Abstract
Streptococcus pneumoniae Rx1 is capable of repairing lesions caused by DNA-damaging agents in an error-free manner but lacks a UV-inducible error-prone repair system due to the absence of chromosomally encoded UmuDC-like proteins. We have identified an operon-like structure 8 kb from the left end of the pneumococcal conjugative transposon Tn5252 that confers SOS function in the host cells. DNA sequence analysis of this region revealed the presence of four open reading frames (ORFs). The deduced amino acid sequence of one of them, ORF13, which is capable of encoding a protein of 49.7 kDa, showed significant homology to UmuC, MucB, and other proteins involved in the SOS response. The carboxy-terminal region of another, ORF14, which is predicted to encode a 26-kDa polypeptide, shared similarity with UmuD- and MucA-like proteins that carry the amino acid residues recognized by the activated RecA* protein for proteolytic cleavage. The presence of plasmids carrying subcloned DNA from this region was found to restore UV-inducible mutagenic repair of chromosomal DNA in Escherichia coli cells defective in error-prone repair as well as in pneumococcus and Enterococcus faecalis UV202. Mutations within ORF13 abolished UV-induced mutagenesis but did not affect the conjugal transposition of the element.
Biological mechanisms to repair UV-damaged DNA are widespread among many, if not all, organisms (25). Escherichia coli is the best-studied model prokaryote to elucidate the various pathways leading to the repair of the genetic material and survival of the cells (25). Besides nucleotide excision repair involving the products of the uvrA, uvrB, and uvrC genes, error-prone repair in association with the umuDC gene products is also known to occur (9). The latter process, the SOS response that results in the increase of replication errors, is triggered by the activated RecA* protein, which facilitates the autocleavage of the UmuD protein to yield the active UmuD′ C-terminal fragment as with the LexA repressor (3). The processed UmuD′C complex is thought to bind to DNA polymerases, altering the fidelity of nucleotide incorporation at abasic sites leading to increased mutagenesis (25). The human pathogen Streptococcus pneumoniae also carries the genes involved in the excision repair of UV-damaged DNA (6, 16). However, the SOS response, which results in mutagenic repair of UV-damaged DNA, has been shown to be absent in this organism (6).
We have been studying the biology of the conjugative transposon Tn5252, which was originally detected in the chromosome of S. pneumoniae BM6001 (1). It is a 47-kb element carrying a chloramphenicol resistance determinant and seems to have a propensity to accumulate a variety of heterologous DNA segments and disseminate them into a number of streptococcal species (1). The element is capable of self-transfer, and it site specifically integrates in the genome of the host cells (30). The entire transposon has been cloned in fragments in E. coli, and a detailed physical map has been obtained (31). Mutational, DNA sequencing, and functional analyses of the genes and their products revealed the clustering of many of the transfer-related genes at the terminal regions of the element (10). However, about 20 kb of DNA in the central region of Tn5252 seem apparently devoid of transfer functions. DNA sequencing analysis showed that the deduced amino acid sequence of two of the four open reading frames (ORFs) within an operon-like structure at the left terminal region of the transposon showed significant similarities to UmuDC homologs from a number of gram-positive and gram-negative bacteria. Here we present our studies relating to the genetic and physical characterization of this operon, which confers resistance to UV light by error-prone repair.
MATERIALS AND METHODS
Bacterial strains, media, transformation, and conjugation.
Bacterial strains and plasmids used in this study are listed in Table 1. The growth of streptococcal cultures, conjugation, competence regimen, and plating techniques have been described previously (1). Recombinant plasmids were generated in recombination-deficient E. coli JM109 by transformation in accordance with the method of Hanahan (8). Bacterial cells carrying pAT29 or derivatives were grown in media containing spectinomycin (150 μg/ml). The antibiotics used for UV-induced mutagenesis were optochin (5 μg/ml) and fusidic acid (20 μg/ml). For scoring of streptococcal transconjugants, optochin (20 μg/ml), erythromycin (5 μg/ml), and streptomycin (200 μg/ml) were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. pneumoniae | ||
| Rx1 | Nonencapsulated laboratory strain | 1 |
| DP1110 | UVs | W. R. Guild |
| SP1000 | str-1 fus Tn5252 | 1 |
| SP1254 | str-1 Tn5252ΩEm Tra+ | 10 |
| SP1291 | str-1 Tn5252ΩEm | This study |
| SP1311 | str-1 derivative of DP1110 | This study |
| SP1317 | SP1311(pSJ142 Spcr) | This study |
| SP1323 | SP1311(pAT29) | This study |
| SP1324 | SP1311(pSJ142Δ0.5-kb XbaI fragment) | This study |
| SP1402 | SP1311::Tn5252 | This study |
| SP1405 | SP1311::Tn5252ΩEm | This study |
| S. pyogenes | ||
| ATCC 19615 | opt | Stillwater Medical Center |
| E. faecalis | ||
| JH2-2 | Wild type, plasmid free | 32 |
| UV202 | fus rif UVs | 32 |
| SF5002 | UV202::Tn5252 | 1 |
| SF5004 | UV202(pSJ142, Spcr) | This study |
| E. coli | ||
| JM109 | F′ lacIq Δ(lacZ)M15 proA+B+/Δ(lac-proAB) recA1 | 33 |
| AB1157 | his-4 | 11 |
| RM1140 | Same as AB1157 but umuC36 | R. V. Miller |
| Plasmids | ||
| pAT29 (6.7 kb) | lacZ Spcr | 29 |
| pDR6 | KS(+)::4.5-kb DNA fragment (uvr operon of Tn5252) | This study |
| pKM101 | R46 derivative carrying the mucAB operon | 17 |
| pSJ142 | pAT29::4.5-kb fragment DNA (uvr operon of Tn5252) | This study |
| pSE117 | pBR322 derivative::E. coli umuCD operon | 15 |
| pVA891 (5.9 kb) | Cm Em | 14 |
DNA procedures.
DNA restriction and modification enzymes were used according to the recommendations of the suppliers. Chromosomal DNAs from streptococci were isolated as described previously (1). Plasmid DNA from E. coli was prepared by using the Wizard Prep kit (Promega). DNA hybridizations were done by the method of Southern (27) with a GeneScreen Plus membrane (Du Pont, New England Nuclear) as support.
DNA sequencing and computer analysis of DNA sequences.
Relevant DNA fragments were subcloned into the pBluescript plasmid vectors SK(+) and KS(+) (Stratagene). Apart from the universal primers, synthetic oligonucleotide primers used in DNA sequencing were made at the Oklahoma State University Core Facility. The dideoxy chain termination DNA sequencing was performed at the Core Facility by using a 373 Strech automatic sequencer (Perkin-Elmer, Applied Biosystems Division) with fluorescent chain terminators. Sequence assembly and analyses were performed by using MacVector 3.5 software.
UV irradiation.
Cells were grown to early exponential phase (approximately 108 CFU/ml) in an appropriate broth at 37°C and harvested by centrifugation at 5,000 × g in a Sorvall RC-5B centrifuge (DuPont Instruments). The cells were washed twice and resuspended in prechilled phosphate-buffered saline buffer containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (pH 7.4). Three milliliters of the cell suspension was placed in a sterile plastic petri dish (60 by 15 mm) and exposed to various doses of UV radiation from a germicidal bulb (model XX-15F; Spectronics Corporation) with a peak emission of 256 nm. Doses of UV radiation were determined with a UVX radiometer (Ultra Violet Products). Survivors were counted by plating appropriate dilutions of cells on agar plates. All manipulations and incubations subsequent to UV irradiation were carried out in the dark or under amber light in order to minimize photoreactivation.
UV-induced mutagenesis.
Appropriate dilutions of E. coli cells were plated immediately following UV irradiation on minimal medium plating lacking histidine to score for revertants. UV-treated S. pneumoniae cells were diluted 20-fold in CATPG medium (1), grown to 2 × 108 CFU/ml at 37°C in the dark, and plated on appropriate selective plates to score for survivors and drug-resistant mutants. When needed, the cells were stored at −80°C in 10% glycerol. The following day, the cells were thawed in an ice water bath, diluted, and plated.
Nucleotide sequence accession number.
The sequence reported here was assigned GenBank accession no. L29324.
RESULTS
Features of the nucleotide sequence.
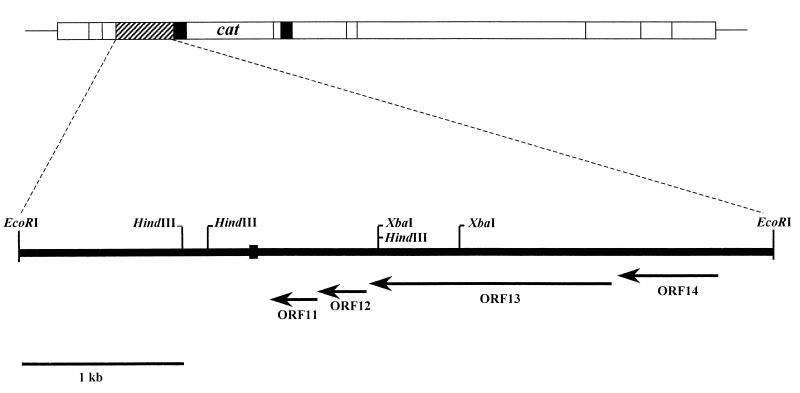
Previously, it had been shown that the genes encoding DNA processing functions, such as site-specific recombination and DNA relaxation during conjugal transfer of Tn5252, were present at the left terminal region of the element (10). To identify other possible transfer-related genes nearby, the DNA sequence of a 3.3-kb fragment of DNA on the right side of the DNA relaxase (28) was determined. For DNA sequencing, a 4.55-kb EcoRI fragment from this region was cloned into pSK+ to create pDR6. Fig. 1 shows the organization of the region in Tn5252 reported here. The G+C content of the sequenced region was 33.6%. The sequence revealed the presence of four ORFs, ORF14, ORF13, ORF12, and ORF11, transcribed in the same orientation as a single unit with a putative ribosome-binding site (RBS) placed upstream of each at the appropriate distance. A gram-positive consensus promoter-like sequence (7) was noted about 140 bases upstream of the translational start site of ORF14. Twenty-one bases upstream of the −35 region, a 14-bp inverted imperfect repeat (12 of 14) was present. In the RBS region of ORF14, another inverted imperfect repeat (17 of 21) with a ΔG value of −14.7 kcal/mol was located. ORF12 and ORF11, which have the capacity to encode proteins with molecular masses of 11 and 11.7 kDa, respectively, were found to overlap by eight bases. ORF14 and ORF13 encoded proteins with molecular masses of 26 and 49.7 kDa, respectively.
FIG. 1.
Restriction map and the predicted gene organization of the uvr operon of the 47.5-kb Tn5252. The EcoRI site at the right end is about 8.5 kb from the left end of the element. Relevant restriction sites are shown. Thin line, chromosomal DNA; box, transposon DNA; crosshatched box, the 4.5-kb DNA containing the uvr genes; black boxes, direct repeats of insertion sequence-like sequences. The location of the cat is shown. The vertical lines in the transposon DNA indicate EcoRI sites. Subclones derived from the 4.5-kb EcoRI fragment of DNA shown in the lower panel with relevant restriction endonuclease sites and a nested set of deletion derivatives obtained following exonuclease III and S1 treatments were used to determine the sequence data from both the strands. The directions and lengths of the potential ORFs are shown at bottom.
The amino acid sequences of ORF12 and ORF11 did not display significant similarity to any proteins in the GenBank database. However, the amino acid sequence of ORF13 showed highly significant similarity to proteins involved in the SOS response in gram-positive and gram-negative bacteria (Fig. 2). These included the UmuC of E. coli and Salmonella typhimurium (25, 26); MucB produced by the plasmid pKM101 (17); their homologs in Bacillus subtilis (34); and the UvrA protein encoded by pAD1, a pheromone-responsive conjugative plasmid, from Enterococcus faecalis (19). UmuC and MucB are proteins encoded by the umuDC and mucAB operons, respectively, which form part of the SOS regulon.
FIG. 2.
Multiple sequence alignment of the predicted product from ORF13 and its homologs ORFU from Lactococcus lactis plasmid pNP10 (5), uvrA of E. faecalis (19), UV-damage repair protein of B. subtilis (accession no. Z99115), rumB of IncJ plasmid R391 from E. coli (12), samB from S. typhimurium (18), mucB of R46 plasmid from S. typhimurium (17), and the chromosomal umuC from S. typhimurium (26) and E. coli (20). Conserved amino acids are shaded in black and conservative substitutions are shaded in gray.
Comparison of the amino acid sequence of ORF14 revealed a high level of similarity to a variety of transcriptional regulators of phages and gram-positive bacteria. In particular, the similarity was significant around the three conserved domains involved in the RecA-mediated cleavage of many of these proteins (20). Several other residues conserved in the LexA family of proteins and phage repressors (2) were also present in ORF14. The product of the gene frp, which is predicted to be the regulator of the fructosyltransferase expression in Streptococcus mutans (24), showed the highest level of similarity to ORF14 (66% identity; 75% similarity). The similarity was very pronounced among the C-terminal residues of the proteins.
UV-induced killing.
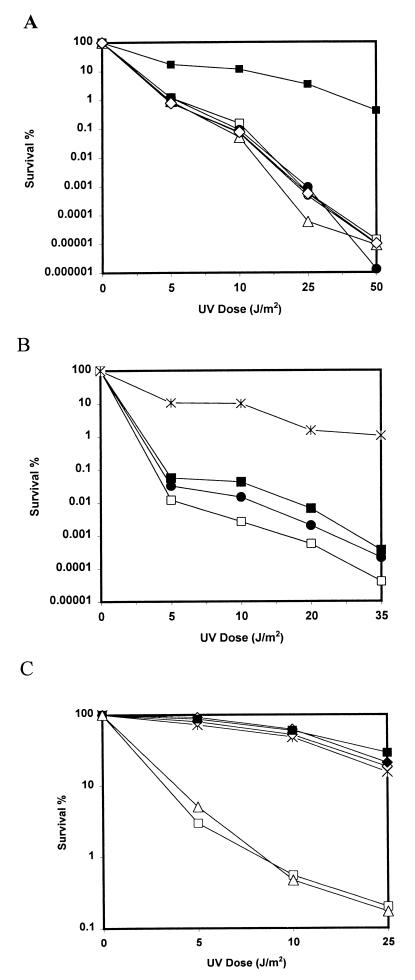
To determine whether the operon carrying ORF13 was involved in the enhancement of survival of the host cell upon UV treatment, the 4.55-kb EcoRI fragment carrying this region was cloned into the Streptococcus-E. coli shuttle multicopy plasmid pAT29 to create pSJ142. The recombinant plasmid was introduced into E. faecalis UV202 cells by electroporation and into S. pneumoniae SP1311 (UVs) and E. coli AB1157 and RM1140 via transformation. Comparison of UV sensitivity levels at 20 J/m2 consistently showed a twofold increase in UV protection of SF5002 carrying Tn5252 with a single copy of ORF13, indicating the involvement of the element in the protection. At this dose, UV resistance increased about 10-fold in SF5004 cells carrying pSJ142 compared to that of the host cells without the plasmid (Fig. 3b), even though at lower doses this level of protection was less evident. On the other hand, in the highly UV-sensitive pneumococcal strain SP1311, the presence of pSJ142 conferred about 3,000-fold protection, whereas no significant protection was detectable when ORF13 was present as a single copy within Tn5252 as a part of the chromosome (Fig. 3A). When a 0.5-kb XbaI fragment internal to ORF13 was deleted from within pSJ142, the UV survival disappeared, indicating a definitive role for ORF13 in the observed protection. Similar results were observed with E. coli host cells, indicating that the streptococcal UV-resistant genes could complement the function in gram-negative bacteria (Fig. 3C).
FIG. 3.
Effect of UV irradiation on survival of three organisms. (A) S. pneumoniae strains: □, SP1311; ■, SP1317; ○, SP1402; ●, SP1405; ▵, SP1323; ◊, SP1324. (B) E. faecalis strains: ×, JH2-2; □, UV202; ●, SF5002; ■, SF5004. (C) E. coli strains: ×, AB1157; □, RM1140; ◊, RM1140(pSE117); ⧫, RM1140(pKM101); ■, RM1140(pSJ142); ▵, RM1140(pAT29). The UV survival curves were obtained as indicated in Materials and Methods, and the numbers are averages obtained from at least two independent experiments.
SOS response.
Even though pneumococci are capable of dark repair of UV-damaged DNA (6, 16), the distinct lack of an SOS system involving error-prone repair has been documented (6). To determine whether the operon containing ORF13 and conferring UV resistance was due to error-prone polymerization of DNA, pneumococcal cells carrying pSJ142 were exposed to various doses of UV and screened for the appearance of mutations in various genetic loci. The results are given in Table 2. For both the markers (resistance to optochin and to fusidic acid) a significant number of mutants appeared compared to the cells not exposed to UV. For reasons that are not clear, the number of optochin-resistant mutants obtained was 20-fold higher than those of the fusidic acid resistance.
TABLE 2.
Effect of the uvr operon of Tn5252 on UV-induced mutagenesis
| Strain | UV dose (J/m2) | % Survival | No. of cells per 108 survivors carrying the indicated mutationa
|
||
|---|---|---|---|---|---|
| Fusr | Optr | His+ | |||
| S. pneumoniae | |||||
| SP1311 | 0 | 100 | 0 | 0 | |
| 10 | 0.152 | 50 | 0 | ||
| 25 | 0.00059 | 40 | 40 | ||
| SP1317 | 0 | 100 | 20 | 30 | |
| 10 | 11.56 | 310 | 4,420 | ||
| 25 | 3.4 | 6,220 | 113,000 | ||
| E. coli | |||||
| AB1157 | 0 | 100 | 0 | ||
| 5 | 72 | 18 | |||
| 10 | 48 | 52 | |||
| 25 | 15.5 | 189 | |||
| RM1140 | 0 | 100 | 0 | ||
| 5 | 3 | 0 | |||
| 10 | 0.55 | 0 | |||
| 25 | 0.2 | 0 | |||
| RM1140(pSE117) | 0 | 100 | 0 | ||
| 5 | 80 | 40 | |||
| 10 | 52 | 187 | |||
| 25 | 18.5 | 310 | |||
| RM1140(pKM101) | 0 | 100 | 0 | ||
| 5 | 92 | 302 | |||
| 10 | 62 | 706 | |||
| 25 | 21 | 880 | |||
| RM1140(pSJ142) | 0 | 100 | 0 | ||
| 5 | 87 | 160 | |||
| 10 | 60 | 264 | |||
| 25 | 29 | 950 | |||
| RM1140(pAT29) | 0 | 100 | 0 | ||
| 5 | 5.1 | 0 | |||
| 10 | 0.47 | 0 | |||
| 25 | 0.17 | 0 | |||
Fusr, resistance to fusidic acid; Optr, resistance to optochin; His+, histidine prototrophic revertants.
To determine whether the uvr operon of Tn5252 could complement the SOS function in E. coli, cells carrying pSJ142 were screened for his+ revertants following various levels of UV treatment on minimal medium plates lacking histidine. E. coli AB1157 yielded a substantial number of mutants compared to RM1140 carrying the umuC36 mutations. As expected, SOS function was restored in RM1140 upon the introduction of multicopy plasmids carrying either the native umuDC operon (in pSE117 [15]) or the mucAB operon (in pKM101 [17]). Interestingly, the level of UV mutagenesis was highest with RM1140 cells carrying pSJ142, whereas no revertants among the cells carrying the vector plasmid alone were scored.
The role of ORF13 in the conjugal transfer of Tn5252.
To assess the relevance of ORF13 in the conjugal transfer of the element, a pneumococcal mutant strain was created. The E. coli plasmid pVA891 carries a streptococcal erythromycin resistance (Emr) determinant that is expressed in pneumococci when the plasmid is integrated into the chromosome (14). The 0.5-kb XbaI fragment in ORF13 in pSJ142 was replaced with XbaI-digested pVA891. The resulting ligated molecule was linearized with PstI that cleaves at the vector pAT29, which is part of pSJ142. The digested DNA was introduced into competent SP1000 cells carrying Tn5252 (1). Due to the flanking homology provided by ORF13 DNA, the heterologous pVA891 was expected to be inserted in this homology-dependent event (10).
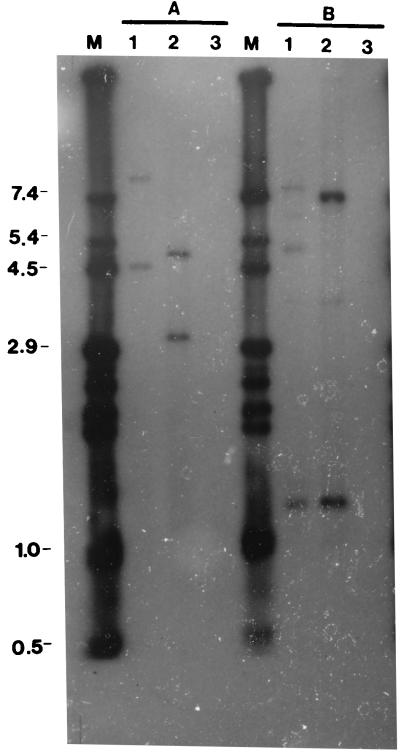
Chromosomal DNA from one of the resulting Emr transformants, SP1291, was analyzed by Southern hybridization using pDR6 as a probe to determine whether the insertion had taken place. The probe did not react with wild-type Rx1 cells, which did not carry Tn5252 (Fig. 4). Due to the presence of a single ClaI site within this region, the probe was expected to react with two fragments of 3.2 and 5 kb in SP1000 (Rx1::Tn5252). As expected, the probe reacted with two ClaI fragments of these sizes. Also, the probe reacted with three HindIII fragments of SP1000 DNA of 6.84, 1.34, and 0.98 kb, the latter appearing more clearly in the original autoradiogram. Replacement of the 0.5-kb XbaI fragment with pVA891 in SP1291 was expected to result in the probe hybridizing to two (4.5- and 9.2-kb or 3.4- and 10.3-kb) ClaI fragments and three (1.34-, 5-, and 8.1- or 1.34-, 2.8-, and 10.3-kb) HindIII fragments depending upon the orientation of the insert. The probe reacted with fragments of the former group in each case, indicating the absence of any unexpected rearrangements.
FIG. 4.
Physical analysis of Emr transformants carrying the insertion of pVA891 within ORF13 in Tn5252. Autoradiogram showing Southern hybridization of 32P-labeled pDR6 to ClaI- (A) and HindIII- (B) digested chromosomal DNA from SP1291 (SP1000 carrying a deletion within ORF13) (lane 1), SP1000 (Rx1::Tn5252) (lane 2), and S. pneumoniae Rx1 (lane 3). The indicated sizes correspond to the standards in lane M, which consist of a set of calibrated fragments from pSK(+) or derivative plasmids, all of which react with the probe.
SP1291 cells were used as donors in filter-mating experiments with S. pneumoniae and Streptococcus pyogenes recipient cells to determine the role of ORF13 in conjugation. SP1254 (10) carrying pVA891 inserted at a different locus that does not carry any transfer-related function in the element served as the control. The results (not shown) indicated that there was no significant difference in the transfer frequency of Tn5252 from SP1291 compared to that from SP1254, indicating that ORF13 did not play any significant role in the conjugal transfer of the element.
DISCUSSION
We have determined the nucleotide sequence of a 3.3-kb DNA fragment at the right end of the DNA relaxase gene, the product of which is thought to be involved in the site-specific nicking of the circularized transposon molecule prior to its conjugal transfer. The sequence data revealed the presence of an operon-like region containing consensus promoter-independent and rho-independent transcription terminator-like sequences. Four ORFs were found, two of which were significantly similar to the proteins involved in error-prone repair of UV-damaged DNA. Homology alignment of ORF13 protein of Tn5252 with UmuC homologs from other systems indicated that, while the proteins originating from gram-negative bacteria were more similar to each other, those from gram-positive sources formed a distinct group, indicating the evolutionary divergence of the two types. Plasmids carrying this segment of transposon DNA were able to confer UV-induced mutagenic response and survival following exposure to UV in gram-positive as well as gram-negative bacteria devoid of error-prone repair capability. Deletion of a 0.5-kb DNA fragment from within ORF13 led to the abolition of the observed SOS mutagenic response, demonstrating the involvement of this region of DNA in the repair of UV-damaged genetic material by introduction of mutations. This is the first demonstration of UV-induced SOS response and mutability in pneumococci.
Two ORFs, encoding proteins of about 46 and 15 kDa, have been noted in the most-studied operons of gram-negative bacteria, umu and muc, whereas four ORFs are present within the SOS operon of Tn5252. The role(s), if any, of the products of ORF11 and ORF12 in SOS mutagenesis remains unknown at present. The imp operon of the IncI plasmid TP110 (13) has also been shown to carry an ORF capable of encoding a 9.5-kDa protein with unknown function in addition to the UmuD and MucB homologs. Further studies should establish the role(s) of these proteins in SOS response in pneumococcus.
According to the current model for SOS response, drawn mostly from studies in E. coli, the activated RecA* protein stimulates the autocleavage of the LexA as well as the UmuD to UmuD′. By a mechanism not currently understood, the UmuCD′2 complex then enables the DNA polymerase to continue through the DNA lesions in an error-prone manner (25). The finding that the SOS-related gene products of Tn5252 could be processed in such a way to restore the UV-induced mutability in E. coli cells demonstrates that the structural and mechanistic details of this class of proteins are probably conserved in a wide range of bacterial species. It was also intriguing that the highest similarity to the ORF14 protein, the homolog of UmuD, was the repressor of the fructosyltransferase gene of S. mutans which carries the conserved residues found in the LexA family of proteins and has been implicated as a virulence factor in the development of dental caries.
Surprisingly, plasmids are rare in clinical isolates of S. pneumoniae even though they have the capacity to receive and maintain plasmids from other streptococci via transformation under laboratory conditions. The recent emergence and dissemination in pneumococci of resistance to multiple antibiotics have been chiefly due to the conjugative transposons (4, 22, 23). These novel elements seem to have functionally replaced plasmids in this species. The chloramphenicol resistance determinant in Tn5252 has been shown to be flanked by direct repeats of an insertion sequence-like element (21) which very often leads to its spontaneous “curing” (21). The remaining cryptic element is transfer proficient (1, 21). The ability of Tn5252-like elements to persist under natural conditions even in the absence of antibiotic selection is probably due to other genes not related to the conjugal transfer which may enhance the survival of their hosts. The presence of genes involved in the SOS response in Tn5252 is an example supporting this notion.
ACKNOWLEDGMENTS
We are grateful to Robert V. Miller for providing E. coli AB1157 and RM1140 and the plasmids pSE117 and pKM101 and for his critical reading of the manuscript.
This work was supported by grant MCB9417052 from the National Science Foundation to M.N.V.
REFERENCES
- 1.Ayoubi P, Kilic A O, Vijayakumar M N. Tn5253, the pneumococcal (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol. 1991;173:1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J D, Davidson B E, Hillier A J. Identification of prophage genes expressed in lysogens of the Lactococcus lactis bacteriophage BK5-T. Appl Environ Microbiol. 1995;61:4099–4104. doi: 10.1128/aem.61.11.4099-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis in Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clewell D B, Flannagan S E. The conjugative transposons of gram-positive bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 369–393. [Google Scholar]
- 5.Garvey P, Fitzgerald G F, Hill C. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl Environ Microbiol. 1995;61:4321–4328. doi: 10.1128/aem.61.12.4321-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasc A M, Sicard N, Claverys J P, Sicard A M. Lack of SOS repair in Streptococcus pneumoniae. Mutat Res. 1980;70:157–165. doi: 10.1016/0027-5107(80)90155-4. [DOI] [PubMed] [Google Scholar]
- 7.Graves M C, Rabinowitz C. In vivo and in vitro transcription of the Clostridium pasteurianum ferridoxin gene. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 10.Kiliç A O, Vijayakumar M N, Al-Khaldi S F. Identification and nucleotide sequence analysis of a transfer-related region in the streptococcal conjugative transposon Tn5252. J Bacteriol. 1994;176:5145–5150. doi: 10.1128/jb.176.16.5145-5150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kokjohn T A, Miller R V. Characterization of the Pseudomonas aeruginosa recA analog and its protein product: rec-102 is a mutant allele of the P. aeruginosa PAO recA gene. J Bacteriol. 1987;169:1499–1508. doi: 10.1128/jb.169.4.1499-1508.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulaeva O I, Wooten J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodwick D, Owen D, Strike P. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res. 1990;18:5045–5050. doi: 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macrina F L, Evans R P, Tobian J A, Hartley D L, Clewell D B, Jones K R. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene. 1983;25:145–150. doi: 10.1016/0378-1119(83)90176-2. [DOI] [PubMed] [Google Scholar]
- 15.Marsh L, Walker G C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162:155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin B, Garcia P, Castanie M-P, Glise B, Claverys J-P. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls an SOS regulon. In: Ferretti J J, Gilmore M S, Klaenhammer T R, Brown F, editors. Genetics of streptococci, enterococci, and lactococci. Basel, Switzerland: International Association of Biological Standardization, Karger Press; 1994. pp. 293–300. [PubMed] [Google Scholar]
- 17.McCann J, Spingarn N E, Kobori J, Ames B N. Detection of carcinogens as mutagens: bacterial tester strains with R-factor plasmids. Proc Natl Acad Sci USA. 1975;72:979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nohmi T, Hakura A, Nakai Y, Watanabe M, Murayama S Y, Sofuni T. Salmonella typhimurium has two homologous but different umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J Bacteriol. 1991;173:1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozawa Y, Tanimoto K, Fujimoto S, Tomita H, Ike Y. Cloning and genetic analysis of the UV resistance determinant (uvr) encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pAD1. J Bacteriol. 1997;179:7468–7475. doi: 10.1128/jb.179.23.7468-7475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry K L, Elledge S J, Mitchell B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priebe S D. Genetic and physical mapping of antibiotic resistance elements in the chromosome of Streptococcus pneumoniae. Ph.D. thesis. Durham, N.C: Duke University; 1986. [Google Scholar]
- 22.Salyers A A, Shoemaker N B, Guthrie E P. Recent advances in Bacteroides genetics. Crit Rev Microbiol. 1987;14:49–71. doi: 10.3109/10408418709104435. [DOI] [PubMed] [Google Scholar]
- 23.Scott J R. Conjugative transposons. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 597–614. [Google Scholar]
- 24.Shibata Y, Kuramitsu H. Identification of the Streptococcus mutans frp gene as a potential regulator of fructosyltransferase expression. FEMS Microbiol Lett. 1996;140:49–54. doi: 10.1111/j.1574-6968.1996.tb08313.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C M, Koch W H, Franklin S B, Foster P L, Cebula T A, Eisenstadt E. Sequence analysis and mapping of the Salmonella typhimurium LT2 umuDC operon. J Bacteriol. 1990;172:4964–4978. doi: 10.1128/jb.172.9.4964-4978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:502–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Srinivas P, Kilic A O, Vijayakumar M N. Site-specific nicking in vitro at oriT by the DNA relaxase of Tn5252. Plasmid. 1997;37:42–50. doi: 10.1006/plas.1996.1268. [DOI] [PubMed] [Google Scholar]
- 29.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijayakumar M N, Ayalew S. Nucleotide sequence analysis of the termini and chromosomal locus involved in the site-specific integration of the streptococcal conjugative transposon Tn5252. J Bacteriol. 1992;175:2713–2719. doi: 10.1128/jb.175.9.2713-2719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijayakumar M N, Priebe S D, Guild W R. Structure of a conjugative element in Streptococcus pneumoniae. J Bacteriol. 1986;166:978–984. doi: 10.1128/jb.166.3.978-984.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yagi Y, Clewell D B. Recombination-deficient mutant of Streptococcus faecalis. J Bacteriol. 1980;143:966–970. doi: 10.1128/jb.143.2.966-970.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 34.Yasbin R E, Cheo D, Bol D. DNA repair systems. In: Sonenshein L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C: American Society for Microbiology; 1993. pp. 529–537. [Google Scholar]