a major therapeutic target, because no antibiotic usable in human therapy so far has been able to specifically inhibit GT activity.
The aim of this study is to gain an in-depth understanding of the mechanism of PBPs’ enzymatic activities through a combination of high-resolution three-dimensional structure and functional data. Solubilization of the periplasmic region of S. pneumoniae PBP 2x (18, 21), E. coli PBPs 2 and 3 (1, 6), and Staphylococcus aureus PBP 2a (23) was obtained by recombinant expression of the transmembrane-deleted form of the proteins. All of these proteins belong to the class B high-Mr PBPs. To the contrary, the extracellular region of class A high-Mr S. pneumoniae PBP 1a was apparently not monomeric in solution (unpublished data), and E. coli PBP 1b remained associated with the bacterial membrane even in the absence of the transmembrane anchor (20, 26). Furthermore, low-Mr E. coli PBPs 5 and 6 are also associated with the periplasmic side of the inner membrane via a C-terminal amphiphilic helix (5, 24).
We have expressed and characterized the periplasmic region of S. pneumoniae PBP 2a (PBP 2a*) deprived of its cytoplasmic domain and transmembrane anchor. Our main finding relates to the lipophilic property of the GT domain and its putative association with the bacterial cytoplasmic membrane.
MATERIALS AND METHODS
Materials.
The thiolester substrate analogue S2d (N-benzoyl-d-alanylmercaptoacetic acid) (13) was obtained from L. Christiaens (Université de Liège, Liège, Belgium). 4,4′-Dithiopyridine, trypsin, thrombin, glutathione, penicillin G, cefotaxime, CHAPS {3-[(3-cholamidopropyl)dimethylammonio]-1-propane-sulfonate}, deoxycholic acid (DOC), Triton X-100, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Sigma. IPTG (isopropyl-β-d-thiogalactopyranoside) was purchased from Promega; ampicillin, leupeptin, and aprotinin were obtained from Boehringer Mannheim. A stock solution of lipid 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) from Sigma was kept in chloroform at −20°C under nitrogen gas.
Expression plasmid and culture conditions.
The S. pneumoniae R6 pbp2a sequence had been deposited previously in the EMBL and GenBank databases under the accession no. AJ002292 (11). Cloning of the PBP 2a* gene was described in reference 12. Briefly, the periplasmic region of the PBP 2a gene was PCR amplified by using a DNA genomic preparation of the S. pneumoniae R6 strain. The upstream primer introduced an ATG start codon beginning at Lys 78. The PCR product was cloned into pGEX-2T plasmid (Pharmacia) in frame with glutathione S-transferase (GST), leading to a construct named pJAH144.
Protein expression was performed with E. coli DH5α [φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169]. Cultures were grown in Luria-Bertani medium (10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per liter) (Life Technologies) supplemented with 100 μg of ampicillin per ml when necessary.
Protein solubilization and purification.
A culture (1.6 liters) of DH5α/pJAH144 cells in Luria-Bertani medium containing 100 μg of ampicillin per ml was grown at 37°C until the optical density at 600 nm (OD600) reached 1. The fusion protein synthesis was induced with 0.5 mM IPTG, and the culture continued overnight at 22°C. All of the following steps were carried out at 0 to 4°C. Cells were harvested by centrifugation at 11,300 × g for 15 min and resuspended in 40 ml of a solution of 50 mM Tris-HCl (pH 8.0)–200 mM KCl containing 1 mM PMSF, 1 μg of leupeptin per ml, and 1 μg of aprotinin per ml. The resulting suspension was disrupted by a 2-min sonication step. The cell lysate was then centrifuged at 31,000 × g for 20 min, and the supernatant was discarded. The pellet was resuspended in 20 ml of 50 mM Tris-HCl (pH 8.0)–200 mM KCl–0.5% CHAPS. The suspension was submitted to rotary shaking to allow a more efficient solubilization of the GST-PBP 2a* fusion protein. The supernatant containing the fusion protein obtained from a 15-min centrifugation at 20,800 × g was kept. The solubilization procedure was repeated twice. The resulting supernatant was pooled and was loaded onto a 5-ml glutathione-Sepharose column (Pharmacia). The column was equilibrated and washed with 50 mM Tris-HCl (pH 8.0)–200 mM KCl–0.5% CHAPS. The fusion protein was cleaved while bound to the column in the presence of 50 U of thrombin for 1 h at room temperature. The thrombin activity was then inhibited by 1 mM PMSF.
Gel filtration experiments were performed with a Superdex 200 HR 10/30 column (Pharmacia). The equilibration buffer was 50 mM Tris-HCl (pH 8.0)–200 mM KCl with or without 0.5% CHAPS. The flow rate was 0.5 ml/min. Fractions (0.5 ml) were collected, and 50-μl aliquots were analyzed for [3H]benzylpenicillin binding and fluorography. One hundred twenty and 60 μg of PBP 2a* and TP domain, respectively, were loaded onto the column.
The concentration of PBP 2a* in the presence of detergent was measured by the bicinchoninic acid (BCA) method (Pierce BCA kit) and by OD280.
Protease digestion of PBP 2a*.
Purified PBP 2a* in 50 mM Tris-HCl (pH 8.0)–200 mM KCl–0.5% CHAPS was incubated at various trypsin/PBP 2a* ratios of 1:20 to 1:500 (wt/wt) for 30 min at 37°C. Following incubation, the proteolyzed samples were subjected to a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel for electrophoresis. The TP domain was prepared by trypsin digestion of PBP 2a* at a trypsin/PBP 2a* ratio of 1:100 (wt/wt) for 30 min at 37°C. The protease activity was inhibited by 1 mM PMSF.
Extensive proteolysis of PBP 2a* with thrombin was performed at a thrombin/PBP 2a* ratio of 1:1 (wt/wt) for 30 min at 37°C, before addition of 1 mM PMSF.
PAGE.
Native discontinuous gel electrophoresis was performed on a vertical slab apparatus by using a 3% stacking gel and an 8% separating gel starting from a stock acrylamide solution of 29% acrylamide–1% N,N′-methylenebisacrylamide. The separating gel contained 373 mM Tris-HCl (pH 8.8) and 0.64% glycerol, while the stacking gel was buffered by 127 mM Tris-HCl (pH 6.8). Polymerization was performed with 0.1% N,N′,N′,N′-tetramethylenediamine and 0.036% ammonium persulfate. The electrode buffer was 25 mM Tris-HCl (pH 8.3)–192 mM glycine. The loading sample buffer was composed of 62.5 mM Tris-HCl (pH 6.8), 10% glycerol, 0.1% bromophenol blue. Charge-shift electrophoresis experiments were performed on such native gels. CHAPS detergent was exchanged in PBP 2a* and TP domain solutions for other detergents (0.5% Triton X-100 or 0.5% Triton X-100–0.25% DOC) by 10-fold dilution and concentration of the protein solutions to their original volume with a Microcon-30 unit (Amicon).
Discontinuous SDS-polyacrylamide gel electrophoresis (PAGE) conditions were slightly modified from the native PAGE. The separating gel concentration was 12.5%, and 0.1% SDS was included in both stacking and separating gels. Denaturing agents (2% SDS, 0.65 M β-mercaptoethanol) were added to the sample buffer, and 0.1% SDS was included in the electrode buffer.
N-terminal protein sequencing.
Sample proteins (2 to 10 μg) were boiled in SDS-PAGE loading buffer for 5 min at 100°C and then loaded onto an SDS–12.5% polyacrylamide gel. The proteins were transferred to a Problot membrane (Applied Biosystems) and stained according to the supplier’s instructions. The protein bands were cut from the membrane and sequenced by automated Edman degradation on an Applied Biosystem gas-phase sequencer model 477A with on-line analysis of the phenyl thiohydantoin derivatives.
Determination of kinetic parameters.
PBPs interact with β-lactam antibiotics according to the three-step scheme
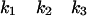 |
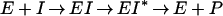 |
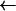 |
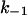 |
where E is the PBP enzyme, I is the β-lactam antibiotic, EI is the Michaelis-Menten complex, EI* is the acyl-enzyme covalent complex, and P is the product of the reaction (degraded β-lactam antibiotic). K, the dissociation constant of the enzyme-substrate complex, is equal to k−1/k1.
The k2/K parameter, accounting for the acylation step efficiency, was determined by monitoring the decrease in the intrinsic fluorescence of the protein in the presence of antibiotic at 37°C by using spectrofluorometric measurements coupled with a Biologic SFM3 stopped-flow apparatus. The excitation wavelength specific for tryptophan was 280 nm, and emission was measured in the 305- to 360-nm range. PBP 2a* at a concentration of 2 μM in 10 mM sodium phosphate (pH 7.0)–200 mM KCl was incubated with concentrations of β-lactam antibiotic ranging from 200 μM to 1 mM.
The activity of the TP domain of PBP 2a* was measured in terms of its ability to hydrolyze the S2d molecule, a synthetic thiolester analogue of cell wall stem peptide, according to the method described by Zhao et al. (27). The assays were performed at 37°C. Protein concentrations were determined with the BCA detection kit (Pierce) with bovine serum albumin (BSA) as a standard.
The titration of functional PBP and determination of k3 were performed with [3H]benzylpenicillin (20 Ci/mmol, 1 mCi/ml; Amersham). Purified PBP 2a* (2 μM) in 50 mM Tris-HCl (pH 8.0)–200 mM KCl was incubated at 37°C with different concentrations of [3H]benzylpenicillin for 15 min, at which time, the reaction was completed. The samples were then subjected to SDS-PAGE electrophoresis. [3H]benzylpenicillin bound to proteins was monitored by two different procedures. The gel was stained with Coomassie blue, destained, incubated with Amplify (Amersham), dried, and either exposed to a film for 16 h or cut around the protein bands. In the latter case, the radioactivity of the gel slice was measured by using a liquid scintillation analyzer (Packard model 2100TR) after being mixed with 10 ml of LSC cocktail (Picofluor 15; Packard).
The deacylation reaction obeys the following equation: −k3t = ln [EI*]t/[EI*]0, where [EI*]0 is the initial concentration of acyl-enzyme and [EI*]t is its concentration at time t. Determination of k3 was performed as follows. A 2 μM concentration of purified PBP was labelled with 1 μM [3H]benzylpenicillin during 15 min at 37°C. Cold benzylpenicillin (15 mM) was added, and the incubation was continued at 37°C. Samples were removed at various times and loaded onto an SDS gel, and the amount of radioactivity was measured in the protein bands as mentioned above.
Lipid vesicle reconstitution.
DOPC solution in chloroform was dried under argon, and the lipids were then redissolved to a final concentration of 15 mM in 50 mM Tris-HCl (pH 8.0)–200 mM KCl–10% CHAPS. The lipid suspension was subsequently sonicated. The purified PBP 2a* protein, TP domain, and PBP 2a* thrombin-digested product were reconstituted into phospholipid bilayer vesicles by removal of detergent by dialysis. PBP 2a* and TP domain concentrations were adjusted to 1 mg per ml in 50 mM Tris-HCl (pH 8.0)–200 mM KCl–3% CHAPS. The PBP 2a* thrombin-digested product was used at 0.07 mg per ml in the same buffer. DOPC (15 mM) was added to make molar protein/DOPC ratios of 1:50 to 1:1,000. The mixtures (20 to 100 μl) were extensively dialyzed against 50 mM Tris-HCl (pH 8.0)–200 mM KCl for at least 24 h at 4°C. The proteoliposome formation was assayed by PAGE under native conditions.
RESULTS
PBP 2a sequence and topology.
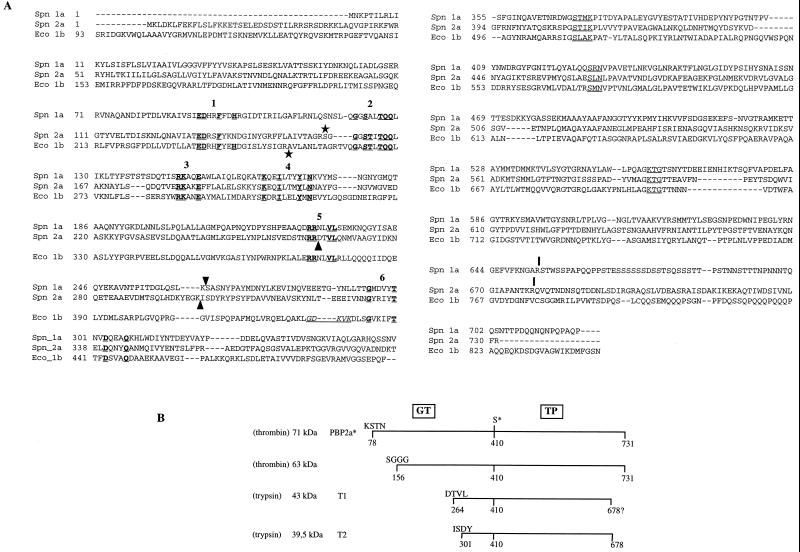
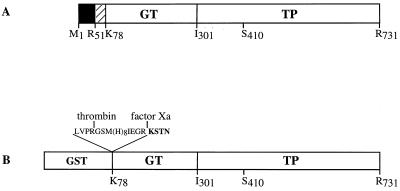
The open reading frame of the pbp2a gene encodes a 731-amino-acid protein which contains all of the features characteristic of class A high-Mr PBPs (Fig. 1A) (11). PBP 2a includes a cytoplasmic N-terminal region (Met 1 to Arg 51) and a hydrophobic 26-amino-acid region (Tyr 52 to Ala 77), which likely functions as a membrane anchor (Fig. 2A). The periplasmic region is divided into two domains: the 25-kDa GT domain and the 50-kDa TP domain. Conserved motifs in the GT and TP domains are indicated in Fig. 1A (11).
FIG. 1.
(A) Alignment sequence of class A high-Mr PBPs S. pneumoniae PBP 1a, PBP 2a (11, 12), and E. coli PBP 1b. The conserved motifs in the GT domain are underlined and in boldface, and are numbered according to the system used in reference 8. The active-site conserved motifs in the TP domain are underlined. Arrowheads indicate the N-terminal positions of the PBP 1a TP domain (Ser 264) (3) and T1 and T2 tryptic products of PBP 2a (this work). The underlined residues in italics correspond to the permissive site found in E. coli PBP 1b delineating the GT and TP junction domains (16). Vertical bars indicate the C terminus of TP domains (reference 3 and this work). Stars indicate the first residue of the detergent-free soluble proteins (reference 26 and this work). (B) Schematic representation of the proteolytic fragments. The molecular mass of each fragment was measured by SDS-PAGE. The N-terminal sequence of the fragments was experimentally determined. The C terminus of T2 is derived from mass spectrometry measurements. The position of the active-site serine 410 is indicated (S*).
FIG. 2.
Schematic diagrams of S. pneumoniae PBP 2a. (A) Topology of the native PBP 2a protein. The solid and hatched boxes indicate the N-terminal cytoplasmic region and the membrane anchor, respectively. (B) Construction of the GST-PBP 2a* fusion protein. The active-site serine 410 is indicated in both figures. The peptide at the GST-PBP 2a* junction includes sequences specific for thrombin and factor Xa cleavage, as well as the polyhistidine tag. The N-terminal sequence of PBP 2a* is presented in boldface.
Expression and purification of PBP 2a*.
The successful expression and characterization of the periplasmic region of S. pneumoniae PBP 2x and PBP 1a (3, 18) prompted us to produce a soluble form of the periplasmic domain of PBP 2a as a GST fusion protein (Fig. 2B). Cleavage sites specific for thrombin and factor Xa have been added between GST and the N terminus of PBP 2a*. A stretch of eight histidines was added following the thrombin cleavage site.
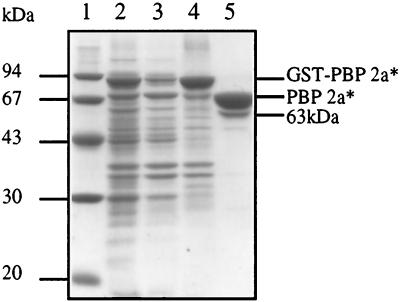
GST-PBP 2a* is expressed in E. coli cells in large amounts as a major 90-kDa species (Fig. 3, lane 2). The fusion molecule partitions into a minor soluble pool and a much larger insoluble fraction (Fig. 3, lanes 3 and 4). Attempts at purifying active PBP 2a* from the soluble fraction were unsuccessful due to the sensitivity of the protein to proteolytic degradation during the purification procedure. Solubilization of the insoluble fraction from the cell debris with urea followed by in vitro refolding led to a heterogeneous nonfunctional GST-PBP 2a*. Solubilization of the insoluble fraction with a series of detergents, including Triton X-100, β-octylglucoside, and CHAPS was efficient, provided that the E. coli cultures were grown at 22°C (Fig. 3, lane 4). A concentration of 0.5% CHAPS was selected for the rest of this work for its high critical micellar concentration (0.25%), allowing easy detergent removal or exchange. GST-PBP 2a* solubilized with CHAPS was purified by affinity chromatography and cleaved with thrombin directly on the glutathione column. The eluted fraction included a dominant species with an apparent molecular mass of 71 kDa, which is in good agreement with the calculated value of PBP 2a* (71.8 kDa), and a 63-kDa minor species (Fig. 3, lane 5). Both molecules were functional with respect to [3H]benzylpenicillin binding (Fig. 4B, lane 2). The N-terminal sequences of the dominant and minor PBP 2a* forms were determined by Edman degradation to be Lys-Ser-Thr-Asn and Ser-Gly-Gly-Gly-Ser-Thr, respectively (Fig. 1B). Therefore, thrombin proteolysis occurred efficiently downstream of the expected site, at the junction between factor Xa cleavage sequence and the N-terminal portion of PBP 2a* (Fig. 1B and 2B). The secondary minor cleavage between Arg 155 and Ser 156 is reminiscent of a similar site within the PBP 1a* sequence (3). After a single chromatography step, the purification yield was around 2 to 3 mg/liter of E. coli culture, with the minor cleavage product acting as the sole source of heterogeneity in the preparation.
FIG. 3.
Analysis of solubilization and purification of PBP 2a*. Proteins were separated by SDS–12.5% PAGE and stained with Coomassie blue. The numbers to the left indicate the sizes of standard molecular mass markers. Lanes: 1, molecular mass markers; 2, total cell proteins; 3, soluble supernatant; 4, 0.5% CHAPS soluble extract; 5, PBP 2a* eluted from glutathione Sepharose after thrombin cleavage.
FIG. 4.
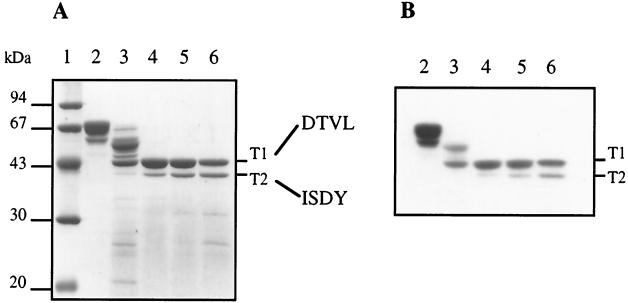
Trypsin cleavage of PBP 2a* leading to TP domain isolation. (A) Proteins were separated by SDS–12.5% PAGE and stained with Coomassie blue. The N-terminal sequence of each product is indicated on the right. (B) Fluorogram of a dried gel from SDS–12.5% PAGE of proteins (2 μg) labelled with [3H]benzylpenicillin prior to the migration. Digestions were performed at 37°C for 30 min. Lanes: 1, molecular mass markers; 2, undigested PBP 2a*; 3, trypsin/PBP 2a* ratio of 1:500 (wt/wt); 4, trypsin/PBP 2a* ratio of 1:100 (wt/wt); 5, trypsin/PBP 2a* ratio of 1:50 (wt/wt); 6, trypsin/PBP 2a* ratio of 1:20 (wt/wt).
Determination of PBP 2a* kinetic parameters.
Quantitative binding of [3H]benzylpenicillin was performed to titrate the active site in PBP 2a*. The protein concentrations were identical, whether determined by active-site titration or by the BCA method (data not shown), showing that all proteins were functional in the preparation. The kinetic properties of PBP 2a* were then determined by using the cephalosporin and penicillin classes of antibiotics and the S2d substrate analogue (Table 1). A second-order rate constant of acylation (k2/K) of 7,700 ± 600 M−1 s−1 was measured with cefotaxime. The very small fluorescent quench of PBP 2a* with benzylpencillin during the 5 s of the experiment impaired accurate determination of k2/K. This shows that benzylpenicillin acylates PBP 2a* with a very low efficiency. This observation is not contradictory with the complete labeling of PBP 2a* by [3H]benzylpenicillin in fluorography experiments when a much longer incubation time (15 min) was used. The efficiency of hydrolysis of S2d thiolester by PBP 2a* is close to values reported for PBP 1a and the PBP 2x of the resistant clinical strain of S. pneumoniae 328, (R-PBP 2x*) (Table 1). The k3 deacylation rate of PBP 2a* for benzylpenicillin is in the same range as those reported for other PBPs (Table 1).
TABLE 1.
Kinetic parameters of S. pneumoniae PBP 2a*
| Protein | Antibiotic
|
Substrate analogue S2d kcat/Km (M−1 s−1)a | Source or reference | ||
|---|---|---|---|---|---|
| Cefotaxime k2/K (M−1 s−1) | Benzylpenicillin k2/K (M−1 s−1) | Benzylpenicillin k3 (s−1) [half-life time; h] | |||
| PBP 2a* | 7,700 (±600)b | <3,800 | 3.2 × 10−5 [6] | 220 (±20)b | This study |
| PBP 1a* | 11,933 | 34,200 | 1.0 × 10−5 [14] | 256 | 3 |
| S-PBP 2x* | 209,000 | 99,000 | 3.5 × 10−5 [5.8] | 2,500 | 18 |
| R-PBP 2x* | 3,300 | <3,800 | 4.2 × 10−5 [4.6] | 20 | 18 |
The kcat/Km value of PBP 1a* for S2d reported in this table takes into account the racemic nature of S2d (twofold increase) compared to the value reported in reference 3
The standard deviation was determined from totally independent measurements.
Limited trypsin proteolysis of PBP 2a* releases the TP domain.
Proteolysis of purified PBP 2a* with a trypsin/protein ratio ranging from 1:100 to 1:20 (wt/wt) results in the production of two major protein fragments, T1 and T2, with molecular masses of 43 and 39.5 kDa, respectively (Fig. 4A). Both fragments are competent for [3H]benzylpenicillin binding (Fig. 4B), showing that they encompass the TP domain. N-terminal sequencing revealed that the T1 fragment starts at position Asp 264, whereas the T2 fragment begins at Ile 301 (Fig. 1 and 4), leading to calculated molecular masses for T1 and T2 of 51,301 and 47,176 kDa, respectively. The calculated mass value and amino acid sequence analysis indicate that trypsin cleavage also occurred at the C terminus of PBP 2a*, probably after Arg 678 for both the T1 and T2 fragments. This cleavage site location has been confirmed by mass spectrometry analysis of fragment T2 (41,236 ± 1.5 Da) (Fig. 1). Conditions limiting the production of the T2 fragment (trypsin/protein ratio of 1:100 [wt/wt]) were used for the remainder of this study.
The lipophilic character of PBP 2a* is due to its GT domain.
The requirement for detergent to recover and purify PBP 2a* from bacterial extracts shows that the molecule or a region of the molecule displays a hydrophobic character. Since the lipid II substrate for the GT activity is membrane bound, this raises the possibility that PBP 2a* is still membrane associated. To test the lipophilic character of functional domains of PBP 2a*, we have relied upon three independent biochemical approaches using full-length PBP 2a* and the TP domain obtained by limited proteolysis.
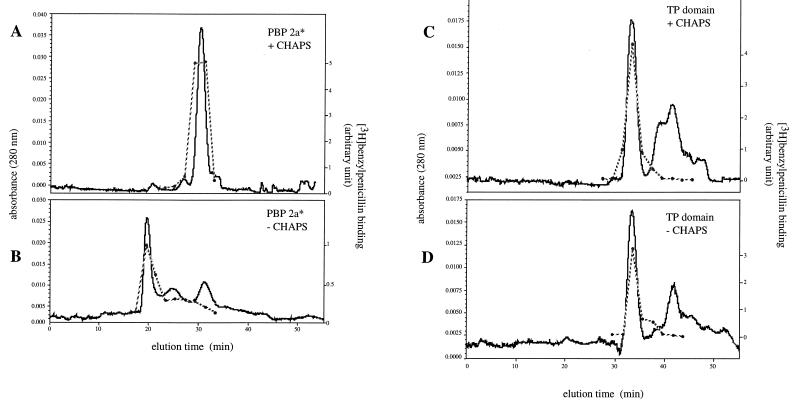
PBP 2a* and TP domain were prepared in the presence of 0.5% CHAPS and analyzed by gel filtration in either the presence or absence of the detergent. Protein fractions from each eluted peak were characterized with respect to their ability to bind to [3H]benzylpenicillin (Fig. 5). In the presence of CHAPS, PBP 2a* elutes as a protein with an apparent molecular mass of 116 kDa (Fig. 5A). This value is larger than the monomeric 72-kDa PBP 2a*, probably due to the formation of a complex with the detergent micelles. In the absence of CHAPS in the column equilibration buffer, PBP 2a* is excluded from the gel matrix (Fig. 5B). Therefore, PBP 2a* requires a detergent to remain soluble. The solubility of the TP domain is independent of the presence of the detergent (compare Fig. 5C and D). Under both conditions, TP protein peaks correspond to a 45- to 48-kDa species compatible with the 43-kDa measured mass for the T1 fragment (Fig. 4). Comparable profiles were obtained when the TP domain was extensively dialyzed to remove CHAPS prior to gel filtration analysis (data not shown). Peptide fragments lacking penicillin binding activity and eluting as a series of slowly migrating peaks (range, 3,600 to 7,600 Da), likely originate from the trypsin cleavage of the GT domain (Fig. 5C and D). Overall, these results indicate that the solubility of TP domain is detergent independent, while PBP 2a* requires CHAPS for its solubility.
FIG. 5.
Gel filtration of PBP 2a* and the TP domain in the presence or absence of 0.5% CHAPS. Chromatography conditions were described in Materials and Methods. Each peak aliquot was assayed for [3H]benzylpenicillin binding (dotted line). (A and B) PBP 2a*. (C and D) TP domain.
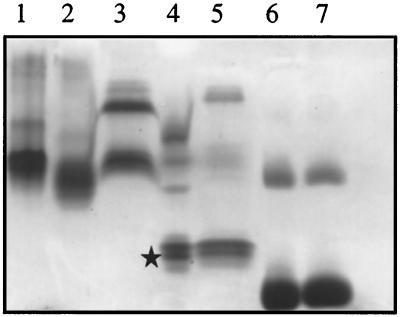
The detergent-binding property of PBP 2a* and the TP domain was assayed by using a modified version of the charge-shift electrophoresis technique described in reference 26. The migration of proteins that bind detergents is altered upon variation of the ionic composition of the detergents, whereas the electrophoretic mobility of soluble proteins is not altered by the presence of detergent. The zwitterionic detergent CHAPS present in the preparation of PBP 2a* and the TP domain was exchanged against Triton X-100 (a nonionic detergent) or a mixture of Triton X-100 and DOC (an anionic detergent) prior to analysis on a native polyacrylamide gel. The migration of PBP 2a* is extensively shifted in the presence of Triton X-100–DOC compared to the migration in the presence of Triton X-100 or CHAPS (compare lanes 1 to 3 in Fig. 6). Under identical experimental conditions, the migration of the TP domain remains unchanged, for the major species (compare Fig. 6, lanes 4 and 5), a behavior also found for the soluble protein, BSA, used as a control (Fig. 6, lanes 6 and 7). This experiment shows that the GT domain is required for the binding of detergents to PBP 2a*.
FIG. 6.
Charge-shift migration on native polyacrylamide gel. Proteins (5 μg) in the presence of 0.5% Triton X-100 or in a mixture of 0.5% Triton X-100–0.25% DOC were separated by native 8% PAGE and stained with Coomassie blue. Lanes: 1 to 3, PBP 2a* in 0.5% CHAPS, 0.5% Triton X-100, and 0.5% Triton X-100–0.25% DOC, respectively; 4, TP domain (star) in 0.5% Triton X-100; 5, TP domain in 0.5% Triton X-100–0.25% DOC; 6, BSA in 0.5% Triton X-100; 7, BSA in 0.5% Triton X-100–0.25% DOC.
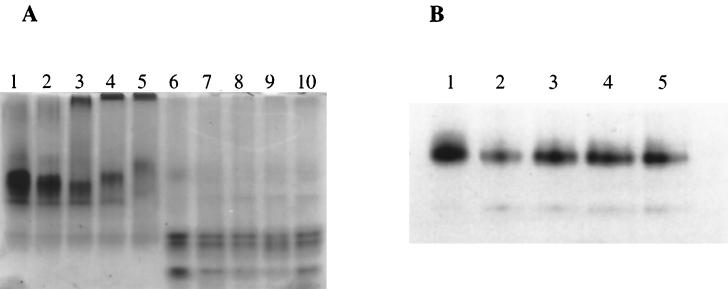
The requirement for detergent during solubilization of PBP 2a* implies that the molecule is membrane associated in S. pneumoniae. To further test the lipophilic character of PBP 2a*, we have compared the abilities of PBP 2a* and the TP domain to incorporate into lipid vesicles. The phospholipid DOPC was added at various protein/lipid molar ratios, and the mixtures were extensively dialyzed to remove the detergent. The efficiency of liposome formation under these experimental conditions was verified by electron microscopy (data not shown). PBP 2a* within reconstituted liposomes exhibited a modification of electrophoretic migration in a native gel which was correlated with the PBP 2a*/lipid molar ratios (Fig. 7A, lanes 3 to 5). As the relative lipid concentration increased, the migration of PBP 2a* became considerably and progressively retarded. The more slowly migrating species was absent when the dialysis was conducted in the absence of lipids (Fig. 7A, compare lane 2 with lanes 3 to 5); under these experimental conditions, PBP 2a* was not aggregated as it entered the gel (Fig. 7A, lane 2). This control shows that retarded migration does not originate from aggregation of PBP 2a* following removal of the detergent, but corresponds to a specific association of PBP 2a* with DOPC. Contrary to PBP 2a*, the electrophoretic behavior of the TP domain was not affected by the presence of lipid (Fig. 7A, lanes 6 to 10). This observation confirms the lipophilic character of the GT domain within PBP 2a*. The GT domain of PBP 2a* reconstituted in lipid vesicles was still sensitive to trypsin digestion (data not shown). This shows that the GT domain is not embedded in the vesicle bilayer but interacts with the lipids via a small region.
FIG. 7.
Reconstitution of proteins in lipid vesicles. Proteins were reconstituted into lipid vesicles as described in Materials and Methods. The vesicles were analyzed by native 8% PAGE. (A) PBP 2a* and the TP domain. Proteins (10 μg) were stained with Coomassie blue. Lanes: 1, PBP 2a* in 3% CHAPS; 2 to 5, PBP 2a* in the presence of molar protein/lipid ratios of 0, 1/50, 1/200, and 1/500, respectively; 6, TP in 3% CHAPS; 7 to 10, TP in the presence of molar protein/lipid ratios of 0, 1/50, 1/200, and 1/500, respectively. (B) PBP 2a* thrombin-digested product. Protein detection (0.7 μg) was performed by [3H]benzylpenicillin labelling. Lanes: 1, thrombin-digested PBP 2a* in 3% CHAPS; 2 to 5, thrombin-digested PBP 2a* in the presence of molar protein/lipid ratios of 0, 1/200, 1/500, and 1/1,000, respectively.
Extensive thrombin digestion of PBP 2a* (protease/PBP 2a* ratio of 1:1 [wt/wt]) releases a major 63-kDa species starting at Ser 156 according to N-terminal sequencing (Fig. 3, lane 5). This shorter form of PBP 2a* offered us the opportunity to delineate more accurately the lipophilic region of PBP 2a*. The ability of the 63-kDa species to bind lipids was monitored by native gel electrophoresis. The functional protein was revealed by [3H]benzylpenicillin labelling (Fig. 7B). The migration of the 63-kDa species was unchanged whether the protein was preincubated with lipids (Fig. 7B, lanes 3 to 5) or not (Fig. 7B, lanes 1 and 2). Thus the membrane association site in PBP 2a* is likely to reside between residues Lys 78 (start of the putative periplasmic region) and Ser 156, for a span of 78 amino acids (Fig. 1).
DISCUSSION
Recent genetic studies have suggested that S. pneumoniae PBP 2a is important in cell viability (12). Moreover, the enzyme plays a role as a cefotaxime resistance determinant (11). Even though this dual character identifies PBP 2a as a potential target for antibiotherapy, the molecule has not yet been studied from a biochemical and structural point of view. Therefore, we have expressed in E. coli cells, purified, and characterized the recombinant periplasmic region of PBP 2a (PBP 2a*), which encompasses the GT and TP domains.
The hydrolytic activity of PBP 2a* with the S2d thiolester is comparable to that of PBP 1a* (3) and PBP 2x from S. pneumoniae clinical isolate 328 (R-PBP 2x*), but is about 20-fold lower than the activity of the R6 S. pneumoniae strain (S-PBP 2x*) (18). This pattern of activity correlates well with the acylation efficiencies toward cefotaxime. Extensive incubation of PBP 2a* with [3H]benzylpenicillin leads to complete acylation of the protein. However, binding of benzylpenicillin to PBP 2a* was so inefficient, that we were unable to measure its acylation rate by stopped-flow fluorometry accurately. This behavior is reminiscent of that of R-PBP 2x* (18). Therefore, it is unlikely that PBP 2a participates in benzylpenicillin resistance. This is further supported by the observation that the PBP 2a sequences from penicillin-resistant clinical isolate 328 and susceptible strain R6 are identical (12). The involvement of PBP 2a in resistance to other β-lactams cannot be excluded, since the molecule was found to be a resistance determinant following selection with cefotaxime, a cephalosporin β-lactam (11).
The functional TP domain, which is the target for β-lactams, was delineated by limited proteolysis. This led to the identification of the PBP 2a* interdomain hinge region (linking the GT and TP domains), which is located in a region similar to that of S. pneumoniae PBP 1a* (3) (Fig. 1A). Insertion mutations in E. coli PBP 1b (16) were used to propose a boundary between the GT and TP domains, located 20 amino acids downstream from the N-terminal TP domains of S. pneumoniae PBP 1a and 2a (Fig. 1A). These results, obtained by different experimental approaches, strengthened the proposed GT/TP domain boundary localization in these three class A PBPs. In a recent review discussing the domain structure of PBPs (8), Goffin and Ghuysen localized the intermodule junction in motif 6 (Fig. 1A) by analogy with the S. pneumoniae PBP 2x structure (21). As shown in Fig. 1A, the N termini of the PBP 1a and PBP 2a TP domains, defined by trypsin digestion, are located between motifs 5 and 6.
The initial attempts to produce PBP 2a* in E. coli at 37°C led to a highly aggregated protein, which could not be recovered as a functional protein after solubilization with urea followed by in vitro refolding. Reduction of the temperature of the E. coli culture and solubilization of the PBP 2a* fusion with nondenaturing detergents allowed the recovery of a protein with TP activity. Treatment of insoluble PBP 2a* with a high salt concentration failed to solubilize the protein, an indication that hydrophobic interactions prevail over electrostatic interactions in PBP 2a* insolubility. Using three independent methods, we have mapped the region of association with detergents and DOPC lipid to the GT domain of PBP 2a*. We have localized the hydrophobic region in the 78 N-terminal residues of the GT domain, between Lys 78 and Ser 156. Remarkably, this finding is similar to that of class A PBP 1b from E. coli (20). Wang et al. (26) have located in E. coli PBP 1b a second membrane association site, distinct from the transmembrane stretch, within the first 163 amino acids of the GT domain, from position 88 (the start of the putative periplasmic domain) to position 251 (Fig. 1A). Gel filtration revealed that recombinant class A S. pneumoniae PBP 1a* forms small aggregates. This property is also linked to the GT domain, since the TP domain isolated by trypsin digestion remains monomeric (unpublished data). We have also reported that the presence of moenomycin, an amphiphilic GT-specific inhibitor, increased PBP 1a* solubility (3).
Taken together, these observations underline the general lipophilic properties of the GT domain of class A PBPs. Computational analyses of the GT sequence did not predict hydrophobic stretches or amphiphilic helices, which could account for the membrane association of the domain. This putative membrane association of class A PBPs GT domain is, however, consistent with the transmembrane location of its lipid II substrate.
ACKNOWLEDGMENTS
This work was supported by the Infectious Diseases Division, Lilly Research Laboratories.
We thank C. Vénien-Bryan for electron microscopy, J.-P. Andrieu for N-terminal sequence determination, and Y. Pétillot for mass spectroscopy analysis.
Footnotes
This is publication no. 630 of the Institut de Biologie Structurale Jean-Pierre Ebel.
REFERENCES
- 1.Adachi H, Ohta T, Matsuzawa H. A water-soluble form of penicillin-binding protein 2 of Escherichia coli constructed by site-directed mutagenesis. FEBS Lett. 1987;226:150–154. doi: 10.1016/0014-5793(87)80569-0. [DOI] [PubMed] [Google Scholar]
- 2.Coffey T J, Daniels M, McDougal L K, Dowson C G, Tenover F C, Spratt B G. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1995;39:1306–1313. doi: 10.1128/aac.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Guilmi A M, Mouz N, Andrieu J-P, Hoskins J, Jaskunas S R, Gagnon J, Dideberg O, Vernet T. Identification, purification, and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniae penicillin-binding protein 1a. J Bacteriol. 1998;180:5652–5659. doi: 10.1128/jb.180.21.5652-5659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira L C, Schwarz U, Keck W, Charlier P, Dideberg O, Ghuysen J M. Properties and crystallization of a genetically engineered, water-soluble derivative of penicillin-binding protein 5 of Escherichia coli K12. Eur J Biochem. 1988;171:11–16. doi: 10.1111/j.1432-1033.1988.tb13751.x. [DOI] [PubMed] [Google Scholar]
- 6.Fraipont C, Adam M, Nguyen-Disteche M, Keck W, Van Beeumen J, Ayala J A, Granier B, Hara H, Ghuysen J M. Engineering and overexpression of periplasmic forms of the penicillin-binding protein 3 of Escherichia coli. Biochem J. 1994;298:189–195. doi: 10.1042/bj2980189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghuysen J M. Molecular structures of penicillin-binding proteins and beta-lactamases. Trends Microbiol. 1994;2:372–380. doi: 10.1016/0966-842x(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 8.Goffin C, Ghuysen J-M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40:829–834. doi: 10.1128/aac.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakenbeck R, Briese T, Ellerbrok H, Laible G, Martin C, Metelmann C, Schier H-M, Tornette S. Targets of β-lactams in Streptococcus pneumoniae. In: Actor P, Daneo-Moore L, Higgins M L, Salton M R J, Shockman G D, editors. Antibiotic inhibition of bacterial cell surface assembly and function. Washington, D.C: American Society for Microbiology; 1988. pp. 390–399. [Google Scholar]
- 11.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoskins, J. A., G. Zhao, T. I. Meier, P. Matsushima, D. L. Mullen, J. Tang, T. I. Nicas, and R. S. Jaskunas. Unpublished observations.
- 13.Jamin M, Hakenbeck R, Frere J M. Penicillin binding protein 2x as a major contributor to intrinsic beta-lactam resistance of Streptococcus pneumoniae. FEBS Lett. 1993;331:101–104. doi: 10.1016/0014-5793(93)80305-e. [DOI] [PubMed] [Google Scholar]
- 14.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 15.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2002. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 16.Lefevre F, Rémy M H, Masson J-M. Topographical and functional investigation of Escherichia coli penicillin-binding protein 1b by alanine stretch scanning mutagenesis. J Bacteriol. 1997;179:4761–4767. doi: 10.1128/jb.179.15.4761-4767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin C, Sibold C, Hakenbeck R. Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 1992;11:3831–3836. doi: 10.1002/j.1460-2075.1992.tb05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouz N, Gordon E, Di Guilmi A M, Petit I, Petillot Y, Dupont Y, Hakenbeck R, Vernet T, Dideberg O. Identification of a structural determinant for resistance to beta-lactam antibiotics in Gram-positive bacteria. Proc Natl Acad Sci USA. 1998;95:13403–13406. doi: 10.1073/pnas.95.23.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 20.Nicholas R A, Lamson D R, Schultz D E. Penicillin-binding protein 1B from Escherichia coli contains a membrane association site in addition to its transmembrane anchor. J Biol Chem. 1993;268:5632–5641. [PubMed] [Google Scholar]
- 21.Pares S, Mouz N, Petillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 22.Reichmann P, Konig A, Marton A, Hakenbeck R. Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb Drug Resist. 1996;2:177–181. doi: 10.1089/mdr.1996.2.177. [DOI] [PubMed] [Google Scholar]
- 23.Roychoudhury S, Dotzlaf J E, Ghag S, Yeh W K. Purification, properties, and kinetics of enzymatic acylation with beta-lactams of soluble penicillin-binding protein 2a. A major factor in methicillin-resistant Staphylococcus aureus. J Biol Chem. 1994;269:12067–12073. [PubMed] [Google Scholar]
- 24.Siligardi G, Harris F, Phoenix D A. Alpha-helical conformation in the C-terminal anchoring domains of E. coli penicillin-binding proteins 4, 5 and 6. Biochim Biophys Acta. 1997;1329:278–284. doi: 10.1016/s0005-2736(97)00117-x. [DOI] [PubMed] [Google Scholar]
- 25.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang C C, Schultz D E, Nicholas R A. Localization of a putative second membrane association site in penicillin-binding protein 1B of Escherichia coli. Biochem J. 1996;316:149–156. doi: 10.1042/bj3160149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao G, Yeh W-K, Carnahan R H, Flokowitsch J, Meier T I, Alborn W E, Jr, Becker G W, Jaskunas S R. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J Bacteriol. 1997;179:4901–4908. doi: 10.1128/jb.179.15.4901-4908.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]