Abstract
Chromatin insulators are DNA-protein complexes that can prevent the spread of repressive chromatin and block communication between enhancers and promoters to regulate gene expression. In Drosophila, the gypsy chromatin insulator complex consists of three core proteins: CP190, Su(Hw), and Mod(mdg4)67.2. These factors concentrate at nuclear foci termed insulator bodies, and changes in insulator body localization have been observed in mutants defective for insulator function. Here, we identified NURF301/E(bx), a nucleosome remodeling factor, as a novel regulator of gypsy insulator body localization through a high-throughput RNAi imaging screen. NURF301 promotes gypsy-dependent insulator barrier activity and physically interacts with gypsy insulator proteins. Using ChIP-seq, we found that NURF301 co-localizes with insulator proteins genome-wide, and NURF301 promotes chromatin association of Su(Hw) and CP190 at gypsy insulator binding sites. These effects correlate with NURF301-dependent nucleosome repositioning. At the same time, CP190 and Su(Hw) both facilitate recruitment of NURF301 to chromatin. Finally, Oligopaint FISH combined with immunofluorescence revealed that NURF301 promotes 3D contact between insulator bodies and gypsy insulator DNA binding sites, and NURF301 is required for proper nuclear positioning of gypsy binding sites. Our data provide new insights into how a nucleosome remodeling factor and insulator proteins cooperatively contribute to nuclear organization.
INTRODUCTION
In eukaryotes, genomic information is efficiently packaged in three-dimensional (3D) space to allow for proper gene expression and DNA replication. At the smallest scale, the genome is organized into 147 bases of DNA wrapped around nucleosomes and further arranged into kilobases to megabases of higher-order structures of loops, topologically associating domains (TADs), compartments, and chromosome territories (reviewed in Jerkovic et al., 2000; Rowley and Corces, 2018) (1,2). This 3D genome organization facilitates the interaction between distant DNA elements and is needed for accurate regulation of gene expression.
Insulators can function as chromatin boundaries between active and inactive chromatin and facilitate chromatin looping through insulator protein-DNA interactions. Insulators can also stabilize contacts between distant regulatory elements or block the communication of enhancers with promoters through loop formation (3). In vertebrates, CTC binding factor (CTCF) is the only factor currently known that possesses insulator activity (reviewed in Chen and Lei, 2019) (4). In contrast to vertebrates, multiple DNA-binding proteins in Drosophila have been shown to exhibit insulator activity, and each of these proteins recruits the universal insulator protein Centrosomal protein 190 (CP190) to chromatin (5–10). CP190 and several of its DNA-binding interactors, such as CTCF, BEAF-32, M1BP, and ZIPIC, frequently localize to gene promoters and are enriched at TAD borders (11–14). However, the Su(Hw) class of insulator complexes (herein referred to as the gypsy insulator) is mainly situated distal to both gene promoters and TAD borders (12,13).
The gypsy insulator complex consists of three core proteins, including Su(Hw), Mod(mdg4)67.2 and CP190. Su(Hw) harbors a cluster of 12 zinc fingers (15) and dictates the DNA-binding specificity of the gypsy insulator complex. Both CP190 and Mod(mdg4)67.2 are recruited by Su(Hw) to chromatin, and CP190 and Mod(mdg4)67.2 can stabilize chromatin association of Su(Hw) (6,16). CP190 and Mod(mdg4)67.2 both contain BTB/POZ domains, which can mediate homotypic and heterotypic protein-protein interactions including multimerization. Several associated factors have been identified that affect the function of the gypsy insulator, including E(y)2, dTopors, Rm62, Shep, Rump, CLAMP, HIPP1, and M1BP (17–24), but more interactors likely remain to be discovered and may provide further insight into insulator function.
Core gypsy insulator proteins colocalize at large nuclear foci known as insulator bodies in interphase cells. Insulator bodies can be induced or enlarged by certain stress conditions, such as osmostress (25). It has been proposed that insulator proteins may accumulate in insulator bodies to facilitate subsequent assembly on chromatin, and this presumptive depot could protect insulator proteins from degradation and/or facilitate quick response to stimulus (25–28). Similar to other visually defined structures, the functional significance of insulator bodies is not fully understood. However, changes in insulator body formation or number have been observed in mutants defective for gypsy insulator function (6,17-19,21,22,24,29).
NURF (Nucleosome Remodeling Factor) is a chromatin remodeling complex that requires ATP to mediate nucleosome sliding. By changing the accessibility of DNA, NURF can regulate the recruitment of transcription factors or other proteins to chromatin to regulate gene expression (30–33). Drosophila NURF has four subunits, including the catalytic subunit ATPase ISWI (34), a WD-40 repeat protein NURF55 (35), an inorganic pyrophosphatase NURF38 (36), and NURF301 (37). NURF301 is the largest subunit, and it is highly conserved with human bromodomain and PHD finger transcription factor (BPTF) (30,37,38). In Nurf301 null mutant flies, the +1 and nucleosomes up to 1.2 kb from the 5’ end drift toward transcription start sites (TSSs) of active genes (39). The specificity of NURF301 activity appears to be dictated by physical interaction with transcription factors (eg. GAF, EcR, and Ken repressor), which can recruit NURF301 to chromatin (30,37,40,41). With respect to chromatin insulator complexes, NURF301 has been shown to physically interact with CP190, colocalize with it on chromatin (42), and promote gypsy enhancer-blocking activity (39,43). NURF301 is also required for enhancer-blocking activity of the CP190-dependent SF1, Fab-7, and Fab-8 insulators (42,43). However, it is not known whether NURF301 affects CP190 or other insulator protein recruitment to chromatin or whether it is involved in insulator-dependent 3D nuclear organization.
In this study, we first performed a high-content RNAi imaging screen for proper gypsy insulator body localization in a cultured cell line and identified the NURF301 chromatin remodeling factor as one of the top hits. We found that NURF301 interacts physically with the gypsy insulator complex and promotes gypsy insulator barrier activity. ChIP-seq analysis revealed that NURF301 overlaps extensively with insulator proteins throughout the genome, particularly with CP190. Depletion of NURF301 results in reduced Su(Hw) and CP190 chromatin association with gypsy binding sites, and altered insulator protein binding correlates with changes in nucleosome positioning in Nurf301 mutant cells. Furthermore, depletion of CP190 or Su(Hw) also disrupts recruitment of NURF301 to chromatin. Consistent with loss of Su(Hw) and CP190 chromatin association with gypsy binding sites, 3D contact and overlap between insulator bodies and specific gypsy binding site DNA are also significantly decreased. Moreover, the overall nuclear volume occupied by gypsy binding sites increases as does their intermingling with non-gypsy binding DNA sequences on the same chromosome after depletion of NURF301. Our findings identify a cooperative relationship between a nucleosome remodeling factor and insulator proteins with respect to chromatin binding, promotion of chromatin insulator activity, and maintenance of nuclear organization.
MATERIALS AND METHODS
Drosophila strains
Fly lines were maintained on standard cornmeal medium at 25°C. We used lines expressing dsRNA against su(Hw) (10724 GD) and Nurf301 (CG32346 GD) from the Vienna Drosophila RNAi Center. Act5C-Gal4, Mef2-Gal4, and l(3)31–1-Gal4 driver lines were obtained from the Bloomington Drosophila Stock Center. UAS-luciferase constructs were inserted into the attP3 landing site using phiC31 site-specific integration (44). Protein extracts from second instar larvae were used for western blotting.
Cell lines
Kc167 cells were grown in CCM3 media (Thermo Scientific HyClone). Cells were maintained in monolayer at 25°C. Gateway cloning was performed to produce the pAWG-mod(mdg4)67.2 vector, which contains the EGFP gene fused to the C-terminus of mod(mdg4)67.2 cDNA. EcoRV (NEB) and SphI (NEB) were used to remove the Act5C promoter. The Ubiquitin promoter was amplified from the pBKS-Ubi-hygro vector (primers are in Supplementary Table S5), and this product was ligated into the digested pAWG-mod(mdg4)67.2 vector. After co-transfection with pCoBlast vector (ThermoFisher), 10 μg/ml blasticidin was used to select stably integrated Mod(mdg4)67.2-GFP cells. Individual clones were selected using limiting serial dilution into untransfected feeder cells, and one selected line was used for screening and subsequent analyses.
Immunofluorescence
Cells were centrifuged at 500 × g for 5 min and washed once in PBS. 1 × 105 cells were applied to poly-l-lysine-coated slides (Tekdon Inc.) at RT for 10 min and then fixed in 4% paraformaldehyde (PFA) in PBS at RT for 10 min. For glass slides subjected to cold stress, slides were transferred to a 4°C cold room for 30 min before fixation. For slides subjected to osmotic stress, 250 mM NaCl was added for 15 min before fixation. We tested different time points on both slides and 384-well plates to determine the most appropriate time for cold shock. Optimal cold shock times were 30 min for slides and 2.5 h for 384-well microplates.
Cells were washed twice with PBS, permeabilized with 0.5% Triton X-100 for 10 min, and washed again. Cells were blocked in 5% bovine serum albumin in PBST (0.1% Triton X-100 in PBS) for 30 min at RT. After blocking, cells were incubated with primary antibody for 1 h at RT and then washed 3 times with PBST, following the incubation of secondary antibodies for 1 h at RT and washed again. Finally, cells were stained in 2 μg/ml DAPI (Molecular Probes) prepared in PBS and mounted using ProLong Diamond (Life Technologies).
RNAi screening
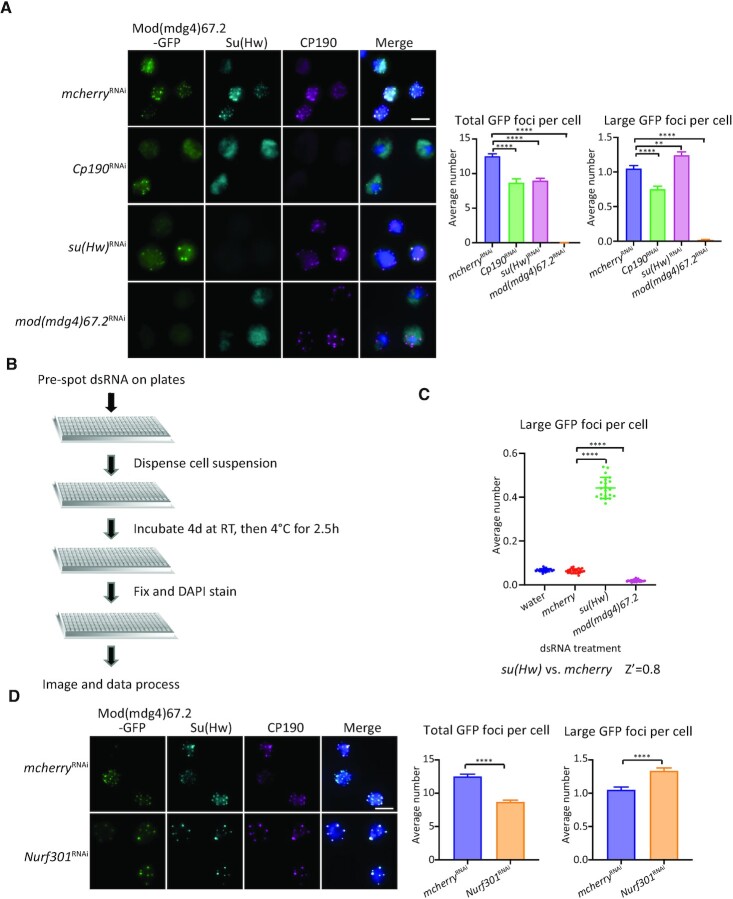
The transcription factor (TF) sub-library (DRSC 2.0) was purchased from the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School. It is comprised of 9 × 384-well plates, containing 993 genes. At least two unique dsRNAs target each gene in independent wells. Cp190, su(Hw), and mod(mdg4)67.2 are also in the TF library. In our screen, we used su(Hw) dsRNA knockdown as a positive control, mcherry knockdown as a negative control, and mod(mdg4)67.2 knockdown as a control for knockdown efficiency. Six wells of each 384-well library plate were filled with mcherry, su(Hw) or mod(mdg4)67.2 dsRNA (0.25 μg/5 μl/well), or water, using an Echo525 acoustic liquid dispenser (Beckman Coulter). Then, 1.0 × 105 Mod(mdg4)67.2-GFP cells in 40 μl were dispensed into 384-well plates using a Multidrop Dispenser (ThermoFisher). Cells were soaked with dsRNA at 25°C for 4 days. Plates were then cold-shocked by placing them in a 4°C room for 2.5 h before fixation. 40 μl of 8% paraformaldehyde (PFA, Electron Microscopy Sciences) was added to the plates using the Bluewasher washer/dispenser (Blue Cat Bio) and fixed for 10 min. After 2× washes in PBS, cells were stained with 2 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) for 10 min and sealed with Alumaseal before storage at 4°C. Plates were imaged on a CellVoyager CV7000 (Yokogawa) confocal spinning disk high-throughput microscope, and images were processed using Columbus software (PekinElmer). Parameters for large foci were determined manually before applying machine learning in Columbus. Well-level data from two biological replicates of the screen were analyzed using the cellHTS2 R package (45) (V2.44.0). Z’ score was used to optimize parameters for high-throughput screening. Z’ is a statistical measure for assay quality. By calculating the standard deviation from the mean of the control data, it shows the separation between the positive and negative controls to evaluate overall assay quality. A maximum of 1.0 indicates an ideal assay, and >0.5 indicates a good, reliable assay (46). Z scores for each RNAi treatment in the library were calculated to rank them based on their phenotypic effect measured as the number of large insulator bodies. The morphology of insulator bodies in cells treated and fixed in 384-well plates is different from those attached to glass slides. On slides, foci are much more distinct, and there is more diffuse GFP signal in 384-well plates after cold stress. The observed differences are likely due to different efficiencies of heat transfer for plastic versus glass materials. We observed Z score values of large insulator bodies also for RNAi treatments (such as PHDP, SMR, Ets65A, and L(1)10Bb) that caused cytotoxic or cytostatic effects, as measured by a smaller Z score value for the number of cells per well (Supplementary Tables S1 and S2). These dsRNA treatments were not considered bona fide hits in the phenotypic screen and were de-prioritized for downstream validation.
DsRNA knockdowns
DsRNA primers against NURF301 were designed based on recommendations from the DRSC. Templates were PCR-amplified from cDNA of Kc cells using primers containing the T7 promoter sequence (listed in Supplementary Table S5). DsRNAs were synthesized using the MEGAscript T7 in vitro transcription kit (Ambion) using PCR templates and then purified on NucAway spin columns (ThermoFisher). 7 × 106 Kc cells were transfected with 5 μg of dsRNA against Nurf301, Cp190, su(Hw), or mod(mdg4)67.2 or mcherry (control) using Amaxa cell line Nucleofector kit V (Lonza) and electroporated using G-30 program. Cells were incubated for 4 days at 25°C before collection, and knockdown efficiency was confirmed by western blotting.
Antibodies
For western blotting, rabbit anti-CP190 (47) (1:10 000; laboratory made), guinea pig anti-Su(Hw) (48) (1:1000; laboratory made), rabbit anti-Mod(mdg4)67.2 (49) (1:1000; laboratory made), rabbit anti-NURF301 (1:1000; Novus Biologicals), rabbit anti-GFP (1:2500, Abcam) and mouse anti-Lamin B (ADL67.2, 1:10 000, DSHB), and mouse anti-α-Tubulin (1:50 000; Sigma) were used. For immunostaining, rabbit anti-CP190 (22) (1:2000 for IF, 1:1000 for IF and FISH; laboratory made), guinea pig anti-Su(Hw) (48) (1:1000; laboratory made), rabbit anti-Mod(mdg4)67.2 (49) (1:1000; laboratory made), rabbit anti-NURF301 (1:700; Novus Biologicals), and mouse anti-Lamin B (ADL67.2, 1:1000, DSHB) were used. Secondary goat antibodies labeled with AlexaFluor 488, AlexaFluor 546, or AlexaFluor 647 (Molecular Probes) were used at 1:1000.
Luciferase insulator barrier activity assay
Insulator barrier activity by luciferase assay was carried out as described previously using Bright-Glo™ Luciferase Assay System (Promega) (18,22). Luciferase signal was quantified using a Spectramax II Gemini EM plate reader (Molecular Devices). Luciferase levels were measured for twelve individual whole third instar male or female for all genotypes indicated in a single panel simultaneously. Luciferase values were normalized to total protein of each larva determined by BCA reagent (Thermo Scientific). The relative luciferase activity of a population of a single genotype was aggregated into a box and whisker plot. Populations were compared with one-way ANOVA followed by a Tukey HSD post-hoc test to obtain p-values for each pairwise comparison.
Co-immunoprecipitation
Embryonic nuclear extract was prepared from 20 g of mixed stage (0–24 h) Drosophila embryos as described previously (21). Nuclei were lysed with 5 ml HBSMT nuclear lysis buffer (50 mM HEPES, 150 m M NaCl, 1 M KCl, 3 mM MgCl2, 0.3% Triton X-100 at pH 7, 1 mM PMSF and Roche cOmplete protease inhibitor) and sonicated for 10 cycles with 10 s on and 10 s off. The soluble fraction of extracts was collected by centrifugation. 25 μl of Protein G or Protein A Sepharose beads (GE Healthcare) were washed three times with nuclear lysis buffer for immunoprecipitation depending on either antibody raised in guinea pig or in rabbit, respectively. 5 μl of rabbit anti-NURF301, 5 μl rabbit IgG (Santa Cruz), 3 μl guinea pig normal serum (Covance Research Products), 3 μl guinea pig anti-Su(Hw), 3 μl rabbit normal serum (Covance Research Products), 3 μl rabbit anti-CP190, or 3 μl rabbit anti-Mod(mdg4)67.2 (rabbit) were incubated with sepharose beads for 1 h at 4°C, and unbound antibodies were removed by centrifugation at 1500 xg for 2 min. Beads were washed three times with 0.2 M sodium borate, pH 9, and then crosslinked with 20 mM DMP in sodium borate for 30 min at RT. Beads were collected by centrifugation, quenched with ethanolamine for 2 h, and washed three times with lysis buffer. After crosslinking, 500 μg of nuclear extract was used for each immunoprecipitation and incubated with antibody-bound beads overnight at 4°C. The next day, beads were collected by centrifugation and washed three times with nuclear lysis buffer. Samples were eluted with SDS sample buffer by boiling, separated using SDS-PAGE, transferred to nitrocellulose membrane in 10 mM CAPS, pH 11, and detected by western blotting.
Chromatin fractionation
Chromatin fractionation was performed as previously described with minor modifications (50,51). Approximately 1–2 × 107 Kc cells were washed twice with PBS. Cells were lysed for 15 min on ice in cold CSKI buffer (10 mM PIPES pH 6.8, 1 mM EDTA, 100 mM NaCl, 300 mM sucrose, 0.5% (v/v) Triton X-100, 1 mM MgCl2 supplemented with 1 Mini Complete tablet (Roche) per 10 ml lysis buffer. Lysate was centrifuged at 500 × g at 4°C for 5 min, and the supernatants containing cytoplasmic and soluble nuclear fraction were collected. The pellets were washed twice in CSKI buffer and then resuspended in RIPA buffer (150 mM Tris–HCl pH 8, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% (w/v) SDS, 1% (v/v) NP-40). All fractions were analyzed by western blotting. We performed this experiment using four biological replicates and quantified proteins using ImageJ (52).
IP and mass spectrometry
Nuclear extracts from 18 g of mixed stage (0–24 h) Drosophila embryos (21) were lysed in 5 ml HBSMT including 1 mM PMSF and Complete protease inhibitor cocktail (Roche). Immunoprecipitation was performed using previously described methods, and six replicates were pooled together for mass spectrometry. After IP, beads were washed once with HBSMT nuclear lysis buffer, twice with HBSM buffer, and eluted with 1% sodium dodecanoate for mass spectrometry.
Proteins were analyzed using tandem HPLC-mass spectrometry at the NIDDK Mass Spectrometry Facility. Mass from eluted peptides was queried in the UniProt database, and results were analyzed using MaxQuant.
Generation of Oligopaint FISH probes
FISH libraries for ChX and Ch3L were designed using the dm6 genome and multiplexed using the Oligominer pipeline as previously described (53) with two or three sub-library barcodes: (i) gypsy forward domains (gypsyF), (ii) gypsy reverse control domains (gypsyR), (iii) domain-specific barcodes to allow for amplification of single gypsy forward or reverse control domains (Ch3L only). The following criteria were used to call domains for the gypsy sub-libraries: (a) Su(Hw), Mod(mdg4)67.2 and CP190 ChIP-seq colocalization in Kc (19); (b) Su(Hw) and Mod(mdg4)67.2 colocalization in BG3 (22); (c) colocalization is defined as at least 30% of each peak region overlapping for each comparison and (d) peak was called in modENCODE data (54). For ChX paints, 2500 bp were added up and downstream to ensure domains were over 5 kb. For Ch3L paints, 15 000 bp were added up and downstream of domains to ensure domains were over 30 kb. For the reverse control sub-libraries, the chromosomal coordinates were reversed in one dimension by subtracting from the total bp of ChX or Ch3L. For example, on Ch3L, the gypsy forward domain has chromosomal coordinates 171083 (start)-181532 (stop), and Ch3L has 22997477 bp total. To obtain the reverse coordinates for this domain, the start and stop coordinates were subtracted from 22997477, giving a reverse domain location on ch3L of 22815945 (start)–22826394 (stop). Any oligos in the reverse control paints that overlapped with gypsyF paints were omitted from the libraries, resulting in the reverse control paints containing only non-gypsy chromatin. Coordinates for all probes can be found in Supplementary Tables S6 to S9. Whole chromosome Oligopaints for Ch2L, 2R and X were a gift from E. Joyce and label the same domains as previously described (55).
IF/FISH with Oligopaints
IF was performed as described above. After IF, slides were post-fixed in 4% PFA for 10 min and washed for 2 × 5 min in PBS-T before proceeding with FISH. FISH was performed as previously described (55). Slides were washed 1 × 5 min in 2 × SSCT (0.3 M NaCl, 0.03 M sodium citrate, 0.1% Tween-20), 1 × 5 min in 2 × SSCT/50% formamide at RT, pre-denatured in 2 × SSCT/50% formamide at 92°C for 2.5 min, then in 2 × SSCT/50% formamide at 60°C for 20 min. 100 pmol of primary Oligopaint probe was mixed in hybridization buffer (10% dextran sulfate/2× SSCT/50% formamide/4% polyvinylsulfonic acid (PVSA)) with a final volume of 25 μl. After spotting of probe, slides were covered with a coverslip, sealed with rubber cement, and denatured for 2.5 min at 92°C before transferring to a 37°C humidified chamber overnight. The next day, slides were washed in 2 × SSCT 15 min at 60°C, 15 min at RT, and 5 min at RT in 0.2 × SSC. Secondary fluorophore probes in hybridization buffer (10 pmol/25 μl) were added to slides, covered with a coverslip, and sealed with rubber cement before incubating in 37°C humidified chamber for 2 h. Slides were washed in 2 × SSCT at 60°C for 15 min, RT for 15 min, and 0.2 × SSC at RT for 5 min. All slides were washed with DAPI (1:10 000 in PBS) for 10 min and washed 2 × 5 min in PBS before mounting in Prolong Diamond (Life Technologies).
Imaging, quantification and data analysis
Images for gypsy insulator IF and FISH were captured at RT on a Leica DMi 6000B widefield fluorescence microscope using a 1.4 NA 63× objective and Leica DFC9000 sCMOS Monochrome Camera. Cells were grown at 25°C, and samples in control and different knockdowns were processed in parallel. For the quantification of Mod(mdg4)67.2-GFP foci, total GFP foci and large GFP foci were considered. We used the TANGO 3D-segmentation plugin for Fiji (56) to call total foci and large foci (>0.5 μm3). Three biological replicates were performed, and at least 500 cells in each condition were analyzed. NURF301 and Lamin B IF images were taken on a Zeiss LSM 880 Airyscan with 63×/1.4 oil Plan-APO objective. Using the Zeiss 32 channel Airyscan detector, SR mode was applied in acquiring the images. DAPI, CY3, CY5, and FITC filter cubes were used for image acquisition.
Images of FISH were deconvolved using Huygens Professional software (Scientific Volume Imaging, Hilversum, Netherlands). After deconvolution, images were segmented using TANGO. All nuclei were segmented using the ‘Hysteresis’ algorithm. For whole chromosome paints, segmentation was performed using the ‘Hysteresis’ algorithm. Foci were segmented using the ‘Spot Detector 3D’ algorithm. All measurements were obtained using TANGO. If there is any voxel (3D pixel) colocalization between FISH probe and CP190 IF signals, we defined this as contact between these objects (57). For each replicate, we quantified the contact frequency by determining whether contact is true or false in each cell in the population. We summarized data from at least four biological replicates, and each dot represents the average of >300 cells from one replicate. Statistical tests were performed using Prism 8 software by GraphPad. Tiff files for example images were created in ImageJ.
ChIP and ChIP-seq library preparation
2–3 × 107 cells were fixed by adding 1% formaldehyde directly to cells in culture medium for 10 min at RT with gentle agitation. 0.125 M glycine was used to quench formaldehyde with gentle agitation for 5 min. Cells were centrifugated at 500 × g for 5 min and washed twice with ice-cold PBS. Chromatin preparation and chromatin immunoprecipitation (ChIP) were performed following earlier described methods (18). Two immunoprecipitation (IP) samples were pooled for library preparation. Libraries were prepared according to the manufacturer's protocol with TruSeq adapters (Illumina). All samples were sequenced with HiSeq2500 (Illumina) using 50 bp single-end sequencing at the NIDDK Genomics Core Facility.
ChIP-seq data analysis
FASTQ files of sequenced single-end 50 bp reads were trimmed using cutadapt v3.4 (58) with arguments ‘–quality-cutoff 20’, ‘-a AGATCGGAAGAGC’, ‘–minimum-length 25’ and ‘–overlap 10’. Then trimmed reads were mapped to the Flybase r6-28 dm6 genome assembly with Bowtie2 v2.4.2 (59) using default arguments. Multimapping reads were removed from mapped reads using samtools v1.12 (60) view command with the argument -q 20. Duplicates were removed from mapped, uniquely mapping reads with picard MarkDuplicates v2.25.2 (http://broadinstitute.github.io/picard/index.html). MACS2 v2.2.7.1 (61) (https://github.com/taoliu/MACS) was used to call peaks by providing replicate IPs and inputs as multiple BAMs, effectively calling peaks on pooled/merged samples and using additional arguments ‘-f BAM’, ‘–gsize = dm’, ‘-q 0.000001’, ‘–mfold 3 100’ (the latter to include a larger set of preliminary peaks for fragment size estimation). Source codes were modified based on Github (https://github.com/lcdb/lcdb-wf/tree/master/workflows/chipseq).
FlyBase release 6.28 annotations were used to annotate peaks as follows. Exons were defined as any exon from any transcript of any gene. Introns were defined as the space between exons derived in a per-transcript manner by using the gffutils (https://github.com/daler/gffutils) method FeatureDB.create_introns. Promoters were defined as the TSS of each transcript plus 1500 bp upstream. Intergenic regions were defined as all regions between gene bodies. Shared co-occupied peaks were determined using pybedtools BedTool.intersect with the v = True or u = True argument, respectively. Each set of peaks was intersected with the annotations using this hierarchy ‘promoter > exon > intron > intergenic’ where a peak was classified according to the highest priority feature. Here, a peak simultaneously intersecting a promoter of one isoform and an intron of a different isoform would be classified as ‘promoter’. To compare percentages across annotated peaks in different types of peaks (all NURF301, all CP190, all Su(Hw), shared NURF301 and CP190 peaks, shared NURF301, and all gypsy sites) the number of peaks in each class was divided by the total number of peaks of that type.
Binary heatmaps were generated using pybedtools v 0.8.2 (62,63). Since peaks for one protein can potentially overlap multiple peaks for other proteins, the output represents unique genomic regions as determined by bedtools multiinter with the -cluster argument. Therefore, as a result, when considering the multi-way overlap with other proteins, the sum of unique genomic regions for a protein is not guaranteed to sum to the total number of called peaks for that protein.
Pairwise comparison for co-localization of different factors was performed using the BEDTools ‘jaccard’ command (62,63). The heatmap was clustered with the scipy.cluster.hierarchy module, using ‘euclidean’ as the distance metric and ‘Ward’ as the clustering method. As the Jaccard statistic is independent of the order of comparison and is symmetric across the diagonal, only the upper triangle is shown.
Heat maps were generated using deeptools (v3.5.1). ChIP reads were normalized to reads of input samples and mapped in a 1.5 kb window centered on the CP190, Su(Hw), or NURF301 summit.
Differential ChIP-seq
To detect differential ChIP-seq binding, we used the Diffbind v3.2/R package (64) using the config object ‘data.frame(RunParallel = FALSE, DataType = DBA_DATA_GRANGES, AnalysisMethod = DBA_ DESEQ2, bCorPlot = FALSE, bUsePval = FALSE, minQCth = 15, fragmentSize = 0)’ and otherwise used defaults. Input files consisted of the final peak calls mentioned above and the IP and input BAM files for each replicate as described above with multimappers and duplicates removed. Then the final results were exported with the dba.report function with parameters ‘th = 1, normalize = DBA_NORM_LIB’ and final differentially gained or lost peaks were those that had a log2 fold change of >0 or <0, respectively, and FDR < 0.05.
ChIP quantitative PCR
ChIP DNA samples were prepared from pre-immune IP, Su(Hw) IP, CP190 IP, and NURF301 IP from Kc cells transfected with dsRNA against mcherry, su(Hw), Cp190, or Nurf301. We used normal serum as a negative control for Su(Hw) and CP190 and rabbit IgG for NURF301. Applied Biosystems real-time PCR machine (Thermo Fisher) was used to amplify ChIP DNA samples using SYBR Green reaction systems (Applied Biosystems, primers are in Supplementary Table S5). Experiments were performed using two independent biological replicates and four technical replicates for each sample. The P-values were calculated by Student's t-test.
Fisher's exact tests
Two-sided FET was executed in R to calculate P-value and odds ratio (24). For Figure 4C, this test was performed for CP190 peaks that are decreased/not decreased in NURF301-depleted cells versus overlapping/not overlapping with Su(Hw) binding in control conditions. For Supplementary Figure S6E, this test was performed for CP190 peaks decreased/not decreased in Su(Hw)-depleted cells versus decreased/not decreased in NURF301-depleted cells. For Supplementary Figure S7, this test was performed for a particular set of peaks that are increased/not increased in Nurf301 knockdown condition versus overlapping/not overlapping with shifted nucleosomes. The same analysis was repeated for decreased/not decreased or changed/unchanged peaks.
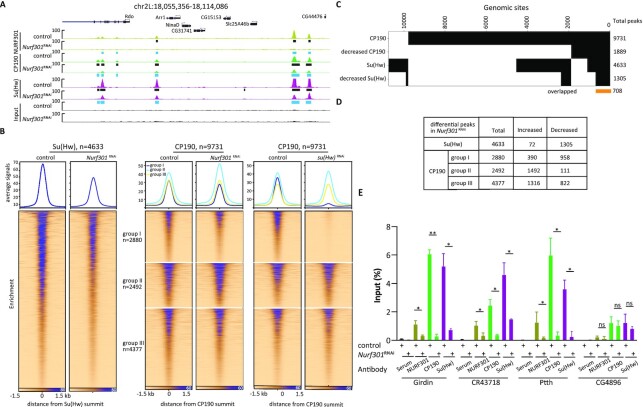
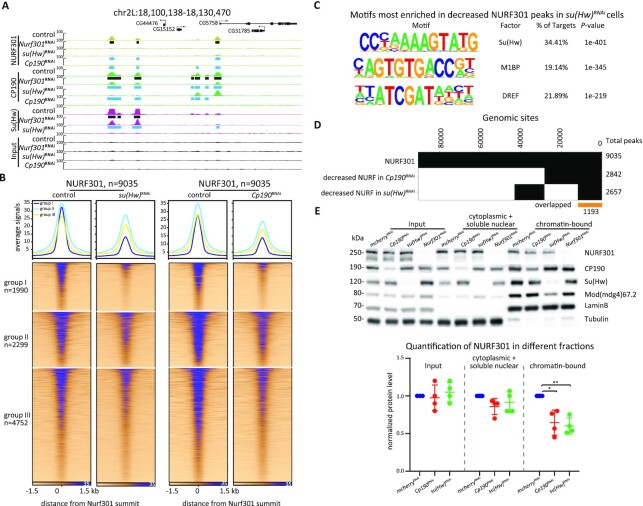
Figure 4.
Depletion of NURF301 decreases the binding of gypsy core components throughout the genome. (A) Screenshot of ChIP-seq profiles of CP190, Su(Hw), and NURF301 in control cells and Nurf301 knockdown cells. Cells were grown at 25°C. Peaks in control samples are indicated by black rectangles, and decreased peaks determined by the Diffbind algorithm (FDR < 0.05) in knockdown samples are indicated by blue rectangles. Three biological replicates were performed for each condition. (B) Su(Hw) and CP190 ChIP-seq signal at called peaks centered on their summit, which are sorted by descending signal in control cells. Left panel is Su(Hw), right panel is CP190 divided into three groups based on whether it overlaps with Su(Hw) (group I), is located near TAD borders (group II), and remaining sites (group III). Corresponding signals are shown for control cells (left) or after NURF301 or Su(Hw) depletion (right). (C) Binary heatmap of CP190, Su(Hw), and their respective decreased peaks after NURF301 depletion. At 708 genomic sites, both CP190 and Su(Hw) are decreased in Nurf301 knockdown cells. (D) Total and differential peaks of Su(Hw) and CP190 in three groups in (B) after depletion of NURF301. (E) Differentially bound CP190 and Su(Hw) ChIP-seq peaks in Nurf301 knockdown were validated by ChIP-qPCR. Chromatin association of CP190 and Su(Hw) at Girdin, CR43718, and Ptth was reduced after depletion of NURF301. Normal serum is used as a negative control for ChIP, and CG4896 is used as a negative control site, at which binding of CP190 and Su(Hw) are not affected by NURF301. Data are from two independent biological replicates and measured using four technical replicates for each sample. The p-values were calculated using Student's t-test. ns: not significant, * P < 0.05, **P < 0.01.
MNase-seq data re-analysis
MNase-seq raw reads from Drosophila larval hemocyte cells (39) were downloaded and mapped to the dm6 genome assembly using bowtie2 (59). Only uniquely mapping reads were kept (mapq > 10) and processed with DANPOS2 (65). This algorithm was applied to call nucleosome positions and nucleosomes for which position on the DNA has shifted in Nurf301 mutant larval hemocytes compared to wild type. ‘jd’ was set to 80, and other parameters were default. Bedtools merge (-d 150) was used to merge adjacent changed nucleosomes.
Motif analysis
To search de novo motifs that are overrepresented, we used HOMER (66) (v4.11) with default settings in the findMotifsGenome.pl script. The highest-scoring sequences with p-value and percentage in targets are presented.
RESULTS
Identification of NURF301 in a high-content RNAi imaging screen for factors that affect the localization of insulator bodies
The proper formation of insulator bodies is highly correlated with gypsy insulator function and therefore serves as a useful phenotypic readout for loss of gypsy insulator activity. To visualize insulator bodies in live cells, we selected stably transfected clones of an embryonic Kc167 hemocyte cell line expressing C-terminal GFP-tagged Mod(mdg4)67.2. In these clones, Mod(mdg4)67.2-GFP is expressed at a similar level to endogenous Mod(mdg4)67.2, and it physically interacts with endogenous CP190 and Su(Hw) core gypsy insulator proteins (Supplementary Figure S1). Furthermore, as judged by confocal fluorescence microscopy, Mod(mdg4)67.2-GFP colocalizes with CP190 and Su(Hw), similar to endogenous Mod(mdg4)67.2 in untransfected Kc167 cells (Supplementary Figure S2).
To validate that Mod(mdg4)67.2-GFP foci can be used to monitor insulator body formation, we depleted individual gypsy insulator proteins by dsRNA transfection and used Mod(mdg4)67.2-GFP localization as a readout for insulator bodies. The efficiency of knockdowns was verified by western blotting, and depletion of any individual gypsy insulator core protein does not affect the expression level of the other gypsy proteins (Supplementary Figure S3A). Mod(mdg4)67.2-GFP, CP190, and Su(Hw) colocalized in the mcherry dsRNA treated cells (control cells), and as expected, no GFP foci were detected after depletion of Mod(mdg4)67.2 (Figure 1A). GFP foci number was also reduced in both Cp190 and su(Hw) knockdown cells, verifying that insulator bodies are disrupted after depletion of either core gypsy insulator protein. We observed that GFP foci became larger in su(Hw) knockdown cells. When only large (>0.5 μm3) GFP-labelled insulator bodies are considered, only knockdown of su(Hw) caused an increase in foci number (Figure 1A). Staining of Su(Hw) and CP190 in parallel indicated colocalization of Mod(mdg4)67.2-GFP with residual insulator bodies after knockdown of either component.
Figure 1.
NURF301 affects the formation of gypsy insulator bodies. (A) Localization of insulator bodies is disrupted after depletion of individual gypsy insulator proteins. Right panels are quantification of average total GFP foci number per cell and average number of large foci (volume > 0.5 μm3) per cell imaged on glass slides. Cells were grown at 25°C. (B) Workflow of high-content RNAi imaging screen: Pre-spot dsRNA on plates, dispense cell suspension, incubate 4 days at RT then 2.5 h at 4°C, fix and DAPI stain, image, and process data. (C) Average large foci number per nucleus after each dsRNA treatment. Z’ factor = 0.8, indicating a good assay. (D) Verification on glass slides that NURF301 knockdown can alter the formation of gypsy insulator bodies. Cells were grown at 25°C, and these samples were processed in parallel with cells in (A). Quantification of the average total foci and large foci number are shown on the right. Scale bars: 5 μm. Comparisons in groups are all compared with mcherry dsRNA knockdown control cells. Three biological replicates were performed with n > 500 cells examined in each condition. Two-tailed unpaired t-test was used, and error bars show standard deviation. **P < 0.01 and ****P < 0.0001.
We then performed a high content imaging screen in 384-well plates in Mod(mdg4)67.2-GFP cells treated with the Drosophila RNAi Screening Center (DRSC) transcription factor sub-library, which targets 993 genes in the Drosophila genome. In the process of optimizing the assay for cell growth and imaging in 384-well plates, we found that subjecting cells to cold stress, but not osmotic stress, also induces the formation of large and distinct insulator bodies amenable for detection using automated analysis (Supplementary Figure S3B, Supplementary video 1–3). Importantly, cold stress does not affect levels of core gypsy insulator proteins or their nuclear/cytoplasmic distribution (Supplementary Figure S3C). For screening, cells were incubated with dsRNA for 4 days at RT and then cold shocked to induce the formation of more prominent insulator bodies (Figure 1B). Under these conditions, the majority of cells in the mcherry knockdown (negative control) and water-treated wells displayed a diffuse distribution of GFP signal in the nucleus in 384-well plates (Supplementary Figure S4A). GFP signal was greatly diminished after knockdown of mod(mdg4)67.2. In su(Hw) knockdown cells, GFP foci became larger and more distinct, in agreement with our preliminary assays (Figure 1A and Supplementary Figure S4A). To verify suitable controls for our RNAi screen, we calculated a Z’ score, where a maximum Z’ score of 1.0 indicates an ideal assay and >0.5 indicates a good, reliable assay (46). We verified that su(Hw) knockdown is a good positive control for our screen, with a Z’ score of 0.8 (Figure 1C). As a negative control, the number of distinct foci for Mod(mdg4)67.2-depleted cells is very low (Figure 1C). We also observed a slightly but significantly reduced large foci number in Cp190 knockdown cells (Supplementary Table S1 and Figure S4B).
From our phenotyping RNAi screen, we identified RNAi treatments that lead to an increase in large GFP foci number (Supplementary Tables S1 and S2). These targets include the known insulator factor Pita, CG3847, and NURF301/E(bx). Pita is a zinc-finger insulator protein that interacts with the BTB domain of CP190 (7,67). CG3847 is predicted to function as a transcription factor regulating wing development and has a C2H2 zinc finger that can bind to DNA (68). We chose to focus on the NURF301 nucleosome remodeller for subsequent analysis because of the strength of the observed effect after its knockdown (Supplementary Table S1 and Supplementary Figure S4B). Follow-up analysis with cells grown at RT and imaged on slides showed that NURF301 is homogenously distributed in nuclei and not concentrated at insulator foci (Supplementary Figure S4C). Moreover, in Nurf301 knockdown cells, the total GFP foci number per nucleus is significantly decreased, but the number of large (>0.5 μm3) Mod(mdg4)67.2-GFP foci is significantly increased (Figure 1D). Therefore, depletion of NURF301 disrupts the localization of gypsy insulator bodies similar to knockdown of su(Hw). Importantly, NURF301 does not affect expression levels of insulator proteins (Supplementary Figure S4D). These results confirm that NURF301 can affect the formation of gypsy insulator bodies.
NURF301 is required for gypsy insulator function
NURF301 has been shown to contribute to gypsy-dependent enhancer-blocking activity but has never been examined for its effect on its barrier activity. To investigate the role of NURF301 in gypsy-dependent barrier activity, we performed a quantitative luciferase-based assay using different tissue-specific Gal4 drivers in order to knockdown Nurf301 in flies. To validate the Nurf301 RNAi hairpin expressing line, we performed western blot analysis after knockdown using a ubiquitous Act5C-Gal4 driver in second instar larvae (Figure 2B). We observed lethality at the third instar larval stage upon depletion of NURF301 using either ubiquitously expressed Act5C-Gal4 or da-Gal4 drivers, or a CNS-enriched l(3)31-1-Gal4 driver. Depletion of NURF301 using a muscle specific Mef2-Gal4 driver did not affect viability, so it was used for further analysis.
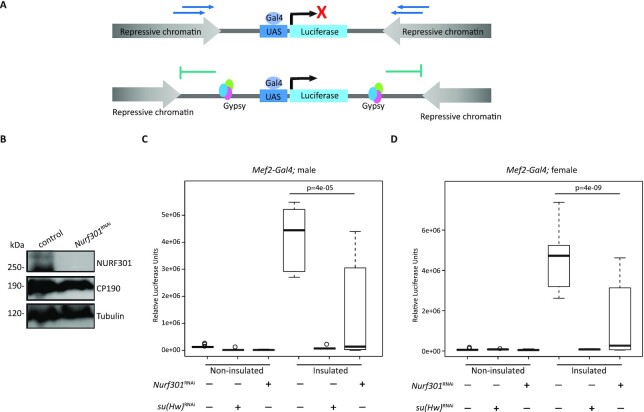
Figure 2.
NURF301 promotes gypsy insulator barrier activity. (A) Schematic diagram of in vivo insulator barrier assay. In the non-insulated UAS-luciferase line, spreading of repressive chromatin leads to low expression of luciferase. In the insulated line, presence of the gypsy insulator acts as a barrier and allows for high expression of luciferase. (B) Western blotting of male second instar larval extracts for NURF301, CP190, and Tubulin (loading control) in control flies and NURF301RNAi knockdown flies using the Act5C-Gal4 driver grown at 25°C. (C, D). Relative luciferase activity of insulated or non-insulated male (C) and female (D) third instar larvae of control or with NURF301RNAi driven by Mef2-Gal4 driver. Su(Hw)RNAi is used as a positive control. For each genotype, n = 12 individual larvae, with each experiment performed twice with similar results. One-way ANOVA followed by Tukey HSD post hoc test was used to calculate P-values for pairwise comparisons. Box represents the 25–75th percentiles, and the median is indicated. The whiskers show minimum and maximum values. P-values are indicated.
In the barrier assay, a luciferase transgene reporter with upstream activation sites (UAS) is either insulated by flanking Su(Hw)-binding sites or non-insulated and inserted into a defined attP site within the genome (22,44). Gal4 expression drives both the luciferase reporter and specific knockdown (Figure 2A). Luciferase is highly expressed in the insulated compared to non-insulated control, due to the impediment to repressive chromatin spreading in uninsulated conditions. As a positive control, depletion of Su(Hw) using Mef2-Gal4 caused loss of barrier function and drastic reduction of luciferase expression in the insulated line. Knockdown of Nurf301 showed a significant decrease of luciferase activity compared to the insulated control line for both males and females (Figure 2C and D). This quantitative assay supports a role for NURF301 in promoting gypsy insulator barrier activity.
NURF301 interacts physically and co-localizes throughout the genome with gypsy insulator components
We next tested whether there is any physical interaction between NURF301 and gypsy insulator components. We performed immunoaffinity purification from Drosophila embryonic nuclear extracts using an anti-NURF301 antibody coupled with quantitative mass spectrometry (IP-MS). We found that NURF301 physically interacts predominantly with many insulator/architectural proteins, such as the core gypsy insulator proteins, Pita, M1BP, GAF, and Ibf1/2 (Supplementary Table S3). NURF301 was also identified in both anti-CP190 and anti-Su(Hw) IP-MS from embryonic nuclear extracts (24). We confirmed the interaction between NURF301 and each of the core gypsy components by co-immunoprecipitation from embryonic nuclear extracts using antibodies recognizing CP190, Su(Hw), or Mod(mdg4)67.2 followed by western blotting (Figure 3A).
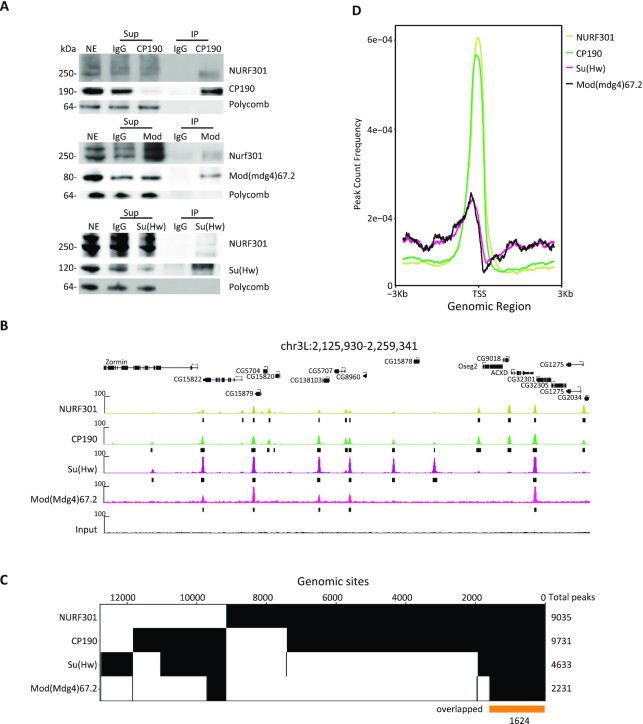
Figure 3.
NURF301 interacts physically with gypsy insulator proteins, and these factors co-localize genome-wide. (A) Co-immunoprecipitation of NURF301 with core gypsy components. Nuclear extracts (NE) are from embryos aged 0–24 h collected at RT, which is the same material used for IP followed by mass spectrometry. NE was immunoprecipitated with each antibody or with normal serum (IgG) as indicated. Unbound supernatant (Sup) and bound (IP) fractions are also shown. Polycomb (Pc) is used as a negative control. (B) Representative screenshot of ChIP-seq profiles shows NURF301 co-localizes extensively with gypsy core components in Kc cells grown at 25°C. Peaks called by MACS2 are indicated by black bars. Three biological replicates were examined for each sample. (C) Binary heatmap of CP190, NURF301, Su(Hw), and Mod(mdg4)67.2 binding sites in Kc cells, ordered by supervised hierarchical clustering. Each column represents a single independent genomic location, and a mark in a row indicates presence of the indicated factor. Total peaks of each factor are shown on the right. 7423 genomic sites co-localize between NURF301 and CP190, and 1624 genomic sites (yellow bar) were bound by all four factors. (D) Average signals of CP190 and NURF301 peaks, but not of Su(Hw) and Mod(mdg4)67.2, accumulate at the TSS.
To investigate the extent to which NURF301 co-localizes with gypsy insulator proteins on chromatin, we performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) for NURF301, CP190, Su(Hw), and Mod(mdg4)67.2 in Kc cells. We identified 9035 NURF301, 9731 CP190, 4633 Su(Hw), and 2231 Mod(mdg4)67.2 peaks in Kc cells (Figure 3B). To validate the specificity of the NURF301 antibody, we compared our NURF301 peaks with Ebx-eGFP ChIP-seq performed by the modERN project using anti-GFP antibody in Drosophila embryos (69). There are 8108 Ebx-eGFP peaks in embryo, and 90% of these peaks were also identified in our NURF301 ChIP-seq (Supplementary Figure S5A), indicating high correspondence between our data and previous results. We also profiled the binding of Mod(mdg4)67.2-GFP using anti-GFP in Kc-Mod(mdg4)67.2-GFP cells compared to endogenous Mod(mdg4)67.2 in untransfected Kc cells. We found that 3182 genomic sites were bound by GFP, corresponding to 94% (2142 of 2231) of endogenous Mod(mdg4)67.2 peaks. Moreover, 2153 of these Mod(mdg4)67.2-GFP peaks also colocalize with Su(Hw) (Supplementary Figure S5B).
In untransfected Kc cells, 7423 genomic sites are shared between NURF301 and CP190, which is equivalent to 76% of CP190 peaks and 82% of NURF301 peaks. Furthermore, 1990 Su(Hw) (43%) and 1653 Mod(mdg4)67.2 (74%) peaks colocalize with NURF301 (Figure 3C). Among these genomic sites, 1624 sites are co-occupied by NURF301, CP190, Su(Hw) and Mod(mdg4)67.2. These results indicate that NURF301 colocalizes with gypsy insulator proteins on chromatin, especially with CP190 (Figure 3B and C). There is also high overlap between NURF301 and other insulator factors M1BP, Beaf32, CTCF, Pita, Zipic, Ibf1, and Ibf2 (Supplementary Figure S5C). Pairwise comparisons of colocalization using Jaccard analysis indicates that NURF301 co-localizes particularly well with CP190, Su(Hw), Mod(mgd4), BEAF-32, M1BP, and Zipic (Supplementary Figure S5D). In addition, NURF301 and CP190 co-occupied sites are enriched in promoter regions (Figure 3D). In contrast, Su(Hw) and Mod(mdg4)67.2 are relatively evenly distributed throughout the genome, and only approximately one-quarter of NURF301/gypsy co-localized sites are enriched at promoters (Supplementary Figure S5E).
NURF301 aids recruitment of Su(Hw) to chromatin and also affects binding of CP190 to gypsy sites
To determine whether NURF301 promotes the recruitment of gypsy insulator proteins to chromatin, we performed ChIP-seq to examine core gypsy components after knockdown of Nurf301. After depletion, NURF301 ChIP-seq signal is greatly decreased (Figure 4A). We also found that Su(Hw) binding diminished across the genome after Nurf301 knockdown (Figure 4A and B). The DiffBind (v3.2) algorithm (64) was applied to identify differential peaks between control and Nurf301 knockdown cells. We identified 1305 (28%) decreased and 90 increased Su(Hw) peaks (false discovery rate (FDR) < 0.05, Figure 4C). Among these decreased Su(Hw) peaks, 720 peaks overlap with NURF301 binding in the control condition (Supplementary Figure S6A), supporting that these effects are likely direct.
Although the average signal of CP190 is increased after depletion of NURF301 (Supplementary Figure S6B), we found that there are changes in both directions, with 3198 (33%) increased and 1891 (19%) decreased CP190 peaks. There are 3182 increased and 921 decreased CP190 peaks that overlap with NURF301 binding in the control condition (Supplementary Figure S6A), again suggesting direct effects of NURF301 depletion. We found that decreased CP190 peaks are enriched for the Su(Hw)-binding motif (Supplementary Figure S6C), and a strong correlation was identified between decreased CP190 peaks after depletion of NURF301 compared to Su(Hw) occupancy in control cells (Figure 4C, Fisher's exact test (FET), odds ratio = 2.8, P < 2.2e−16). In contrast, increased CP190 peaks are enriched for M1BP and DREF binding motifs, which tend to be located near TSSs and TAD borders (Supplementary Figure S6C). Both DREF and BEAF-32 share almost identical binding specificities (Hart et al., 1999).
Since CP190 is a component of many different insulator complexes, we divided CP190 peaks into three separate groups in order to more deeply interrogate changes after NURF301 depletion. The first group colocalizes with Su(Hw) (group I, n = 2880), and these sites are mostly promoter-distal sites further from TAD borders (Supplementary Figures S5E and S6D). The second group is comprised of non-Su(Hw)-overlapping CP190 sites that are close to TAD borders (± 1 kb, group II, n = 2492). The third group contains all remaining CP190 sites (group III, n = 4377). After depletion of NURF301, the average signal of CP190 group I decreases while the signal of group II increases, and group III remains mostly unchanged (Figure 4B), which is consistent with the changes of CP190 peaks in groups (Figure 4D). We validated a subset of decreased peaks by directed ChIP-qPCR. At these sites, binding of both CP190 and Su(Hw) are significantly reduced after NURF301 depletion (Figure 4E). To assess whether NURF301 may play a similar role to Su(Hw) in recruiting CP190 to chromatin, we profiled CP190 in su(Hw) knockdown cells and verified that CP190 binding is essentially abolished specifically at Su(Hw) sites after Su(Hw) depletion (Figure 4B). The sets of respective decreased CP190 peaks in either Nurf301 or su(Hw) knockdown cells are highly overlapping (Supplementary Figure S6E, FET, odds ratio = 3.3, P < 2.2e−16). Our results suggest that Su(Hw) partially depends on NURF301 to be recruited to chromatin, and these findings are consistent with the possibility that loss of Su(Hw) after NURF301 depletion subsequently results in loss of CP190 binding. On the other hand, the binding of CP190 near TAD borders is reinforced in the absence of NURF301, which is not observed after Su(Hw)-depletion.
Loss of Su(Hw) binding in NURF301-depleted cells corresponds to changes in nucleosome positioning
Since NURF301 is a nucleosome remodeller, we wanted to determine whether and how changes in nucleosome positioning coincide with changes in Su(Hw) or CP190 binding after depletion of NURF301. Previously it was shown by MNase-seq on hemocytes derived from Nurf301 null mutant third instar larvae that loss of NURF301 leads to nucleosomes becoming more tightly packed at Su(Hw)/CP190 co-occupied sites (39). We thus compared these MNase-seq data to our ChIP-seq data to investigate the relationship between affected Su(Hw) and CP190 chromatin binding sites to nucleosomes with shifted DNA position after losing NURF301.
We found that NURF301-dependent Su(Hw) binding is modestly enriched for overlap with shifted nucleosomes (50% decreased Su(Hw) peaks, Supplementary Figure S7A, FET, odds ratio = 1.2, P = 8.2e−4). For CP190, NURF301-dependent binding is actually depleted for overlap with shifted nucleosomes (39% decreased CP190 peaks, Supplementary Figure S7B, FET, odds ratio = 0.4, P < 2.2e−16). However, CP190 peaks that increased after NURF301-depletion are significantly enriched for overlap with shifted nucleosome positioning (Supplementary Figure S7B, FET, odds ratio = 1.8, P < 2.2e−16). We next compared the MNase signal density at affected and unaffected Su(Hw) or CP190 ChIP-seq sites after depletion of NURF301. For both Su(Hw) and CP190 decreased peaks, we found an average increase in nucleosome occupancy over peak summits in Nurf301 null mutants (Supplementary Figure S7C and D), suggesting that NURF301 may help create nucleosome-free regions at these sites to promote Su(Hw) and CP190 binding. For increased CP190 peaks in Nurf301 knockdown cells, a larger nucleosome free region is visible in Nurf301 null mutants, perhaps due to a larger protein complex footprint. Lesser changes in nucleosome positioning were detected in unchanged Su(Hw) and CP190 sites. These analyses support the conclusion that NURF301-dependent binding of Su(Hw) and CP190 to chromatin is functionally related to NURF remodeling activity.
Su(Hw) and CP190 promote the binding of NURF301 to chromatin
To determine whether insulator proteins have any impact on NURF301 binding, we profiled NURF301 by ChIP-seq after CP190 or Su(Hw) depletion. NURF301 protein levels were not affected by knockdown of insulator proteins (Supplementary Figure S8A). However, when assessing chromatin-associated NURF301, we found 2842 (31%) and 2657 (29%) decreased NURF301 peaks in Su(Hw)- and CP190-depleted cells, respectively, and few increased NURF301 peaks in either knockdown (Figure 5A). Similar to our analysis for CP190 binding after depletion of NURF301 and Su(Hw), we divided NURF301 peaks into three groups, depending on whether they colocalize with Su(Hw) (group I), do not colocalize with Su(Hw) and are near TAD borders (group II), or neither (group III). Knockdown of either Su(Hw) or CP190 resulted in substantial loss of NURF301 from all three groups, but reduction of signal is most apparent in group I (Figure 5B). Consistent with a major effect on group I sites, decreased NURF301 binding sites after knockdown of either Su(Hw) or CP190 are enriched for the Su(Hw) motif (Figure 5C and Supplementary Figure S8B). Finally, we observed 1193 commonly decreased NURF301 peaks after Su(Hw) and CP190 depletion (Figure 5D). As a secondary test, we used Triton X-100 extraction to measure the amount of NURF301 protein levels in the chromatin-bound versus nuclear soluble plus cytoplasmic fraction after Su(Hw) or CP190 (Figure 5E). We found that NURF301 levels are reduced in the chromatin-bound fraction after depletion of CP190 or Su(Hw), consistent with our ChIP-seq results. These findings indicate that both Su(Hw) and CP190 are required for proper NURF301 recruitment throughout the genome.
Figure 5.
Su(Hw) assists the recruitment of NURF301 at a subset of sites. (A) Representative screenshot of decreased NURF301 after knockdown of Nurf301, su(Hw), or Cp190 in Kc cells. Cells were grown at 25°C. Peaks called in control samples are indicated by black rectangles, and decreased peaks in knockdown conditions are shown as blue rectangles. Three biological replicates were performed for each condition. (B) ChIP-seq signals for NURF301 are classified into three groups based on overlap with Su(Hw) (group I), location near TAD borders (group II), and remaining sites (group III) in control and Su(Hw)- or CP190-depleted cells. (C) Motif enrichment of decreased NURF301 peaks after depletion of Su(Hw). (D) Binary heatmap of NURF301 and decreased NURF301 after depletion of Su(Hw) or CP190. (E) Protein levels of NURF301, CP190, Su(Hw), and Mod(mdg4)67.2 in total cell lysate, cytoplasmic and soluble nuclear fraction, and chromatin-bound fractions of control and knockdown cells, as indicated. LaminB and Tubulin were blotted as a control for a chromatin-bound and cytoplasmic protein, respectively. Quantification of NURF301 protein level is graphed at the bottom. Data are from four biological replicates and paired t-test was used. *P < 0.05, **P < 0.01.
Depletion of NURF301 results in loss of CP190 localization specifically to gypsy sites in the nucleus
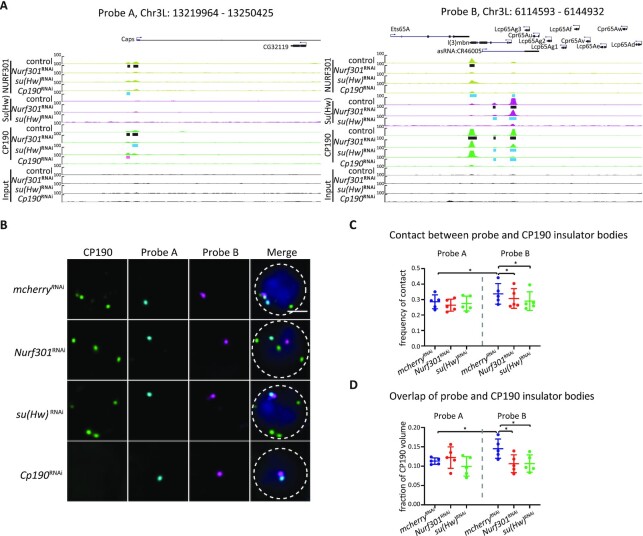
We next wanted to explore whether NURF301 affects the interaction of gypsy insulator bodies with gypsy insulator complex DNA binding sites in the nucleus. To investigate this possibility, we visualized four sub-chromosomal loci on Chr3L in pairs using Oligopaint FISH (probes A–D; Supplementary Table S4) combined with immunofluorescence to detect CP190. Probes A and C label genomic regions with no gypsy insulator binding sites and were used as controls. Probes B and D label genomic regions with multiple CP190/Su(Hw)/Mod(mdg4)67.2/NURF301 binding sites (gypsy probes; Figure 6A and Supplementary Figure S9A). We performed IF/FISH for probes A and B or probes C and D (Figure 6B, Supplementary Figure S9B) and quantified the contact frequency in the cell population and overlap volume (voxel colocalization) in individual cells between insulator bodies and each probe. We observed that on average, insulator bodies more often contact probes containing gypsy binding sites than non-gypsy binding site probes in control cells (Figure 6C, D and Supplementary Figure S9C, D). The population contact frequencies between insulator bodies and gypsy binding site probes ranged from 24% to 44%, while the contact frequencies between insulator bodies and non-gypsy binding sites ranged from 22% to 35%. Contact between insulator bodies and gypsy binding site probes was significantly decreased in the population after depletion of NURF301 or Su(Hw), but no difference was observed for contact between insulator bodies and control probes after either depletion (Figure 6C and Supplementary Figure S9C). Importantly, even when contact is made, the overlap volume in individual cells is reduced specifically between gypsy binding probes and insulator bodies after Nurf301 and su(Hw) knockdowns, and no significant change in the population average overlap of control probes and insulator bodies was observed (Figure 6D and Supplementary Figure S9D–F). These results support the possibility that CP190 protein is released from gypsy binding sites after depletion of either NURF301 or Su(Hw).
Figure 6.
NURF301 specifically affects the co-localization of insulator bodies with gypsy insulator DNA binding sites in the nucleus. (A) Screenshot of the ChIP-seq profiles of CP190/Su(Hw)/Mod(mdg4)67.2/NURF301 in 30 kb probe regions. Black bars represent peaks identified by MACS2 in control Kc cells. Pink and blue rectangles indicate increased and decreased peaks in knockdown samples, respectively. Probe A was used as a control, which has no CP190/Su(Hw)/Mod(mdg4)67.2 colocalized sites (gypsy sites). Probe B has multiple sites with enriched signals of CP190/Su(Hw)/Mod(mdg4)67.2. (B) Representative images of CP190 immunostaining and Oligopaint FISH signals in Kc cells grown at 25°C after dsRNA treatment as indicated. Scale bar represents 2 μm. (C) Percentage of cells displaying contact between CP190 and respective probes. Data are from five biological replicates, each dot represents one biological replicate, n > 300 cells per replicate. (D) For cells showing contact in (C), average overlapped volume between CP190 and probes relative to CP190 volume. Overlap value was normalized to the volume of CP190 to exclude the bias of increased size of insulator bodies in Nurf301 and su(Hw) knockdown cells. Paired t-test was used, and error bars show standard deviation. ns, not significant; *P < 0.05 as indicated.
NURF301 specifically alters the 3D organization of gypsy insulator binding sites
As NURF301 depletion not only alters insulator body localization but also disrupts the contact of insulator bodies and insulator protein binding sites, we wondered whether NURF301 knockdown affects the overall 3D spatial organization of gypsy DNA binding sites. Therefore, we designed Oligopaints to specifically detect all DNA sequences co-occupied by CP190, Su(Hw), and Mod(mdg4)67.2 on Chr3L and ChrX as defined by ChIP-seq in Kc cells (19,22) and designated these paints ‘gypsyF’ (Figure 7A and Supplementary Figure S10A). As a control, we used the one-dimensional (1D) reverse of the chromosomal coordinates for the gypsyF paints, resulting in the same 1D distribution of domains but labeling only non-gypsy insulator chromatin (gypsyR paints). We also verified that the volume of gypsyF and gypsyR paints are similar in control cells, consistent with a similar amount of DNA being painted by the two probe sets (Figure 7A–C and Supplementary Figure S10A–C).
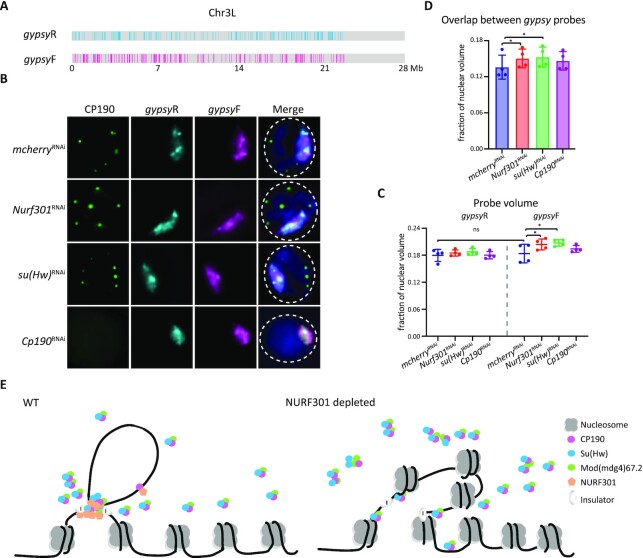
Figure 7.
Depletion of NURF301 specifically alters 3D arrangement of gypsy insulator binding sites. (A) Schematic of gypsy probes on Chr3L. The co-occupied sites of Su(Hw)/CP190/Mod(mdg4)67.2 are designated as gypsyF probe, and the 1D reverse of non-gypsy sites are designated as control probe (gypsyR). The DNA amount labeled by gypsyF or gypsyR paint is ∼5 Mb. (B) Representative images of CP190 IF and signals of gypsyF and gypsyR probes in dsRNA treated Kc cells. Cells were cultured at 25°C. Images are maximal projections of 26 Z-stacks. Nuclear edge is indicated with dashed line. Scale bar: 2 μm. (C) Probe volume relative to the fraction of nuclear volume. Left panel is control gypsyR probe, right is gypsyF probe. (D) Intermixing volume of gypsyF and gypsyR paints relative to nuclear volume. Data are from four biological replicates and a dot represents each replicate, n > 300 cells in each sample. Paired t-test was used, and error bars show standard deviation. ns, not significant; *P < 0.05 as indicated. (E) Nucleosome remodeller NURF301 cooperates with Su(Hw) to promote stable insulator complex binding at gypsy insulator sites to establish higher-order chromatin structure. After depletion of NURF301, Su(Hw) and CP190 cannot properly bind to gypsy binding sites, thus impairing gypsy insulator function and influencing chromatin organization.
To test if there are changes to the 3D organization of gypsy binding sites chromosome-wide after knockdown of NURF301 and insulator proteins, we quantified the volume of gypsyF and R paints relative to nuclear volume before and after knockdown (Figure 7C). We observed no significant change in gypsyR paint volume for either chr3L or chrX in Kc cells treated with dsRNA against Nurf301, su(Hw), or Cp190 (Figure 7C and Supplementary Figure S10C). However, we observed a significant increase in gypsyF paint volume in NURF301 and Su(Hw)-depleted cells. No significant change was observed for gypsyF paint volume in CP190-depleted cells (Figure 7C and Supplementary Figure S10C). These results might suggest that the 3D structure of chromatin containing gypsy sites becomes decondensed in NURF301 and Su(Hw)-depleted cells.
If gypsyF-painted regions, which occupy ∼18% of the chromosome, become decondensed after NURF301 or Su(Hw) knockdown, we would expect the volume of nuclei and whole chromosome territories (CTs) to increase accordingly. To test this possibility, we performed Oligopaint FISH using previously published whole chromosome paints labeling chromosomes 2L, 2R, and X in Kc cells (Supplementary Figure S11A, S11B and Supplementary Table S4) (55). We found that nuclear volume was unchanged by all dsRNA treatments compared to mcherry (Supplementary Figure S11C). Furthermore, no significant differences in CT volume or overlap were observed (Supplementary Figure S11D and S11E). These results suggest that gypsy binding sites are not simply becoming decondensed after depletion of NURF301 or Su(Hw). Instead, the increased volume of gypsyF paints likely indicates a declustering of gypsy domains within the CT. Our results with the whole chromosome paints further suggest that depletion of NURF301 or gypsy insulator proteins does not lead to global changes in 3D genome organization. Therefore, our findings suggest that NURF301 and Su(Hw) depletion specifically alters the 3D organization of gypsy insulator binding sites.
To test our model that depletion of NURF301 or Su(Hw) specifically alters the clustering of gypsy domains within a CT, we quantified the overlap between gypsyF and gypsyR probe sets for single chromosomes. If domains labeled by gypsyF paints are becoming declustered but CT volume is not increasing, we would predict an increase in overlap between the gypsyF and gypsy R paints. Indeed, we observed significantly increased overlap between gypsyF and R paints after depletion of NURF301 or Su(Hw), concomitant with the increase of gypsyF volume (Figure 7D and Supplementary Figure S10D). This result supports a model where depletion of NURF301 or Su(Hw) changes the sub-chromosomal 3D organization of domains previously occupied by the gypsy insulator complex. Together, these FISH-based studies indicate that NURF301 plays a crucial role in regulating the 3D organization of gypsy chromatin domains and recruiting insulator bodies to gypsy sites.
DISCUSSION
Here, we identified NURF301 as a factor required for proper gypsy insulator body localization using a high throughput visual RNAi screen. NURF301 promotes gypsy-dependent barrier activity and physically interacts with gypsy insulator core components. Furthermore, NURF301 colocalizes with gypsy insulator core components genome-wide on chromatin. We found that NURF301 and Su(Hw) mutually affect the binding of one another to chromatin, and NURF301 also promotes the recruitment of CP190 to gypsy binding sites. These effects generally correspond to NURF301-dependent changes in nucleosome remodeling. Finally, NURF301 is specifically required for the proper 3D arrangement of gypsy insulator binding sites and their partitioning within a chromosome territory.
NURF301 and Su(Hw) appear to function cooperatively, and each factor promotes the binding of the other at endogenous gypsy binding sites throughout the genome (Figure 7E). More than one-fourth of Su(Hw) ChIP peaks decrease after depletion of NURF301, and these decreased peaks on average display increased nucleosome occupancy in Nurf301 null mutants. At the same time, depletion of Su(Hw) also results in partial loss of NURF301 binding, particularly at Su(Hw)-binding sites. Mild loss of NURF301 binding at non-Su(Hw)-binding sites could be an indirect effect on alternative CP190-containing complexes. Our results are consistent with the possibility that Su(Hw) helps recruit NURF301 and the NURF complex to DNA to remodel nucleosomes, which in turn stabilizes Su(Hw) binding. Creation of a nucleosome-free region could promote the ability of the gypsy insulator complex to act as a barrier to interrupt the spread of repressive chromatin, which may require nucleosomes to be adjacent in order to extend efficiently. Indeed, previous studies demonstrated that the nucleosome turnover rate is particularly high at insulator chromatin (12,70,71). Reduced nucleosome occupancy could also result in a change in flexibility or adoption of a particular DNA conformation incompatible with Polycomb-dependent silencing. Interestingly, Su(Hw) binding sites display high regularity of adjacent nucleosome phasing (72), similar to what has been observed for mammalian CTCF (73,74). Previous work showed that NURF301 also promotes gypsy-dependent enhancer blocking activity, but it is unclear how creation of a nucleosome free region affects this particular function. Perhaps increased or stabilized Su(Hw) binding at these sites could affect protein-protein or DNA looping interactions needed to constrain enhancer-promoter interactions. Ultimately, Su(Hw) promotes the recruitment of the universal insulator protein CP190, and CP190 is essential for gypsy insulator activity (6).
Although we also found that NURF301 promotes CP190 binding to endogenous gypsy insulator sites, the functional relationship between NURF301 and CP190 appears more complex than that between NURF301 and Su(Hw). In NURF301-depleted cells, approximately one-third of CP190 ChIP peaks that overlap with Su(Hw) are reduced. Consistent with previous reports (6,54), we found that CP190 binding is drastically and specifically reduced at Su(Hw)-binding sites after Su(Hw)-depletion, indicating an absolute requirement for Su(Hw) to recruit CP190 to endogenous gypsy insulator sites. Therefore, loss of Su(Hw) binding after NURF301 depletion is likely to also result in loss of CP190 binding. However, in NURF301-depleted cells, CP190 binding not only partially decreases at sites to which Su(Hw) normally binds but also increases at other CP190-binding sites, particularly those located near promoters and TAD borders. This increase cannot simply be due to excess availability of CP190 as a result of loss of recruitment to Su(Hw) sites. Whether this accumulation of CP190 at TAD borders is due to an effect on the specific cofactors recruiting CP190 to these sites is not clear, but motif analysis revealed enrichment of M1BP and BEAF-32 binding motifs. We conclude that NURF301 can either promote or inhibit CP190 binding depending on the specific context, perhaps dependent on which cofactors are present. Depletion of CP190 mainly affects NURF301-binding at some Su(Hw) sites but also disrupts its binding at additional sites. Loss of CP190 has previously been shown to have a destabilizing effect on Su(Hw)-binding (6,54) and could affect additional insulator complexes with which CP190 and NURF301 interact.
Our Co-IP and mass spectrometry data verified physical interaction between NURF301 and all three core gypsy insulator proteins but also identified other classes of insulator proteins associated with NURF301. These additional factors include Pita, BEAF-32, Ibf1, and Ibf2, all of which also are known to interact with CP190 (5,7,75). Consistent with NURF301 interacting with multiple insulator complexes, it has been shown previously that NURF301 can promote Fab-7, Fab-8,SF1, and gypsy-dependent enhancer blocking activity (39,42,43). Fab-8 and SF1 insulators are known to be CTCF- but not Su(Hw)-dependent (8,9,76), suggesting that recruitment of nucleosome remodeling activity may be a general feature of chromatin insulator complexes. For each insulator sequence, it was shown that nucleosome occupancy increases at these insulator sequences when NURF301 is depleted, suggesting that NURF301-dependent remodeling activity does occur at these sites. However, one study found that NURF301 and ISWI actually attenuate Fab-8 enhancer blocking activity and also suggested that NURF301 and ISWI may affect the gypsy insulator outside the context of the NURF complex (43). Interestingly, previous immunoaffinity purifications of embryonic nuclear extracts using either anti-CP190 or anti-Su(Hw) coupled with quantitative mass spectrometry identified NURF301 as an abundant interactor, but the other three components of the NURF complex, including Iswi the catalytic subunit, were observed at much lower levels or were completely absent (24). Furthermore, ISWI, Caf1-55, and Nurf-38 were each assayed using two independent dsRNAs as part of the DRSC transcription factor library in our visual screen for insulator body mislocalization, but none of these other NURF subunits were scored as positive hits (Supplementary Table S1). It remains a possibility that NURF301 may affect gypsy insulator complex formation at least partially outside the context of NURF.
The functional significance of gypsy insulator bodies is still a matter of debate. We found that cold stress, like osmotic stress (25), can enhance the formation of insulator bodies. However, the mechanism by which either stress is detected by cells and transmitted to insulator bodies is completely unknown. We took advantage of this phenomenon to perform a visual screen and followed up candidates under normal growth conditions. Upon their initial discovery, it was suggested that insulator bodies could act as hubs to scaffold chromatin loops throughout the genome (77). Later work proposed that insulator bodies may act as intranuclear storage sites for insulator proteins that are not bound to chromatin (26). Addressing this question directly for the first time, our study examined the colocalization of insulator bodies with endogenous gypsy insulator DNA binding sites by DNA FISH with Oligopaints. Importantly, we found that insulator bodies more frequently contact genomic domains containing gypsy binding sites than non-gypsy chromatin on the same chromosome. Similarly, when contact is made, the resulting 3D overlap volume in individual cells between insulator bodies and domains containing gypsy binding sites is significantly larger than the overlap between insulator bodies and non-gypsy sites. These results indicate that insulator bodies do likely correspond to chromatin bound insulator complexes, but the cell-to-cell variability in both contact and overlap between insulator bodies and our FISH probes suggests that not all gypsy DNA binding sites are located in insulator bodies in all cells. Higher resolution studies will be needed to fully address this question. Furthermore, depletion of either Su(Hw) or NURF301 reduces both contact and overlap of insulator bodies with these gypsy insulator binding sites, consistent with chromatin binding being required for insulator body proximity to DNA. Aberrantly large insulator bodies after knockdown of either factor may result from release of insulator proteins from gypsy binding sites and increased multimerization of insulator proteins. Interestingly, when we examined whole chromosome gypsy insulator binding site paints, we found that the entire volume occupied by these probes but not control reverse probes increased in Su(Hw) and NURF301-depleted cells. This finding implies that gypsy insulator complexes promote the clustering of gypsy sites within CTs, perhaps by promoting looping between binding sites. Moreover, intermixing between these two probe sets increased while overall CT volume was unaffected. These results suggest that gypsy insulator complexes help partition the genome into distinct 3D structures within a CT, consistent with their role in demarcating distinct transcriptional domains.
Taken together, our results support a model in which gypsy insulator bodies are functionally relevant structures that correspond at least to some extent to insulator proteins bound to DNA (Figure 7E). Our results are also consistent with the possibility that insulator bodies may function as storage depots for insulator proteins to facilitate rapid loading or disassociation of insulator proteins at these sites, a process that may be regulated under stress conditions. We found that Su(Hw) cooperatively interacts with NURF301 to create nucleosome free regions at gypsy binding sites to further recruit CP190. Together, these factors promote gypsy insulator function and gypsy-dependent nuclear organization. Further work is needed to address the precise biochemical mechanism of NURF-dependent nucleosome remodeling with respect to gypsy and potentially other insulator complexes.
DATA AVAILABILITY
All other relevant data of this study are available from the corresponding author upon reasonable request. The accession numbers for the raw data FASTQ files, processed files, and BigWig files for all sequencing data generated in this study and deposited in NCBI GEO are GSE198361. Bed file for Ebx-eGFP ChIP-seq in Drosophila embryos was obtained from modERN (doi:10.17989/ENCSR559AJG). In this study, all other publicly available sequencing data in Kc167 analyzed are BEAF-32 GSM762845, Ibf1 GSM2133766, Ibf2 GSM2133767, Pita GSM2133768, ZIPIC GSM2133769, M1BP GSM4231397, and CTCF GSM1535983. Mass spectrometry data files have been deposited in the PRIDE repository with dataset identifiers PXD032165 for NURF301.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Nguyen and E. Joyce for CT Oligopaints and members of the Lei laboratory for critical reading of the manuscript.
Author contributions: Conceptualization: S.C., E.P.L.; methodology: S.C.; plasmid cloning and transfection: P.J.M., E.P.L.; clonal selection: N.M.; high-content imaging screen: G.P., S.C.; mass-spec: S.C.; Oligopaints: L.F.R., S.C.; MNase data analysis: G.-Y. Y.; bioinformatics analysis: S.C.; validation: S.C., E.P.L.; formal analysis: S.C., E.P.L.; investigation: S.C.; writing – original draft: S.C., E.P.L.; supervision: E.P.L.; funding acquisition: E.P.L.
Contributor Information
Shue Chen, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Biochemistry and Genetics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Leah F Rosin, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Biochemistry and Genetics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Gianluca Pegoraro, High-Throughput Imaging Facility (HiTIF), Laboratory of Receptor Biology and Gene Expression, Center for Cancer Research, National Cancer Institute, Bethesda, MD, USA.
Nellie Moshkovich, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Cellular and Developmental Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Patrick J Murphy, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Cellular and Developmental Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Guoyun Yu, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Cellular and Developmental Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
Elissa P Lei, Nuclear Organization and Gene Expression Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA; Laboratory of Biochemistry and Genetics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Programs of the National Institute of Diabetes and Digestive and Kidney Diseases [DK015602 to E.P.L.]; National Cancer Institute [1-ZIC-BC011567-01 to HiTIF]; National Institutes of Health. Funding for open access charge: Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jerkovic I., Szabo Q., Bantignies F., Cavalli G.. Higher-order chromosomal structures mediate genome function. J. Mol. Biol. 2020; 432:676–681. [DOI] [PubMed] [Google Scholar]
- 2. Rowley M.J., Corces V.G.. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018; 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chetverina D., Fujioka M., Erokhin M., Georgiev P., Jaynes J.B., Schedl P.. Boundaries of loop domains (insulators): determinants of chromosome form and function in multicellular eukaryotes. Bioessays. 2017; 39: 10.1002/bies.201600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen D., Lei E.P.. Function and regulation of chromatin insulators in dynamic genome organization. Curr. Opin. Cell. Biol. 2019; 58:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuartero S., Fresan U., Reina O., Planet E., Espinas M.L.. Ibf1 and ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014; 33:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pai C.Y., Lei E.P., Ghosh D., Corces V.G.. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004; 16:737–748. [DOI] [PubMed] [Google Scholar]
- 7. Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M.K., Kirsch R., Bonchuk A., Fedotova A.et al.. Two new insulator proteins, pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015; 25:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moon H., Filippova G., Loukinov D., Pugacheva E., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R.et al.. CTCF is conserved from drosophila to humans and confers enhancer blocking of the fab-8 insulator. EMBO Rep. 2005; 6:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerasimova T.I., Lei E.P., Bushey A.M., Corces V.G.. Coordinated control of dCTCF and gypsy chromatin insulators in drosophila. Mol. Cell. 2007; 28:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bushey A.M., Ramos E., Corces V.G.. Three subclasses of a drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009; 23:1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hou C., Li L., Qin Z.S., Corces V.G.. Gene density, transcription, and insulators contribute to the partition of the drosophila genome into physical domains. Mol. Cell. 2012; 48:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Negre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A.et al.. A comprehensive map of insulator elements for the drosophila genome. PLoS Genet. 2010; 6:e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramirez F., Bhardwaj V., Arrigoni L., Lam K.C., Gruning B.A., Villaveces J., Habermann B., Akhtar A., Manke T.. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 2018; 9:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G.. Three-dimensional folding and functional organization principles of the drosophila genome. Cell. 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- 15. Parkhurst S.M., Harrison D.A., Remington M.P., Spana C., Kelley R.L., Coyne R.S., Corces V.G.. The drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988; 2:1205–1215. [DOI] [PubMed] [Google Scholar]
- 16. Ghosh D., Gerasimova T.I., Corces V.G.. Interactions between the su(hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001; 20:2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Capelson M., Corces V.G.. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005; 20:105–116. [DOI] [PubMed] [Google Scholar]
- 18. Bag I., Dale R.K., Palmer C., Lei E.P.. The zinc-finger protein CLAMP promotes gypsy chromatin insulator function in drosophila. J. Cell Sci. 2019; 132:jcs226092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King M.R., Matzat L.H., Dale R.K., Lim S.J., Lei E.P.. The RNA-binding protein rumpelstiltskin antagonizes gypsy chromatin insulator function in a tissue-specific manner. J. Cell Sci. 2014; 127:2956–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurshakova M., Maksimenko O., Golovnin A., Pulina M., Georgieva S., Georgiev P., Krasnov A.. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell. 2007; 27:332–338. [DOI] [PubMed] [Google Scholar]
- 21. Lei E.P., Corces V.G.. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 2006; 38:936–941. [DOI] [PubMed] [Google Scholar]
- 22. Matzat L.H., Dale R.K., Moshkovich N., Lei E.P.. Tissue-specific regulation of chromatin insulator function. PLoS Genet. 2012; 8:e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melnikova L., Molodina V., Erokhin M., Georgiev P., Golovnin A.. HIPP1 stabilizes the interaction between CP190 and su(hw) in the drosophila insulator complex. Sci. Rep. 2019; 9:19102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bag I., Chen S., Rosin L.F., Chen Y., Liu C.Y., Yu G.Y., Lei E.P.. M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity. Nat. Commun. 2021; 12:4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schoborg T., Rickels R., Barrios J., Labrador M.. Chromatin insulator bodies are nuclear structures that form in response to osmotic stress and cell death. J. Cell. Biol. 2013; 202:261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Golovnin A., Volkov I., Georgiev P.. SUMO conjugation is required for the assembly of drosophila su(hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J. Cell Sci. 2012; 125:2064–2074. [DOI] [PubMed] [Google Scholar]
- 27. Golovnin A., Melnikova L., Volkov I., Kostuchenko M., Galkin A.V., Georgiev P.. Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 2008; 9:440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood A.M., Van Bortle K., Ramos E., Takenaka N., Rohrbaugh M., Jones B.C., Jones K.C., Corces V.G.. Regulation of chromatin organization and inducible gene expression by a drosophila insulator. Mol. Cell. 2011; 44:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerasimova T.I., Corces V.G.. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998; 92:511–521. [DOI] [PubMed] [Google Scholar]
- 30. Badenhorst P., Voas M., Rebay I., Wu C.. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002; 16:3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carre C., Ciurciu A., Komonyi O., Jacquier C., Fagegaltier D., Pidoux J., Tricoire H., Tora L., Boros I.M., Antoniewski C.. The drosophila NURF remodelling and the ATAC histone acetylase complexes functionally interact and are required for global chromosome organization. EMBO Rep. 2008; 9:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamiche A., Sandaltzopoulos R., Gdula D.A., Wu C.. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999; 97:833–842. [DOI] [PubMed] [Google Scholar]
- 33. Tsukiyama T., Wu C.. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995; 83:1011–1020. [DOI] [PubMed] [Google Scholar]
- 34. Tsukiyama T., Daniel C., Tamkun J., Wu C.. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995; 83:1021–1026. [DOI] [PubMed] [Google Scholar]
- 35. Martinez-Balbas M.A., Tsukiyama T., Gdula D., Wu C.. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gdula D.A., Sandaltzopoulos R., Tsukiyama T., Ossipow V., Wu C.. Inorganic pyrophosphatase is a component of the drosophila nucleosome remodeling factor complex. Genes Dev. 1998; 12:3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao H., Sandaltzopoulos R., Wang H.M., Hamiche A., Ranallo R., Lee K.M., Fu D., Wu C.. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol. Cell. 2001; 8:531–543. [DOI] [PubMed] [Google Scholar]
- 38. Barak O., Lazzaro M.A., Lane W.S., Speicher D.W., Picketts D.J., Shiekhattar R.. Isolation of human NURF: a regulator of engrailed gene expression. EMBO J. 2003; 22:6089–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwon S.Y., Grisan V., Jang B., Herbert J., Badenhorst P.. Genome-Wide mapping targets of the metazoan chromatin remodeling factor NURF reveals nucleosome remodeling at enhancers, core promoters and gene insulators. PLoS Genet. 2016; 12:e1005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Badenhorst P., Xiao H., Cherbas L., Kwon S.Y., Voas M., Rebay I., Cherbas P., Wu C.. The drosophila nucleosome remodeling factor NURF is required for ecdysteroid signaling and metamorphosis. Genes Dev. 2005; 19:2540–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwon S.Y., Xiao H., Glover B.P., Tjian R., Wu C., Badenhorst P.. The nucleosome remodeling factor (NURF) regulates genes involved in drosophila innate immunity. Dev. Biol. 2008; 316:538–547. [DOI] [PubMed] [Google Scholar]
- 42. Bohla D., Herold M., Panzer I., Buxa M.K., Ali T., Demmers J., Kruger M., Scharfe M., Jarek M., Bartkuhn M.et al.. A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS One. 2014; 9:e107765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li M., Belozerov V.E., Cai H.N.. Modulation of chromatin boundary activities by nucleosome-remodeling activities in drosophila melanogaster. Mol. Cell. Biol. 2010; 30:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Markstein M., Pitsouli C., Villalta C., Celniker S.E., Perrimon N.. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008; 40:476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boutros M., Bras L.P., Huber W.. Analysis of cell-based RNAi screens. Genome Biol. 2006; 7:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J.H., Chung T.D., Oldenburg K.R.. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999; 4:67–73. [DOI] [PubMed] [Google Scholar]
- 47. Moshkovich N., Nisha P., Boyle P.J., Thompson B.A., Dale R.K., Lei E.P.. RNAi-independent role for argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011; 25:1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moshkovich N., Lei E.P.. HP1 recruitment in the absence of argonaute proteins in drosophila. PLoS Genet. 2010; 6:e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Van Bortle K., Nichols M.H., Li L., Ong C.T., Takenaka N., Qin Z.S., Corces V.G.. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014; 15:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He D.C., Nickerson J.A., Penman S.. Core filaments of the nuclear matrix. J. Cell. Biol. 1990; 110:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nazer E., Dale R.K., Chinen M., Radmanesh B., Lei E.P.. Argonaute2 and LaminB modulate gene expression by controlling chromatin topology. PLoS Genet. 2018; 14:e1007276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B.et al.. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fields B.D., Nguyen S.C., Nir G., Kennedy S.. A multiplexed DNA FISH strategy for assessing genome architecture in caenorhabditis elegans. Elife. 2019; 8:e42823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schwartz Y.B., Linder-Basso D., Kharchenko P.V., Tolstorukov M.Y., Kim M., Li H.B., Gorchakov A.A., Minoda A., Shanower G., Alekseyenko A.A.et al.. Nature and function of insulator protein binding sites in the drosophila genome. Genome Res. 2012; 22:2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosin L.F., Nguyen S.C., Joyce E.F.. Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in drosophila nuclei. PLoS Genet. 2018; 14:e1007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ollion J., Cochennec J., Loll F., Escudé C., Boudier T.. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 2013; 29:1840–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rosin L.F., Crocker O., Isenhart R.L., Nguyen S.C., Xu Z., Joyce E.F.. Chromosome territory formation attenuates the translocation potential of cells. Elife. 2019; 8:e49553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011; 2011:17. [Google Scholar]
- 59. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of chip-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dale R.K., Pedersen B.S., Quinlan A.R.. Pybedtools: a flexible python library for manipulating genomic datasets and annotations. Bioinformatics. 2011; 27:3423–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R.et al.. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012; 481:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen K., Xi Y., Pan X., Li Z., Kaestner K., Tyler J., Dent S., He X., Li W.. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013; 23:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sabirov M., Kyrchanova O., Pokholkova G.V., Bonchuk A., Klimenko N., Belova E., Zhimulev I.F., Maksimenko O., Georgiev P.. Mechanism and functional role of the interaction between CP190 and the architectural protein pita in drosophila melanogaster. Epigenetics Chromatin. 2021; 14:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schertel C., Albarca M., Rockel-Bauer C., Kelley N.W., Bischof J., Hens K., van Nimwegen E., Basler K., Deplancke B.. A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating drosophila wing development. Genome Res. 2015; 25:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kudron M.M., Victorsen A., Gevirtzman L., Hillier L.W., Fisher W.W., Vafeados D., Kirkey M., Hammonds A.S., Gersch J., Ammouri H.et al.. The ModERN resource: genome-wide binding profiles for hundreds of drosophila and caenorhabditis elegans transcription factors. Genetics. 2018; 208:937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mito Y., Henikoff J.G., Henikoff S.. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007; 315:1408–1411. [DOI] [PubMed] [Google Scholar]
- 71. Deal R.B., Henikoff J.G., Henikoff S.. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010; 328:1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Baldi S., Jain D.S., Harpprecht L., Zabel A., Scheibe M., Butter F., Straub T., Becker P.B.. Genome-wide rules of nucleosome phasing in drosophila. Mol. Cell. 2018; 72:661–672. [DOI] [PubMed] [Google Scholar]
- 73. Wiechens N., Singh V., Gkikopoulos T., Schofield P., Rocha S., Owen-Hughes T.. The chromatin remodelling enzymes SNF2H and SNF2L position nucleosomes adjacent to CTCF and other transcription factors. PLoS Genet. 2016; 12:e1005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fu Y., Sinha M., Peterson C.L., Weng Z.. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008; 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhao K., Hart C.M., Laemmli U.K.. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995; 81:879–889. [DOI] [PubMed] [Google Scholar]
- 76. Li M., Belozerov V.E., Cai H.N.. Analysis of chromatin boundary activity in drosophila cells. BMC Mol. Biol. 2008; 9:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gerasimova T.I., Byrd K., Corces V.G.. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell. 2000; 6:1025–1035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other relevant data of this study are available from the corresponding author upon reasonable request. The accession numbers for the raw data FASTQ files, processed files, and BigWig files for all sequencing data generated in this study and deposited in NCBI GEO are GSE198361. Bed file for Ebx-eGFP ChIP-seq in Drosophila embryos was obtained from modERN (doi:10.17989/ENCSR559AJG). In this study, all other publicly available sequencing data in Kc167 analyzed are BEAF-32 GSM762845, Ibf1 GSM2133766, Ibf2 GSM2133767, Pita GSM2133768, ZIPIC GSM2133769, M1BP GSM4231397, and CTCF GSM1535983. Mass spectrometry data files have been deposited in the PRIDE repository with dataset identifiers PXD032165 for NURF301.