Abstract
Carotenoid cleavage dioxygenase (CCD) is the key enzyme for carotenoid cleavage, and the products of carotenoid cleavage regulate the ability of plants to stress. In this paper, six CCD genes were obtained from Morus notabilis (Mn) by reverse transcription-polymerase chain reaction (RT-PCR) and we classified them into three subgroups based on gene structures and phylogenetic analysis. The CDS (coding sequence) regions of the six MnCCD genes were 1617, 1620, 1635, 1713, 1746, and 1791 bp in full length, encoding 538, 539, 544, 570, 581, and 596 amino acids, respectively. Then, Pcold–TF-MnCCD plasmids were constructed and independently transferred into E. coli BL21 (DE3), and the MnCCD proteins were successfully expressed by prokaryotic expression with an expected molecular weight of recombinant proteins (∼120 kDa) and high solubility. These results will lay a foundation for the identification of mulberry carotenoid products.
1. Introduction
A carotenoid is a kind of pigment widely distributed in nature [1, 2]. Its cleavage products are also precursors for the synthesis of a variety of active substances, which play an important role in plant stress response, growth, and development [3, 4]. Carotenoid cleavage oxygenase (CCO) is the key enzyme for carotenoid cleavage, including two subfamilies of CCD and 9-cis epoxy carotenoid dioxygenase (NCED) [5]. In Arabidopsis, the CCO family contains a total of nine genes, five of which belong to the NCED subfamily and are involved in the formation of abscisic acid precursors; the other four (AtCCD1, AtCCD4, AtCCD7, and AtCCD8) belong to the CCD subfamily and are involved in the cleavage of various carotenoids [6].
The products of carotenoid cleavage dioxygenase involve multiple biological processes including light sensation and hormone signaling and also contribute to the production of compounds related to smell and color [7, 8]. Moreover, this activity is of great significance in the process of the plant stress response. Studies have shown that carotenoid cleavage products (apocarotenoids) can improve the symbiosis efficiency of mycorrhizal fungi (AM), and AM can coexist with plant roots, thereby improving plant absorption of water and mineral nutrients, reducing plant absorption of heavy metal ions (copper, chromium, etc.), and ultimately helping plants resist stress [9–11]. In Arbuscular mycorrhiza of wheat, corn, and barley, carotenoids were cleaved into C13 and C14 apocarotenoids, and these apocarotenoids can also increase the Mycorrhiza symbiosis efficiency [12, 13]. So far, the CCD genes were subsequently isolated in a variety of plants, such as maize, tobacco, pepper, and saccharum [14–17]. Moreover, the carotene cleavage pathway in many species has been studied very clearly, for example, melon, tomato, osmanthus, and morning glory [18–21]. Specifically, the cleavage substrate and action site of CCD1 in different plants are not the same, but their products are involved in the production of flavors and aromas commonly [22, 23]. CCD2 is currently only found in the crocus plant, Crocus sativus, and CsCCD2 cleaves zeaxanthin and finally generates crocetin dialdehyde [8], so that the stigma of saffron appears yellow, orange, and red [24]. CCD4 has different cleavage sites in different plants. For example, the CCD4 of potatoes cleaved all-trans-βcarotene to produce β-ionone (Market et al., 2015). Also, some plants such as CCD4 of citrus can cleave β-cryptoxanthin and zeaxanthin at the 7,8 (7′,8′) positions to synthesize β-orange pigment, whereas CCD7 and CCD8 are involved in the synthesis of strigolactone [25]. Even though extensive research has been performed on the function of CCD gene, little is known about the enzyme activity characteristics of CCD. The key to studying the characteristics of enzyme activity is to obtain the active CCD protein in vivo. Hence, this study aimed to successfully express the MnCCD proteins from the supernatant, that is, the soluble protein, which can lay a solid foundation for the subsequent enzymatic activity experiments. First, the cDNA of the six MnCCD genes were cloned from the young fresh leaves of mulberry by reverse transcriptase-polymerase chain reaction (RT-PCR). Furthermore, six MnCCD proteins were successfully obtained by prokaryotic expression.
2. Materials and Methods
2.1. Chemicals and Reagents
Phosphate-buffered solution (PBS) was obtained from Shanghai Biological Engineering Co. of China (Shanghai, China). The plant RNA extraction kit, reverse transcription kit, quantitative PCR kit, BCA protein assay kits, and His-tag Protein Purification Kit were purchased from Ruize Biological Technology Co. Ltd. (Nanning, China). Strains of Escherichia coli (E. coli) and Pcold–TF vector were kept in our laboratory.
2.2. Cloning of MnCCD Genes
CCD gene sequences from mulberry (Mn012507, Mn012508, Mn012509, Mn008739, Mn008741, Mn023189, GenBank ID: EXB32777.1, EXB32778.1, EXB32779.1, EXB58585.1, EXB58586.1, and EXC17835.1) were retrieved from the Mulberry Genome Database (https://morus.swu.edu.cn/morusdb/) and NCBI database (https://www.ncbi.nlm.nih.gov), and CCD genes in other plants (GenBank ID: NP_189064.1, NP_188062.1, NP_193569.1, NP_177960.1, etc.) were retrieved from the NCBI database. Multiple sequence alignment was performed with the ClustalX2.1 software. Quantitative PCR and full-length primers for six MnCCD genes were designed using Premier 5.0 (Table 1). Total RNAs were extracted from roots and leaves of mulberry using a plant RNA extraction kit according to the manufacturer's instructions. First-strand cDNA was produced using the reverse transcription kit and used as a template for PCR amplification. The amplification programme used for PCR is as follows: 95°C for 5 min, 32 cycles, 95°C for 30 s, 60°C for 30 s, 72°C for 90 s, and 72°C for 10 min. The products were separated on 1.0% agarose gels and cloned into the pMD19-T vector (TaKaRa, Dalian, China) to verify the sequence. The bioinformatic analysis tools were used with reference to the method of Liu et al. [26]. Putative N-terminal chloroplast transit and conservative domain predictions of MnCCDs were analyzed using the ProtParam tool (https://web.expasy.org/protparam/), ChloroP 1.1 Server (https://www.cbs.dtu.dk/services/ChloroP/), and conserved domains available online (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). A schematic diagram of the gene structure of MnCCDs was drawn using the Exon–Intron Graphic Maker (https://www.wormweb.org/exonintron).
Table 1.
Primers used in this study.
| Gene | Primer (5′-3′) | Used for |
|---|---|---|
| MnCCD1A | ATGGCGGAAGTGGCGTC | RT-PCR |
| GATATTCAGTTTGAGTCCGAGTA | ||
|
| ||
| MnCCD1A | atggagctcggtaccATGGCGGAAGTGGCGTC | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcGATATTCAGTTTGAGTCCGAGTA | ||
|
| ||
| MnCCD1B1 | ATGGCCGAAATAGTGGATGTGAAT | RT-PCR |
| TAGGACACCATTCCCAACATCAA | ||
|
| ||
| MnCCD1B1 | atggagctcggtaccATGGCCGAAATAGTGGATGTGAAT | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcTAGGACACCATTCCCAACATCAA | ||
|
| ||
| MnCCD1B2 | GTGGGACTGTTCAAGGC | RT-PCR |
| CTCTGCTGGGCAAATC | ||
|
| ||
| MnCCD1B2 | atggagctcggtaccATGGCGGAGAAGCCGAGC | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcGAGTTTTGCTTGTTCTTGCAGTTG | ||
|
| ||
| MnCCD4 | ATGGCTTCCTCTTTTTTGGCAC | RT-PCR |
| TCATGCCTGGTGAACCAAGTCC | ||
|
| ||
| MnCCD4 | atggagctcggtaccATGGCTTCCTCTTTTTTGGCAC | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcTCATGCCTGGTGAACCAAGTCC | ||
|
| ||
| Mn008739 | ATGCCACATCACGTCTCCAA | RT-PCR |
| CACCGAGACTGGTATGAAAGCT | ||
|
| ||
| Mn008739 | atggagctcggtaccATGCCACATCACGTCTCCAA | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcCACCGAGACTGGTATGAAAGCT | ||
|
| ||
| Mn008741 | ATGGCATCATCGTATATGGCAT | RT-PCR |
| TGGTGAGATTGGTATGAAAGCT | ||
|
| ||
| Mn008741 | atggagctcggtaccATGGCATCATCGTATATGGCAT | Prokaryotic expression vector construction |
| caggtcgacaagcttgaattcTGGTGAGATTGGTATGAAAGCT | ||
Lowercase letters represent the homology arms containing Kpn I or ECOR I restriction sites.
2.3. Expression and Purification of Recombinant MnCCD Proteins
The coding region of the six MnCCD genes containing the homologous sequence of Pcold–TF vector was cloned with the correct sequenced plasmid as a template. Then, the Pcold–TF vector and the target gene fragment were connected by homologous recombination. The correctly sequenced Pcold–TF-MnCCD plasmids were independently transferred into E. coli BL21 (DE3) and grown to OD about 0.6 for recombinant protein expression.The recombinant protein was expressed and purified with reference to the method of Liu et al. [26].
3. Results
3.1. Isolation of CCDs from M. notabilis
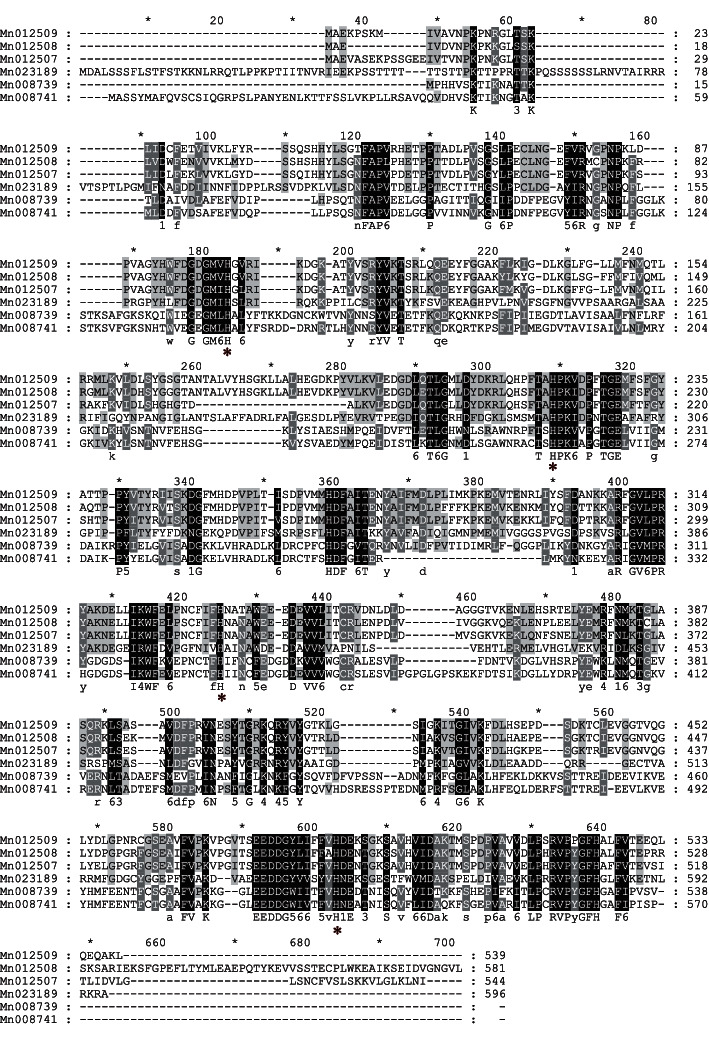
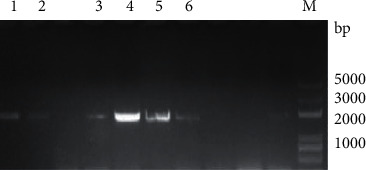
Six CCD genes were obtained in mulberry, and the six cloned MnCCD genes were Mn008739, Mn008741, Mn012507, Mn012508, Mn012509, and Mn023189 and the open reading frames of the six genes were 1617, 1620, 1635, 1713, 1746, and 1791 bp (Figure 1), encoding proteins of 538, 539, 544, 570, 581, and 596 amino acids, respectively (Figure 2).
Figure 2.
PCR amplification of MnCCD gene. M: 5000 bp marker. (1) Mn023189, (2) Mn008741, (3) Mn012508, (4) Mn012509, (6) Mn008739, and (7) Mn012507.
3.2. Bioinformatic Analysis of MnCCDs
The protein sequences of MnCCD proteins were compared and analyzed with reported CCD protein sequences from other species using ClustalX2.1 local software (Figure 2). Then, the related phylogenetic tree was obtained through the software MEGA5 (Figure 3). According to the evolutionary tree, plant carotenoid cleavage dioxygenases were divided into five subfamilies, CCD1, CCD2, CCD4, CCD7, and CCD8. The CCDs in plants were named according to the genetic relationship and homology of Arabidopsis CCDs (AtCCDs). From the phylogenetic tree, it can be seen that Mn012507, Mn012508, and Mn012509 are closely related to AtCCD1, so they were named MnCCD1A, MnCCD1B1, and MnCCD1B2, respectively. Mn023189 has the closest relationship with AtCCD4, so it was named MnCCD4. However, Mn008739, Mn008741, PaCCD1 (Populus alba), and JmCCD1 (Juglans microcarpa) are grouped together separately.
Figure 3.
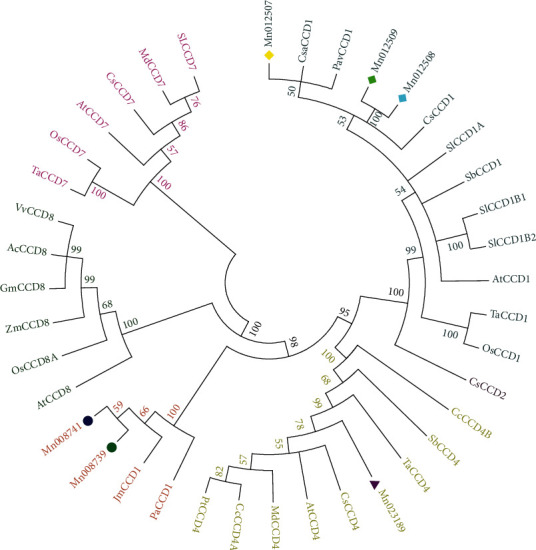
Phylogenetic tree analysis of MnCCDs with the other species. Circle symbols of different colors represent the carotenoid cleavage dioxygenases (CCDs) of mulberry (MnCCDs), and the letter with the same color means the same subfamily. At, Arabidopsis thaliana; Ta, Triticum aestivumLinn; Cs, Camellia sinensis; Os, Oryza sativa; Mn, Morus notabilis Schneid.; Sb, Sorghum bicolor; Cs, Cannabis sativa; Pt, Populus trichocarpa; Pav, Prunus avium; Sl, Solanum lycopersicum; Pa, Populus alba; Jm, Juglans microcarpa; Zm, Zea mays; GenBank numbers: AtCCD1(), AtCCD4(), AtCCD7(), AtCCD8(), TaCCD4 (QEX50885.1), TaCCD1 (QEX50880.1), TaCCD7 (ASW22783.1), SlCCD1A (NP_001234542.1), SlCCD1B1 (NP_001233838.1), SlCCD1B2 (NP_001310512.1), CsCCD1 (AYK03324.1), CsCCD4 (AYK03325.1), SlCCD1A (NP_001234542.1), SlCCD1B1 (NP_001233838.1), SlCCD1B2 (NP_001310512.1), SbCCD1 (AGN03859.1), SbCCD4 (AGN03860.1), CcCCD4B (ABC26012.1), CcCCD4A (ABC26011.1), MdCCD4 (ABY47995.1), MdCCD7 (AUZ82805.1), OsCCD7 (XP_015637185.1), OsCCD1 (ABA99624.2), OsCCD8A (sp|Q93VD5.1), CsCCD1 (XP_030493365.1), PtCCD4 (XP_024457659.1), PavCCD1 (XP_021820758.1), PaCCD1 (XP_034890793.1), JmCCD1 (XP_041018063.1) Mn012507 (EXB32777.1), Mn012508(EXB32778.1), Mn012509 (EXB32779.1), Mn008739 (EXB58585.1), Mn008741 (EXB58586.1), Mn023189 (EXC17835.1).
Mulberry CCD belongs to the RPE65 superfamily and the proteins of the MnCCD subfamily do not contain signal peptides or chloroplast transit peptides (except MnCCD4 and Mn008741), Notably, only MnCCD4 is localized in the chloroplast, the rest of the MnCCDs were localized in the cytoplasm, which may be related to their functional differentiation (Figure 4.).
Figure 4.
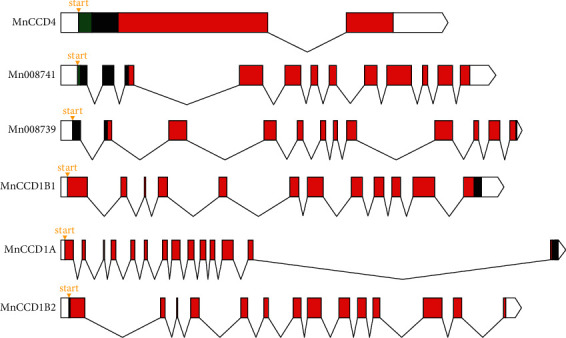
Abridged general view of the MnCCD gene structure. Orange triangle: transcription start site; white rectangles: 5′UTR; green rectangles: chloroplast transit peptide; red rectangles: RPE65 domain; black broken lines: introns; white polygons: 3′UTR.
3.3. Heterologous Expression and Purification of MnCCDs
There was a heavily expressed band at ∼50 kDa on SDS-PAGE in the control group and this is consistent with the predicted molecular weight of the tagged protein. However, the sample groups showed a heavily expressed new band at ∼120 kDa on SDS-PAGE and consistent with the expected molecular weight of the recombinant protein (Figure 5(a)). This result indicates the recombinant protein has been successfully expressed. Moreover, the six MnCCD crude protein samples were purified by nickel affinity chromatography and the eluted fractions were detected by SDS-PAGE electrophoresis and the positions of the purified six target proteins in 300 mM imidazole eluate were consistent with the predicted recombinant MnCCD proteins after Coomassie brilliant blue staining. Notably, there was only one obvious band in the sample group and the molecular weight was ∼120 kDa (Figure 5(b)). This shows that we have successfully purified the active target proteins from the supernatant.
Figure 5.
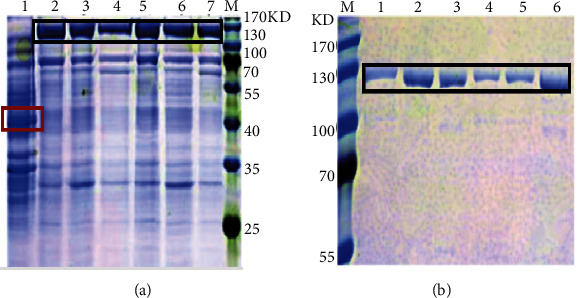
SDS-PAGE analysis of MnCCD proteins expressed in E. coli BL21 (DE3) cells and their purification. M: 180 kDa protein marker. (a) Lane 1: control, lanes 2–7: Mn008741, MnCCD1B1, MnCCD1A, MnCCD4, MnCCD1B2, and Mn008739; (b) lanes 1–6: the purified MnCCD proteins. Target protein bands are marked with black boxes, and the tagged protein band is marked with a red box.
4. Discussion
The number of CCD genes varies in different species. For example, 12, 11, and 10 CCD genes were identified in Pyrus bretschneideri, Fragaria vesca, and Prunus persica, respectively, and the phylogenetic tree clustered these genes into five branches CCD1, CCD4, CCD7, CCD8, and CCD-like [27]. Even in different varieties of the same species, the classification of CCD genes is different. In the case of sugarcane, S. spontaneum has 38 CCD genes and they were divided into seven groups, while the CCDs from R570 were 11 and were classified into five groups, missing CCD4 and CCD7in R570 compared to S. spontaneum [28]. In this study, the phylogenetic tree showed that the candidate MnCCDs clustered into three subfamilies: MnCCD1, MnCCD4, and CCD1-like (Mn008739 and Mn008741). Mn008739 and Mn008741 were clustered with PaCCD1 and JmCCD1, but they were independent of CCD1 of other plants, so Mn008739 and Mn008741 were named MnCCD1-like, as CCD7 and CCD8 were lost in Morus notabilis, this may be related to incomplete genomic information.
Eukaryotic and prokaryotic expression systems are commonly used to express recombinant proteins. Regardless of the expression system, the choice of vector and host cell is critical. Prokaryotic systems are more widely used because of their ability to obtain large quantities of recombinant proteins in a short period of time. The system is mainly based on E. coli, although Bacillus species are increasingly used [29]. Usually, different expression strains can be selected according to different needs. For example, Rosetta (DE3) PLySs is a highly stringent expression strain that can control expression levels and provide tRNAs with rare codons. Moreover, plasmid expression vectors include promoters, multiple cloning sites, terminators, replicons, signal peptides, fusion tags, and selectable markers. According to these characteristics of the vector, there are a variety of plasmids to choose from. Common prokaryotic expression vectors include pBAD, pET, POW3.0, and an expression vector for expressing GST fusion protein. Pet can express foreign proteins at a high level in host bacteria such as E.coli BL21 (DE3). Since the expressed proteins contain calmodulin-binding polypeptides and thrombin cleavage points, they are widely used in the purification industry. In this study, the pET32a vector was chosen as the expression vector, while the six recombinant MnCCD proteins did not express in the supernatant, it only expressed in a large amount in the precipitate, that is, it expressed in the form of inclusion bodies. Afterwards, we optimized the experimental conditions, and the situation remained the same. Finally, we reselected a vector, Pcold–TF, which provides cold-shock inducibility and triggers the expression of factors to improve the correct folding of the protein, thereby enabling soluble expression of recombinant proteins. Fortunately, we successfully expressed the six recombinant MnCCD proteins in the supernatant.
5. Conclusion
We identified six CCD genes from mulberry. Furthermore, we successfully obtained the MnCCD recombinant proteins by prokaryotic expression. The results showed that all MnCCD genes including CCD1-like genes could encode active carotenoid cleavage dioxygenases. These results lay the foundation for the analysis of the cleavage pathway of mulberry carotenoids.
Figure 1.
Multiple sequence alignment of six deduced MnCCD sequences. Conserved histidine residues are marked with a red asterisk.
Acknowledgments
This research was supported by the “High-Yield, High-Quality, and High-Resistance Mulberry Variety Selection and Research on Key Surting Technologies” Guangxi Science and Technology Major Special Project, grant number (Guike AA19182012-2) and China Agriculture Research System of MOF and MARA.
Contributor Information
Dan Liu, Email: 1106961605@qq.com.
Qiang Lin, Email: gxlq67@swu.edu.cn.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Q.L. and D.L. conceived and designed experiments. D.L., C.Q., Y.Z., C.Z., T.L., G.Z., and J.H. performed the experiments. D.L., C.Q., Y.Z., C.Z., T.L., G.Z., and J.H. analyzed the data. D.L. and Y.Z. wrote the paper.
References
- 1.Hou X., Rivers J., León P., Ryan P. M., Barry J. P. Synthesis and function of apocarotenoid signals in plants. Trends in Plant Science . 2016;21(9) doi: 10.1016/j.tplants.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Ureshino K., Nakayama M., Miyajima I. Contribution made by the carotenoid cleavage dioxygenase 4 gene to yellow colour fade in azalea petals. Euphytica . 2016;207(2):401–417. doi: 10.1007/s10681-015-1557-2. [DOI] [Google Scholar]
- 3.Iuchi S., Kobayashi M., Taji T., et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in arabidopsis. Plant Journal for Cell and Molecular Biology . 2010;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnson J. Do carotenoids serve as transmembrane radical channels? Free Radical Biology and Medicine . 2009;47(3):321–323. doi: 10.1016/j.freeradbiomed.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Ohmiya A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnology . 2009;26(4):351–358. doi: 10.5511/plantbiotechnology.26.351. [DOI] [Google Scholar]
- 6.Auldridge M., Block A., Vogel J., et al. Characterization of three members of the arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. The Plant Journal . 2006;45(6):982–993. doi: 10.1111/j.1365-313x.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 7.Huang F., Horváth G., Molnár P., et al. Substrate promiscuity of RdCCD1, a carotenoid cleavage oxygenase from rosa damascena. Phytochemistry . 2009;70(4):457–464. doi: 10.1016/j.phytochem.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Frusciante S., Diretto G., Bruno M., et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proceedings of the National Academy of Sciences . 2014;111(33):12246–12251. doi: 10.1073/pnas.1404629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui L., Guo H., Li W., et al. Study on the characteristics of mycorrhizal colonization in Chinese fir plantations at different ages. Sheng Tai Xue Bao . 2019;39:1926–1934. doi: 10.5846/stxb201809202061. [DOI] [Google Scholar]
- 10.Smith S., Smith F. A., Jakobsen I. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiology . 2003;133(1):16–20. doi: 10.1104/pp.103.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michael H. W., Daniela S., Hans J., Fester T., Strack D. Apocarotenoid biosynthesis in arbuscular mycorrhizal roots: contributions from methylerythritol phosphate pathway isogenes and tools for its manipulation. ChemInform . 2007;68(1):130–138. doi: 10.1016/j.phytochem.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 12.Akiyama K., Matsuzaki K., Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature . 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 13.Walter M., Floß D., Hans J., Fester T., Strack D. Apocarotenoid biosynthesis in arbuscular mycorrhizal roots: contributions from methylerythritol phosphate pathway isogenes and tools for its manipulation. Phytochemistry . 2007;68(1):130–138. doi: 10.1016/j.phytochem.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z., Hans J., Walter M., et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta . 2008;228(5):789–801. doi: 10.1007/s00425-008-0781-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Q., Li Q., Li P., et al. Carotenoid cleavage dioxygenases: identification, expression, and evolutionary analysis of this gene family in tobacco. International Journal of Molecular Sciences . 2019;20(22):p. 5796. doi: 10.3390/ijms20225796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan X., Liu G. Genome-wide identification, phylogenetic relationships, and expression analysis of the carotenoid cleavage oxygenase gene family in pepper. Genetics & Molecular Research Gmr . 2016;15(4) doi: 10.4238/gmr.15048695. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X., Jia D. M., Duan M.-Z., et al. Genome-wide identification and expression profiling of the carotenoid cleavage dioxygenase (CCD) gene family in brassica napus L. PLoS One . 2020;15(9) doi: 10.1371/journal.pone.0238179.e0238179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibdah M., Azulay Y., Portnoy V., et al. Functional characterization of CmCCD1, a carotenoid cleavage dioxygenase from melon. Phytochemistry . 2006;67(15):1579–1589. doi: 10.1016/j.phytochem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Simkin A., Underwood B. A., Auldridge M., et al. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of petunia flowers. Plant Physiology . 2004;136(3):3504–3514. doi: 10.1104/pp.104.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldermann S., Kato M., Kurosawa M., et al. Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of osmanthus fragrans lour. Journal of Experimental Botany . 2010;61(11):2967–2977. doi: 10.1093/jxb/erq123. [DOI] [PubMed] [Google Scholar]
- 21.Simkin A., Schwartz S., Auldridge M., Taylor M. G., Klee H. J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. The Plant Journal . 2004;40(6):882–892. doi: 10.1111/j.1365-313x.2004.02263.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu S., Tian N., Liu Z., Huang J. N., Li J. Cloning and characterization of a carotenoid cleavage dioxygenase from artemisia annua L. Applied Mechanics and Materials . 2011;108:274–281. doi: 10.4028/www.scientific.net/amm.108.274. [DOI] [Google Scholar]
- 23.Kato M., Matsumoto H., Ikoma Y. The role of carotenoid cleavage dioxygenases in the regulation of carotenoid profiles during maturation in citrus fruit. Journal of Experimental Botany . 2006;57(10):2153–2164. doi: 10.1093/jxb/erj172. [DOI] [PubMed] [Google Scholar]
- 24.Ahrazem O., Rubio-Moraga A., Argando A-Picazo J. Intron retention and rhythmic diel pattern regulation of carotenoid cleavage dioxygenase 2 during crocetin biosynthesis in saffron. Plant Molecular Biology . 2016;91:1–20. doi: 10.1007/s11103-016-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikihisa U., Atsushi H., Satoko Y. Inhibition of shoot branching by new terpenoid plant hormones. Nature . 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 26.Liu D., Meng S., Xiang Z., He N., Yang G. Antimicrobial mechanism of reaction products of morus notabilis (mulberry) polyphenol oxidases and chlorogenic acid. Phytochemistry . 2019;163:1–10. doi: 10.1016/j.phytochem.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J., Li J., Zhang J., et al. Genome-wide identification and expression analysis of the carotenoid cleavage oxygenase gene family in five rosaceae species. Plant Molecular Biology Reporter . 2021;1194:1–13. [Google Scholar]
- 28.Su W., Zhang C., Feng J., et al. Genome-wide identification, characterization and expression analysis of the carotenoid cleavage oxygenase (CCO) gene family in saccharum. Plant Physiology and Biochemistry . 2021;162:196–210. doi: 10.1016/j.plaphy.2021.02.041. [DOI] [PubMed] [Google Scholar]
- 29.Porowinska D., Wujak M., Roszek K., Michał K. Prokariotyczne systemy ekspresyjne. Postepy Hig Med Dosw (online) . 2013;67:119–129. doi: 10.5604/17322693.1038351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


