Abstract
Streptomyces coelicolor (Sc) is a model organism of actinobacteria to study morphological differentiation and production of bioactive metabolites. Sc zinc uptake regulator (Zur) affects both processes by controlling zinc homeostasis. It activates transcription by binding to palindromic Zur-box sequences upstream of −35 elements. Here we deciphered the molecular mechanism by which ScZur interacts with promoter DNA and Sc RNA polymerase (RNAP) by cryo-EM structures and biochemical assays. The ScZur-DNA structures reveal a sequential and cooperative binding of three ScZur dimers surrounding a Zur-box spaced 8 nt upstream from a −35 element. The ScRNAPσHrdB-Zur-DNA structures define protein-protein and protein-DNA interactions involved in the principal housekeeping σHrdB-dependent transcription initiation from a noncanonical promoter with a −10 element lacking the critical adenine residue at position −11 and a TTGCCC −35 element deviating from the canonical TTGACA motif. ScZur interacts with the C-terminal domain of ScRNAP α subunit (αCTD) in a complex structure trapped in an active conformation. Key ScZur-αCTD interfacial residues accounting for ScZur-dependent transcription activation were confirmed by mutational studies. Together, our structural and biochemical results provide a comprehensive model for transcription activation of Zur family regulators.
INTRODUCTION
Streptomyces is the largest genus within the Actinobacteria, producing about half of all clinically used antibiotics and other important pharmaceuticals of natural origin (1). Streptomyces bacteria undergo a complex life cycle of morphological differentiation that is closely associated with the bioactive metabolite production. Both processes are subjected to strict regulation at the transcriptional level. Transcription initiation steps catalysed by RNA polymerase (RNAP) are the most common targets for regulation. As observed in other bacteria, a single RNAP core enzyme (α2ββ’ω) is used by streptomycetes to transcribe all genes, whereas most streptomycetes contain one primary σ factor and multiple alternative σ factors to coordinate expression of genes by recognizing alternative promoters (2,3). Streptomyces coelicolor is a model organism of actinobacteria. Among 64 σ factors encoded in S. coelicolor genome, σHrdB is the principal housekeeping σ factor and analogous to σ70 of Escherichia coli. The presence of σHrdB is essential across streptomycetes. ChIP-seq analysis of σHrdB identifies a total of 2137 protein-coding genes in S. coelicolor (4). Due to its importance in transcription regulation, streptomycete RNAP has been genetically and biochemically characterized (5–7), whereas no structural information is available to date.
Transcription factors bind to specific sequences overlapping or near RNAP binding sites of promoters to repress or activate the expression of genes (8). The catabolite activator protein (CAP; also known as the cAMP receptor protein, CRP) is one of the most studied transcription factors (9). In the presence of the allosteric effector cAMP, it activates transcription at >100 promoters by binding to a 22-base pair (bp) palindromic cis element in Escherichia coli, via either class I or class II mechanism. One of the best characterized class I CAP-dependent promoters is the lac promoter, which has a CAP binding site at position −61.5 (10). Transcription activation at the class I CAP-dependent promoter involves protein-protein interactions between CAP and the carboxyl-terminal domain of the α subunit (αCTD) of RNAP, which enhance RNAP binding to the promoter by a recruitment mechanism. At class II CAP-dependent promoters, CAP binds at position −41.5 and forms protein-protein interactions with multiple subunits of RNAP (11,12). Both recruitment and isomerization mechanisms are involved in transcription activation at class II CAP-dependent promoters. A few bacterial transcription factors, such as members of MerR family, bind between the −10 and −35 elements at target promoters and activate transcription by shortening the length of non-optimal −35/−10 spacer (13,14). Although most transcription activators binding to specific DNA sequences of promoters, actinobacteria have unique transcription factors, such as CarD and RbpA, that active transcription regardless of promoter sequences (15,16).
Zinc (Zn) uptake regulator (Zur) is the most prevalent regulator of Zn uptake genes in bacteria, regulating its regulon genes in response to the fluctuations of the intracellular Zn levels (17,18). It belongs to the ferric uptake regulator (Fur) family, which is a family of bacterial proteins that regulate metal ion uptake to maintain metal homeostasis. Zur is first discovered in E. coli as a repressor of znuABC genes encoding high-affinity Zn importer and further in other bacteria including S. coelicolor, Mycobacterium tuberculosis, Xanthomonas campestris etc. (19–22). In the presence of sufficient Zn, the Zn-bound form of Zur acts predominantly as a transcriptional repressor by blocking RNAP binding sites. In Zn deficiency, the Zn-free form of Zur has a low DNA-binding affinity, leading to the transcription of Zn regulon genes. Available structures reveal that Zur proteins act as a homodimer, of which each monomer contains an N-terminal DNA binding domain (DBD), a C-terminal dimerization domain (DD), and a hinge region linking the two domains (19–22). The Zur may contain two or three Zn sites per monomer. According to structural and biochemical analyses of S. coelicolor Zur (ScZur), the C-site with four conserved cysteines for Zn coordination is a common structural site, while the other two sites are involved in regulatory functions (16). Zn binding in regulatory sites affects the orientation of the two DBD domains and regulates DNA binding. In contrast to the well-studied Zn binding in several Zur proteins, Zur–DNA interaction has only been structurally characterized for E. coli Zur (EcZur), which is crystallized in complex with its target DNA, a conserved 30-bp AT-rich sequence called a Zur-box (19). The EcZur binds to the Zur-box as a dimer of dimers, which is stabilized by a pair of salt bridges between neighbouring monomers. Each monomer of EcZur recognizes the DNA sequence of the Zur-box by both base readout and shape readout.
Although Zur proteins usually act as repressors, they may also activate transcription of genes, at least in S. coelicolor and X. campestris, where Zur proteins activate putative Zn exporters of cation diffusion facilitator (CDF) superfamily (23,24). In X. campestris, footprinting identifies a 59-bp GC-rich sequence with a 20-bp inverted repeat in the upstream region of a CDF gene. Intriguingly, the 59-bp GC-rich sequence shows no significant similarity with the Zur-box of X. campestris. In S. coelicolor, the promoter region of zitB gene encoding a CDF exporter contains a 15-bp Zur-box, that is spaced 8 nt upstream from the −35 element (Supplementary Figure S1). However, in the presence of enough Zn, the Zur protects both the Zur-box motif and the further upstream region up to position −138 relative to the transcription start site (TSS) of zitB (24). The size of the protected regions in both cases suggests that oligomeric Zur binding is required for transcription activation. The effectiveness of cis-regulatory elements in Zn-dependent transcription activation is tested in vivo. The regulatory region up to position −60 only allows partial activation of zitB, whereas the regulatory region up to position −228 enables full gene activation.
The Zur-box in the zitB promoter of S. coelicolor is spaced 8 nt upstream from the −35 element (Supplementary Figure S1A), suggesting that the ScZur may activate transcription via the class II activation mechanism (11,12,24). However, the precise mechanism by which ScZur interacts with zitB promoter DNA and ScRNAP to activate transcription remains obscure. In the present study, we determined cryo-EM structures of ScZur-DNA and ScRNAPσHrdB-Zur-DNA complexes. The ScZur-DNA structures unveil sequential trimerization of ScZur surrounding the Zur-box while the ScRNAPσHrdB-Zur-DNA structures demonstrate that ScZur activates ScRNAPσHrdB by contacting αCTD. These results from structural analysis are supported by the Mango-based in vitro transcription assays. Together, these findings provide a comprehensive model for transcription activation of Zur family regulators.
MATERIALS AND METHODS
Plasmids
All primers used for amplification by PCR are listed in Supplementary Table S1. Genomic DNA of S. coelicolor M145 was used as a template. The rpoA, rpoB, rpoC and rpoZ encoding RNAP core enzyme (α2ββ’ω) were cloned into a single plasmid pET28a to get a final vector, pET28a-RNAPcore. Restriction sites XbaI, NdeI, BamHI, EcoRI and HindIII were used for inserting rpoA, rpoB, rpoC and rpoZ genes. In the resulting vector, β and β’ were fused in frame by inserting a 9 amino-acid polylinker between the C-terminal of β and the N-terminal of β’. An 8xHis tag was engineered on the 3’ end of the connected β and β’ gene. Structures of reported bacterial RNAP complexes show that the two terminals are close to each other and the strategy of fusing β and β’ has been used in a heterogeneous expression of Mycobacterium smegmatis RNAP core (25). The dissociable σHrdB that directs RNAP to promoters of housekeeping genes and the Zur monitoring in vivo Zn were inserted into the pET28a vector via NdeI and EcoRI restriction sites to obtain pET28a-hrdB and pET28a-zur respectively.
ScRNAP core purification
The vector pET28a-RNAPcore was transformed into E. coli strain BL21(DE3) for expression. Cultures were incubated at 37°C, transferred to 16°C when reaching an optical density at 600 nm (OD600) of 0.6, and allowed the cultures to cool for 20 minutes before inducing with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 16 h. Cells were harvested, washed, and resuspended in lysis buffer (50 mM Tris (pH 8.0), 500 mM NaCl, 10 mM MgCl2, 10% glycerol (v/v), 1 mM PMSF, 1 mM β-mercaptoethanol, 0.1 mg/ml sodium deoxycholate and 5 mM imidazole). Following sonication on ice, cell debris was removed by centrifugation at 15 000g for 40 min at 4°C. The resulting supernatant was loaded onto nickel-NTA resin equilibrated with lysis buffer, followed by washing with lysis buffer containing 20 mM imidazole (pH 8.0) and eluting with elution buffer (20 mM Tris (pH 8.0), 200 mM NaCl, 5 mM MgCl2, 10% glycerol (v/v) and 200 mM imidazole). The eluted fractions were pooled and loaded onto a HiTrap Heparin HP affinity column pre-balanced in buffer A (20 mM Tris (pH 8.0), 5 mM MgCl2, 10% glycerol (v/v)). The RNAP core was eluted by buffer B (20 mM Tris (pH 8.0), 1 M NaCl, 5 mM MgCl2, 10% glycerol) that was increased from 0 to 100% in 20 ml. The fractions were pooled, concentrate and polished by a Superose 6 gel filtration column equilibrated with size exclusion chromatography (SEC) buffer (20 mM Tris (pH 8.0), 150 mM NaCl, 2 mM MgCl2, 3 mM DTT and 10% glycerol). The resulting protein was concentrated to 3 mg/ml, and then stored at −80°C (Supplementary Figure S2). The RNAP core carrying truncated or mutated α subunit was prepared by same protocol.
σHrdB purification
Escherichia coli strain BL21(DE3) transformed with pET28a-hrdB was incubated at 37°C until the OD600 reached ∼0.6. Expression was induced with 0.3 mM IPTG at 16°C for 16 h. Cells were harvested, washed, and resuspended in the lysis buffer without MgCl2 and sodium deoxycholate. Cells were lysed by using sonication. Supernatant obtained by centrifugation was applied to nickel-NTA resin equilibrated with lysis buffer, washed with lysis buffer containing 20 mM imidazole (pH 8.0) and eluting with elution buffer. The eluted fractions were pooled, loaded onto a HiTrap Heparin HP affinity column pre-balanced in buffer A and eluted by buffer B that was increased from 0 to 100% in 20 ml. The fractions were pooled, concentrate and polished with a Superdex 200 gel filtration column equilibrated with SEC buffer. The resulting protein was concentrated to 6 mg/ml and stored at −80°C (Supplementary Figure S2).
ScZur purification
Escherichia coli strain BL21(DE3) transformed with pET28a-zur was cultured, harvested and lysed as described for σHrdB. The purification protocol was also similar to σHrdB except that Zur was eluted with lysis buffer containing 250 mM imidazole (pH 8.0) and polished with a Superdex 75 gel filtration column equilibrated with SEC buffer with 1 μM ZnSO4. According to Roe et al. (21), the KD of ScZur for Zn is 0.45–0.78 fM. The resulting holo–Zur (∼1.5 mg/ml) was stored at −80°C. To obtain apo–Zur without Zn, the elution fractions from nickel-NTA column was dialyzed into buffer C (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 5% glycerol and 5 mM EDTA) to remove imidazole and nickel. The EDTA was removed by dialyzing against buffer D (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 10% glycerol and 1 mM DTT), concentrated to 1 mg/ml, and stored at −80°C (Supplementary Figure S2). The Zur mutants were prepared by same protocol.
ScRNAPσHrdB holoenzyme
S. coelicolor RNAP core enzyme and σHrdB were incubated in a 1:2 ratio for 1 h on ice. The reaction mixture was loaded onto Superose 6 gel filtration column equilibrated with SEC buffer and eluted with the same buffer. Fractions containing S. coelicolor RNAP-σHrdB holoenzyme were pooled and stored at −80°C (Supplementary Figure S2).
In vitro transcription
The transcription activities of ScRNAPσHrdB in the absence or presence of ScZur are evaluated by measuring the fluorescence intensities of Mango III (26,27). A 296 bp DNA fragment consisting of the zitB promoter (−228 to + 15) and the mango III coding sequence were synthesized and cloned into pUC19 via restriction sites EcoRI and HindIII (Supplementary Figure S1). The resulting plasmid was used as a template to amplify the DNA scaffold carrying the zitB regulatory region up to −68 (TP−68), −100 (TP−100) or −140 (TP−140), and the mango III by using primers listed in Supplementary Table S1. Mango III transcription assays were performed in black 96-well plates. 0 or 1 μM ScZur was mixed with 10 nM nucleic acid scaffold in 100 μl transcription buffer (20 mM Tris–HCl, pH 8.0, 0.1 M KCl, 5 mM MgCl2, 20 μM ZnSO4, 2 mM DTT, 4 U RNaseIn, 1 μM TO1–PEG–biotin, and 5% Glycerol) at room temperature for 10 min, followed by supplementation of 0.1 μM ScRNAPσHrdB and 1 mM NTPs. The reactions were incubated for 10 min at 30°C and stopped by 0.5 ng heparin. Corresponding reactions without NTP were used as blanks. Fluorescence emission intensities were measured using SpectraMax Id5 Multi-Mode Microplate Reader (Molecular Devices) with excitation wavelength of 510 nm and emission wavelength of 535 nm.
Electrophoretic mobility shift assay (EMSA) for ScZur–DNA binding
The dsDNA containing a 5′-conjugated FAM was prepared by annealing complementary oligonucleotides (10 μM each, Supplementary Table S1) in 20 mM Tris pH 8.0 by heating the reaction to 95°C for 10 min and allowing it to cool to room temperature slowly. For Zn-dependent EMSA reactions, 1 ng of labelled dsDNA and 100 nM purified apo-ScZur or mutants were mixed in 10 μl binding buffer (20 mM Tris–HCl (pH 8.0), 100 mM KCl, 2 mM DTT, 0.1 mg/ml BSA, 5% glycerol, 0.1 mg of poly(dI-dC) and 0–40 μM ZnSO4) and incubated at room temperature for 20 min. For Zur-dependent EMSA reactions, 0–800 nM purified ScZur or mutants and 1 ng of labelled dsDNA were mixed in 10 μl binding buffer (20 mM Tris–HCl (pH 8.0), 100 mM KCl, 2 mM DTT, 0.1 mg/ml BSA, 5% glycerol, 0.1 mg of poly(dI-dC) and 20 μM ZnSO4) and incubated at room temperature for 20 min. The binding mixture was then subjected to electrophoresis at 4°C on a 5% polyacrylamide gel at 130 V in TB (89 mM Trizma base, 89 mM boric acid) buffer, followed by a visualization using Amersham Typhoon RGB Biomolecular Imager with Cy2 filters. Signals from all shifted bands were combined to obtain the bound fraction, and the signal from non-shifted band was used for non-bound fraction. Data from three independent experiments were fitted to a Hill equation to obtain the nh and the KD-ave.
Assembly of ScRNAPσHrdB–Zur–DNA complex
The zitB promoter DNA carrying 13-bp (zitB13) or 6-bp (zitB6) non-complementary transcription bubble were prepared by annealing nontemplate (nt) strand (28 μM final) and template (t) strand (30 μM) in annealing buffer (10 mM Tris (pH 8.0), 50 mM NaCl, 10 mM MgCl2). RNA (5′-GUAGG-3′; 42 μM) was supplemented for preparing nucleic-acid scaffold carrying 13-bp non-complementary transcription bubble. The reactions were heated to 95°C for 10 minutes and cooled to room temperature slowly. To assemble ScRNAPσHrdB-Zur-DNA, 270 μl core enzyme of 11 μM and 20 μl σHrdB of 243 μM were added into 700 μl assembling buffer (20 mM HEPES pH 8.0, 50 mM KCl, 5 mM MgCl2, 20 μM ZnSO4, 3 mM DTT) and incubated for 10 min at room temperature, followed by supplementation of 100 μl promoter DNA of 28 μM and incubation of another 10 minutes. The complexes were loaded onto a Superose 6 10/300 GL column (GE Healthcare), eluted by assembling buffer, concentrated to 7 mg/ml and stored at −80°C (Supplementary Figure S2).
Cryo-EM data acquisition
Two batches of data (5550 movies in total) were collected for ScRNAPσHrdB-Zur-zitB13. 3 μl ScRNAPσHrdB-Zur-zitB13 of 0.8 mg/ml were applied to Quantifoil R1.2/1.3 Au 300 mesh grids freshly glow-discharged at 25 mA for 60 s. Grids were blotted for 2 s at 16°C with 100% chamber humidity and then plunge-frozen in liquid ethane in a Vitrobot (FEI, Inc.). 3523 movies were collected by using SerialEM on Titan Krios 300 kV microscopes with Gantan K2 detector operated in counting mode with a pixel size of 1.1 Å/pixel, a defocus ranging from −1.0 to −2.0 μm, and a total dose of 40 e–/Å2 fractionated into 32 frames. 2027 movies were collected by Gantan K3 detector using EPU with the same pixel size, defocus ranging and dose fractionation.
For data acquisition of ScRNAPσHrdB-Zur-zitB6, 3 μl samples of 1.0 mg/ml were used for preparing the grids as described above. 15 232 movies were collected by using SerialEM on Titan Krios 300 kV microscopes with Gantan K2 detectors operated in counting mode. Exposures of a total dose of 40 e–/Å2 were fractionated into 32 frames with a pixel size of 1.05 Å/pixel and a defocus ranging from −1.0 to −2.0 μm.
Cryo-EM data processing
Datasets ofScRNAPσHrdB-Zur-zitB13 acquired with Gantan K2 detector were subjected to motion correction using RELION’s own implementation of the UCSF MotionCor2 program in RELION-3.1 (28). The aligned images were imported into CryoSPARC suite v3.3.1 (29) and subjected to patch CTF estimation, manually curate exposures, and template picker with good 2D classes from blob picker as templates, followed by two rounds of 2D classification of picked particles. Datasets acquired with Gantan K3 detector were processed by the same protocol. Particles of good 2D classes of the two datasets were combined and subjected to the third round of the 2D classification, followed by ab-Initio reconstruction, homogeneous refinement, non-uniform refinement and 3D variability analysis. Good 3D classes were selected, iteratively refined with C1 symmetry, and subjected to local resolution estimation and local filtering to produce final maps. The final particle stacks were subjected to particle subtraction to keep only the signal of ScZur region, followed by masked local refinements to improve the map quality and interpretability (Supplementary Figure S3).
2D class averages corresponding to ScZur-DNA unexpectedly obtained in the 2D classification of picked particles from ScRNAPσHrdB-Zur-zitB13 sample and were used as the templates to pick ScZur-DNA particles from the two datasets respectively in CryoSPARC suite v3.3.1 (29). The picked particles were subjected to two rounds of 2D classification respectively before combination. The combined particles were subjected to the third round of the 2D classification. Particles in good 2D classes were selected and subjected to ab-initio reconstruction, followed by iterative refinement, local resolution estimation, and local filtering to produce final maps (Supplementary Figure S4).
For the datasets of ScRNAPσHrdB-Zur-zitB6, dose-fractionated movies were subjected to patch motion correction, patch CTF estimation, manually curate exposures, and template picker using good 2D classes from ScRNAPσHrdB-Zur-zitB13 as templates, followed by three rounds of 2D classification of picked particles and ab-Initio reconstruction in CryoSPARC suite v3.3.1 (29). Particles in good 2D classes were selected and imported into RELION-3.1 using csparc2star.py module for re-extraction, 3D auto-refinement with the model from ab-Initio reconstruction as a starting reference, CTF refinement and Bayesian polishing (28). The polished particles were imported back into CryoSPARC suite v3.3.1 again for non-uniform refinement, 3D variability analysis and 3D classification. Good 3D classes were selected, iteratively refined with C1 symmetry, and subjected to local resolution estimation and local filtering to produce final maps. The ScZur region was subjected to masked local refinement with subtracted particle to improve the map quality and interpretability (Supplementary Figure S5). The charge density distribution map of a local refinement map was prepared as described by Wang et al. (30).
Cryo-EM model building and refinement
The initial atomic models of ScRNAPσHrdB subunits were generated by SWISS-MODEL (31). The crystal structure of Zn-bound ScZur (21) and the homology model of ScRNAPσHrdB were fitted into the cryo-EM density maps using ChimeraX (32). The model of the promoter DNA and the RNA oligomer were built manually in Coot (33). The coordinates were refined in Phenix with secondary structure restraints, rotamer restraints and Ramachandran restraints (34). The final coordinates were validated using MolProbity (35). The Map versus Model FSCs of five cryo-EM maps and their corresponding atomic models of this study were generated by Phenix (Supplementary Figure S6). The statistics of cryo-EM refinement were summarized in Supplementary Table S2.
RESULTS
Zur activates zitB via Zur-box and further upstream regions
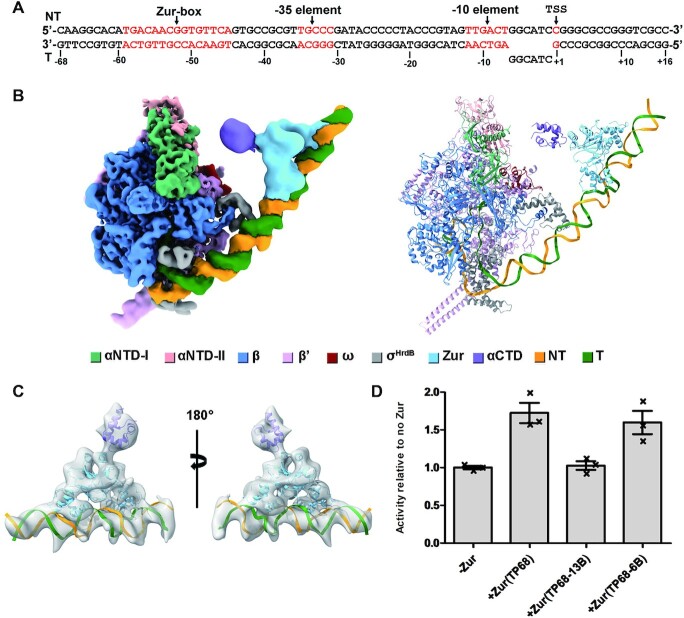
We purified the RNAP core enzyme, and housekeeping σHrdB of S. coelicolor for in vitro run-off transcription reactions. Although the zitB promoter lacks the critical adenine residue at position −11 (Supplementary Figure S1A), it can be recognized by σHrdB in previous in vitro transcription reactions (24). ScZur protects a region of 98 bp (−138 to −40) in a footprint on zitB promoter. In addition to the 7−1−7 Zur-box (−45 to −59), inspection of the 98 bp DNA sequence identified another two palindrome (−67 to −93 nt, and −117 to −133), of which both show similarity with the Zur box (Supplementary Figure S1B). Previous in vivo assays demonstrate that the zitB regulatory region with the Zur-box motif alone can only allow marginal gene activation (∼1.3-fold), whereas the upstream sequence up to −228 nt enables full gene activation (∼6.5-fold) (24). The DNA templates used for the in vitro run-off transcription reactions contain the zitB region from −140, −100 or −68 to + 15 respectively, followed by sequence encoding Mango III (36,37), which is a fluorogenic RNA aptamer used for direct readout of transcription (Figure 1, Supplementary Figure S1A). As expected, the ScRNAP-σHrdB holoenzyme can initiate basal transcription of all three DNA templates. In the presence of the ScZur, transcription was enhanced ∼1.6-fold for the DNA template containing the zitB region from −68 to +15 (TP−68), whereas the level of enhancement observed in transcription reactions of the other two templates (up to −100 or −140, TP−100 or TP−140) was more than 4-fold.
Figure 1.
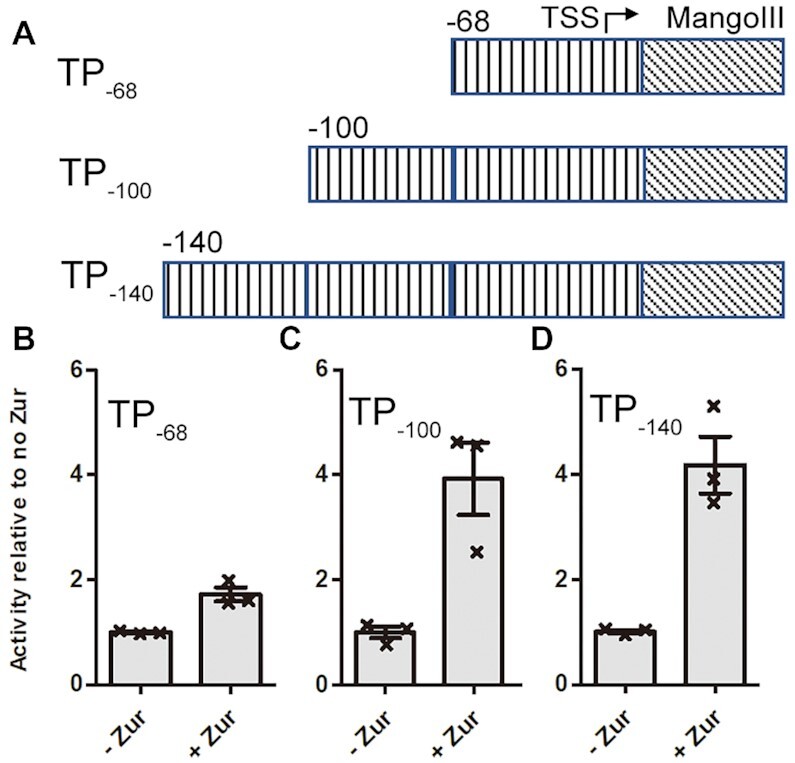
Zur activates transcription by RNAP-σHrdB holoenzyme on the zitB promoter. (A) DNA template used for in vitro run-off transcription reactions. The DNA templates contain the zitB region from −68 (TP−68), −100 (TP−100) or −140 (TP−140) to + 15 respectively, followed by Mango III encoding an RNA fluorogenic aptamer, which becomes fluorescent when bound to TO1-Biotin. (B–D) Transcription is enhanced ∼1.6-fold by Zur for the DNA template TP−68 (B), whereas the level of enhancement for the templates TP−100 (C) and Template TP−140 (D) is more than 4-fold. Data are presented as mean ± SEM from three independent assays.
A 33 bp zitB Zur-box DNA probe (−68 to −36) has been proposed to binding two ScZur dimers according to the estimated molecular weight of the retarded band in previous EMSA assays (18). We observed three retarded bands in the EMSA assay at 20 μM ZnSO4 with ScZur from 0 to 800 nM (Supplementary Figure S7). An average dissociation constant KD-ave of 34.0 ± 0.7 nM and a Hill coefficient nh of 2.3 ± 0.1 were obtained from a Hill Plot. We examined ScZur binding to DNA sequences containing palindromes 1 or 2, using DNA probes from −141 to −109 or from −101 to −69. Oligomeric ScZur binding was observed on both DNA probes at higher concentration (200 nM) compared to the Zur-box DNA probe (6.25 nM) (Supplementary Figure S8). The palindromic 7−1−7 Zur-box contains highly conserved GC base pairs at positions 2, 8 and 14 (Supplementary Figure S1B), which are the signatures of the Zur-box. The modified zitB Zur-box DNA probe (M1) with the three GC base pairs replaced by AT base pairs showed a KD-ave of 97.0 ± 7.7 nM and an nh of 1.6 ± 0.2 (Supplementary Figure S7B). Replacing the corresponding second G by C in EcZur-box results in ∼4 times increase of the KD-ave (from 2.9 ± 0.8 to 11.9 ± 0.4 nM), while replacing the 14th C by G results in ∼13 times increase of the KD-ave (from 2.9 ± 0.8 to 39.1 ± 1.3 nM) (19). Consequently, a zitB Zur-box M2 probe was constructed by introducing C, T and G at positions 2, 8 and 14, showing a KD-ave of 223.1 ± 30.5 nM and an nh of 0.9 ± 0.1 (Supplementary Figure S7C). Introducing T and A at positions 3 and 13 into the M2 probe further decreased the binding of ScZur (Supplementary Figure S7D). No binding of ScZur was observed on the DNA probe downstream of the Zur-box (−29 to +50) (24). We replaced the sequence from −100 to −69 of the TP−100 by the sequence from +18 to +49 of zitB to obtain a hybrid template TP−100H (Supplementary Figure S8C and D). In the presence of ScZur, transcription was enhanced ∼1.7-fold, indicating that Zur binding to the upstream regions is necessary for full activation.
ScZur binding to Zur-box as a trimer of dimers
ScZur shows low to moderate sequence identity to Zur of E. coli (21%), X. campestris (24%) and M. tuberculosis (55%) (19,20,22). The α3 recognition helices of DBD domains are highly conserved, accounting for their Zur-box similarity (Supplementary Figure S9). The structure of Zn-bound ScZur has been reported(21), whereas the DNA-bound structure is still unavailable. The cryo-EM structures of ScZur-promoter complexes were unexpectedly obtained during an initial attempt to assemble ScRNAPσHrdB–Zur–DNA complexes. In the course of cryo-EM data processing, 2D classification revealed the presence of ScZur–DNA complexes. 3D classification of the complexes revealed two structures, which were determined at overall resolutions of 3.4 Å (a trimer of dimers, TOD) and 3.8 Å (a dimer of dimers, DOD) respectively (Supplementary Figure S4). The two structures of ScZur-promoter complexes are essentially the same except that the ScZur dimer close to the upstream terminal of the promoter DNA (DIII) is missing in the DOD structure (Figure 2, Supplementary Figure S10). The three ScZur dimers in TOD complex protect the DNA sequence from −68 to −36, which comprises the 7−1−7 Zur-box (−59 to −45). The ScZur dimer in the middle (DII) sits in the Zur-box and interact with the palindrome by both monomers. In contrast, the other two ScZur dimers (DI and DIII) make contacts with the Zur-box by using only one monomer (Figure 3). Interactions with the Zur-box are indispensable since ScZur binding to DNA downstream of the Zur-box is not observed. The three retarded bands observed in EMSA assays of the zitB Zur-box DNA probe and the TOD and DOD structures suggest a sequential binding of each ScZur dimer. Superimposition of ScZur dimer with EcZur dimer gives an rmsd of 2.4 Å (187 aligned Cα atoms). The first N-terminal helix observed in EcZur is missing in ScZur (Supplementary Figure S9A). The EcZur binds to its cognate DNA as a dimer of dimers instead of the trimer of dimer observed in the ScZur structure (14). The relative orientation between dimers is different. When the two structures are superposed according to DI of the ScZur-promoter complex, the rotation and displacement between the second dimers are 53° and 21 Å respectively (Supplementary Figure S11). Compared to the EcZur−DNA complex, the ScZur binding only results in slight bends in the target DNA. The ScZur-promoter structures reveal direct protein-protein interactions between adjacent dimers. D37 and H41 residues of each DII monomer interact with their counterparts in the neighbouring dimers (Figure 2D). In EMSA analysis, retarded bands of high molecular weights appear as Zn increased, however the enhancement of Zn on oligomeric ScZur binding is partially reduced when D37 and/or H41 are mutated to alanine. Compared to the wild type protein, all mutants display a reduction of transcription activation (Supplementary Figure S12).
Figure 2.
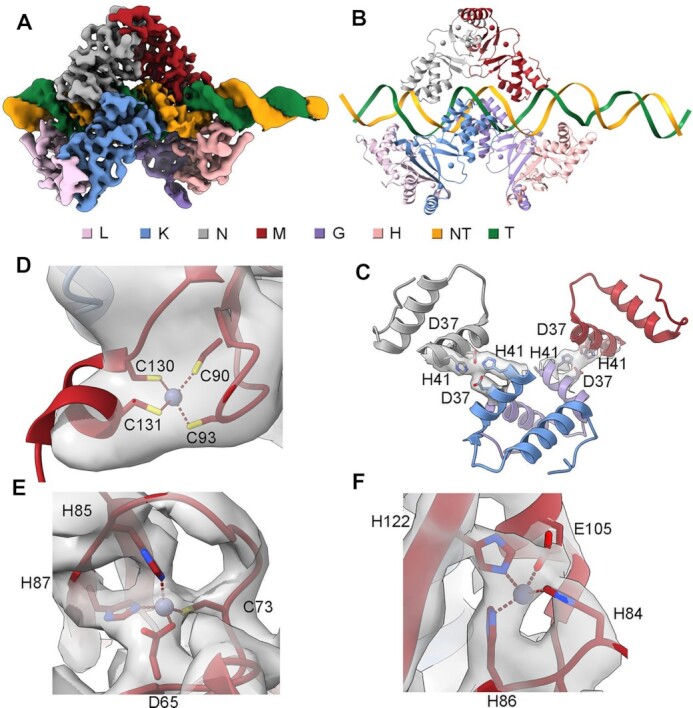
Structures of ScZur–DNA TOD complex. (A) Cryo-EM map of ScZur–DNA TOD complex. (B) Structure of ScZur–DNA TOD. (C) D37 and H41 residues involved in dimer-dimer interactions. (D–F) Density maps for Zn binding sites. ScZur subunits and DNA are colored as in the color key. Zn atoms are shown as spheres. Residues involved in dimer-dimer interaction and Zn binding are shown as sticks. Density maps are shown as transparent surface. The figures were prepared by Chimera X.
Figure 3.
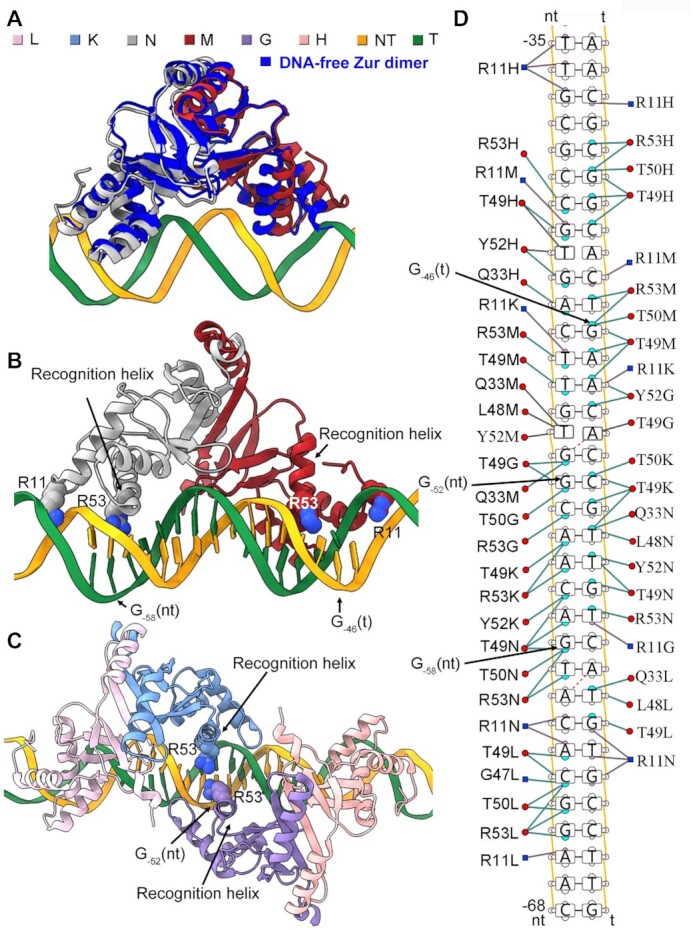
Interaction between ScZur and targeted dsDNA. (A) Superimposition of the DNA-bound ScZur dimer with the DNA-free ScZur dimer. Distance between the α1 helices of the two monomers is increased upon DNA binding. (B) The R53 residues of two DII monomers interact with the GC base pair at positions 2 (G−58(nt)) and 14 (G−46(t)) of the Zur-box respectively. (C) The GC base pair at position 8 (G−52(nt)) is close to the R53 residues of both DI monomer and DIII monomer. (D) Protein-nucleic acid interactions between ScZur dimers and the targeted dsDNA. The figures were prepared by Chimera X and DNAproDB.
Overall conformations of the two monomers of the dimers are essentially the same with an overall rmsd of 1.1 Å. Moderate differences (rmsd 1.6 Å) are revealed by superimposition of the DNA-bound ScZur dimer with the DNA-free ScZur dimer (16). DNA binding results in an increased distance (from 27 to 34 Å) between the N-terminals of the recognition helices of the two monomers (measured from the Ca atoms of L48 residues) (Figure 3A). Otherwise, the α1 N-terminal would conflict with the phosphodiester backbone of the DNA duplex. The ScZur dimer utilizes two recognition helices (Figure 3B), each from one monomer, to make contacts with the DNA in the major groove, by using residues R53, which is conserved in Zur proteins (Supplementary Figure S9). The R53A mutation compromises both the DNA binding and the transcriptional activation of ScZur (Supplementary Figure S13). The R53 residues of two DII monomers interact with the GC base pair at positions 2 (G−58(nt)) and 14 (G−46(t)) respectively (Figure 3B), whereas the single GC base pair at position 8 (G−52(nt)) is close to the R53 residues of both DI monomer and DIII monomer (Figure 3C). Considering the conservative palindromic 7–1−7 structure, the binding mode of a trimer of dimers is likely to be applicable to other Zur-box sites in S. coelicolor. Conserved R11 of the L1 loop is inserted into the minor groove (Figure 3B). In contrast to the R53A mutation, the R11A mutation slightly compromises the DNA binding but significantly decreases the transcriptional activation (Supplementary Figure S13B and H). The R11 side chains bind at the relatively narrow regions of the minor-groove in the ScZur-promoter structures. The minor groove topographies of the unbound DNA were probed by hydroxyl radical cleavage pattern ORChID2, which correlates with the width of the minor groove (38). The analysis revealed that the minor-groove regions that R11 side chains contact are intrinsically narrow in the unbound DNA (Supplementary Figure S14). In many DNA binding proteins, the Arg residue inserting into the minor groove helps recognize a DNA binding site by a shape readout mechanism (39). Analysis by DNAproDB, a web-based tool for protein–DNA complex analysis (40), revealed a large number of interactions between ScZur dimers and promoter dsDNA, involving Q33, L48, T49, T50 and Y52 residues (Figure 3D). Mutating R53, L48, T50 and Y52 impaired the ability of DNA binding of ScZur. In contrast, mutating R11, Q33, and T49 only slightly reduced DNA binding (Supplementary Figure S13). Mutating these residues also decreased activation of ScZur in transcription assays of TP−68, whereas no correlation between transcription activities and DNA-binding abilities of the mutants could be observed. The TP−100 was used in the transcription assays of T49A and Y52A, and the transcription activities correlated with their DNA-binding abilities.
Overall structure of ScRNAPσHrdB–Zur–DNA
Pre-melted promoter DNA fragments are often used to assembly RNAP-promoter open complex (RPo) and RNAP-promoter initial transcribing complex (RPitc) to improve the sample homogeneity and enhance protein–DNA interaction. Initially, we designed an 84-bp zitB promoter DNA fragment comprising a 57-bp (−68 to −12) upstream promoter double-stranded DNA (dsDNA), a 13-bp (−11 to +2) non-complementary transcription bubble, and a 14-bp (+3 to +16) downstream promoter dsDNA (Supplementary Figure S10A). To assemble the RPitc complex, ScZur was incubated with the 84-bp zitB promoter DNA carrying the 13-bp non-complementary bubble (zitB13) followed by supplementation of the ScRNAPσHrdB holoenzyme. We added a 5-nt RNA primer to stabilize the template single-stranded DNA (ssDNA) of the unwound transcription bubble. The RPitc structures carrying one (ScRNAPσHrdB−Zurd1−zitB13) or two ScZur dimers (ScRNAPσHrdB−Zurd2−zitB13) were obtained at 4.1 and 3.7 Å resolution respectively (Supplementary Figure S3). The Zur regions are visible at a lower contour level compared to the RNAP regions in the density maps, suggesting their flexibilities. Interactions between ScZur and ScRNAPσHrdB are not observed. In vitro transcriptional assays were carried out by using a template with the zitB13 promoter carrying a pre-melted 13-bp bubble. The results reveal that the ScZur does not enhance the transcription of the zitB13 promoter (Figure 4), suggesting that the RPitc with a pre-melted 13-bp bubble may be not a target of ScZur activation.
Figure 4.
The overall structure of ScRNAPσHrdB-Zurd1-zitB6. (A) DNA scaffold with a 6-bp pre-melted bubble used in structure determination. The zur-box, −35 element, and −10 element are highlighted in red. (B) The cryo-EM density map and structure model of ScRNAPσHrdB-Zurd1-zitB6. The density map and cartoon representation structure are colored as indicated in the color key. NT, non-template-strand promoter DNA; T, template-strand promoter DNA. The map was generated by merging the consensus map of the full ScRNAPσHrdB-Zurd1-zitB6 complex and the focused map of the Zurd1-DNA subcomplex from the complex (Supplementary Figure S5) in Chimera X. (C) Two views of the charge density distribution map generated from the cryo-EM map of local 3D refinement focused on the ScZurd1 region. (D) ScZur activates transcription from zitB6 promoter with a 6-bp pre-melted bubble instead of zitB13 with a 13-bp pre-melted bubble. Figures of density maps and structures are prepared by Chimera X.
Next, we synthesized a similar 84-bp zitB promoter DNA fragment comprising a pre-melted 6-bp (−6 to −1) bubble (zitB6) (Figure 4A). The transcription of the zitB6 promoter was enhanced by the ScZur in the Mango-based in vitro assays (Figure 4D) and was used to assemble the RPo complexes. 127 088 single particles were obtained from 2D classification and used to reconstruct a map of 3.3–5 Å resolution at the ScRNAPσHrdB and 8–12 Å resolution at the ScZur. The low resolution of the ScZur region compared to the ScRNAPσHrdB region in the density map indicates that the ScZur region is flexible as observed in the ScRNAPσHrdB–Zur–zitB13. The density of the ScZur dimer binding at the upstream terminal of the promoter DNA is visible at a lower contour level compared to the rest portions in the density map. We observed extra density between ScZur and ScRNAP at a lower contour level. The density is close to ScRNAP αNTD, suggesting that it likely belongs to ScRNAP αCTD. We used 3D variability analysis (3DVA), an algorithm that fits a linear subspace model of conformational change to cryo-EM data (41), to visualize the detailed molecular motions of ScRNAPσHrdB−Zur−zitB6 (Movie 1) and to cluster the particles into 10 classes (Supplementary Figure S5D). The ninth class carrying one ScZur dimer (ScRNAPσHrdB−Zurd1−zitB6) is refined to a resolution of 3.8–7 Å at ScRNAPσHrdB and 10–15 Å at ScZur (Figure 4B, Supplementary Figure S5D). Local refinement focused on the ScZurd1 region generated a 11-Å-resolution map. A charge density distribution map tends to be better resolved than the corresponding EM map (30). We then used the 11-Å-resolution map to generate a charge density distribution map of ∼7 Å using Laplacian (30), which allowed more confident docking of ScZur and ScRNAP αCTD components (Figure 4C) and demonstrated direct contact between ScZur and ScRNAP αCTD. Particles of another seven classes were combined and subjected to 3D classification focused on ScZur region, generating density maps of RPo complexes carrying DOD (ScRNAPσHrdB−Zurd2−zitB6) or TOD (ScRNAPσHrdB−Zurd3−zitB6) of ScZur (Supplementary Figure S5D). The densities for the ScRNAP αCTD domains are visible at a lower contour level in both maps. ScRNAP αCTD domains docked in both maps are distant from the ScZur dimers (Figure 5, Supplementary Figure S15), whereas the flexibility of this regions suggests the possibility of ScZur to directly contact ScRNAP αCTD.
Figure 5.
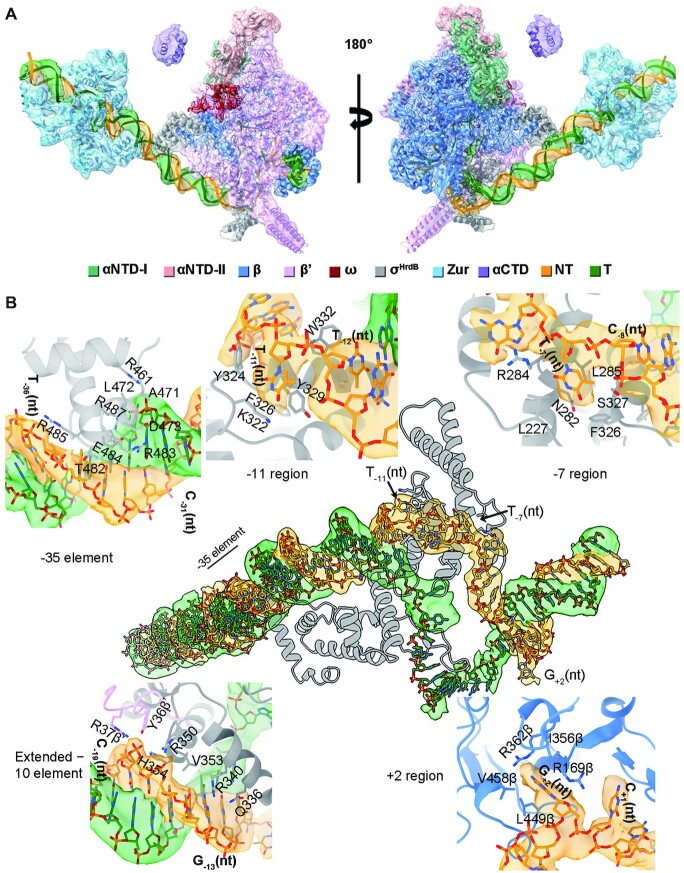
Interactions between ScRNAPσHrdB and promoter DNA. (A)Two views of the density map and structure of ScRNAPσHrdB–Zurd3–zitB6. The density map shown as transparent surface was generated by merging the consensus map of the full ScRNAPσHrdB–Zurd3–zitB6 complex and the focused map of the Zurd3–DNA subcomplex from the complex (Supplementary Figure S5) in Chimera X. The map and cartoon representation structure are colored as indicated in the color key. (B) Interactions between promoter DNA and ScRNAPσHrdB. The density map of DNA is shown as transparent surface. The residues involved in the interactions are shown as sticks. The figures were prepared by Chimera X.
ScRNAPσHrdB–promoter DNA interactions
Among the three ScZur-bound RPo complexes, map quality was highest for ScRNAPσHrdB–Zurd3–zitB6 and therefore subsequent analysis focussed on this structure. The final cryo-EM map was reconstructed using a total of 54 799 single particles and refined to an overall 3.45-Å resolution. The resolution at the ScZur region was ∼ 10 Å and increased to 5.37 Å by local refinement (Supplementary Figure S5D). The 84-bp promoter DNA (−68 to +16), four major regions of the σHrdB subunit (σ1.2, σ2, σ3 and σ4), five subunits of ScRNAP core (α2ββ’ω), three ScZur dimers, and αCTD were docked into the map (Figure 5A). The overall structure of ScRNAP closely resembles the reported actinobacteria RNAP structures of M. tuberculosis RPo (PDB ID: 6VVY) and M. smegmatis RPitc (PDB ID: 5VI5) with rmsd of 1.079 Å (2759 Cα aligned) and 1.047 Å (2636 Cα aligned) respectively (Supplementary Figure S16) (42,43). The σ2 and σ4 of σHrdB occupy the same positions and make the same interactions with RNAP (35–37). Actinobacteria RNAPs contain a lineage-specific insertion of ∼90 residues that folds into two long anti-parallel α-helices at the N-terminal of the β’ subunit (44). In the ScRNAP, the lineage-specific insertion (β’i1) roughly spans residues 141–228. A large non-conserved region (NCR) is inserted between regions 1.2 and 2.1 in housekeeping σ factors, such as E. coli σ70. As observed in σA factors of M. tuberculosis and M. smegmatis, the NCR region of S. coelicolor σHrdB is significantly shorter compared to that of E. coli σ70 (Supplementary Figure S16C and D) (45).
Only 20% of streptomycete promoters contain similar sequences to those recognized by E. coli σ70 (46). The −10 element of zitB promoter (−12 to −7, TTGACT) lacks the critical adenine residue at position −11, while the −35 element (−36 to −31, TTGCCC) deviates from the canonical TTGACA motif (24). The RPo structures reveal numerous interactions between ScRNAPσHrdB and the zitB promoter (Figure 5). ScRNAPσHrdB recognizes the promoter DNA by interactions between the σHrdB and the conserved −10 and −35 DNA elements (Figure 5B). The σ4 binds to the −35 element in its major groove. R483 and E484 extend into the major groove and likely make base-specific polar interactions with nucleotides of −35 element. T482 and R485 make interactions with the phosphate backbone of the non-template strand while R461, A471, L472, D473 and R487 contact the phosphate backbone of template strand. Duplex DNA between the −35 and −10 elements contributes to RNAP-promoter interaction through the phosphodiester backbone of the non-template strand. The −17/−18 ‘Z-element’ interacts with Y36 and R37 of β’ zipper, V353 and H354 of σ3.0, and R350 of σ2.4 (38). S. coelicolor promoters lacking the conserved −35 element often have a conserved GG motif (−13 and −14) (39), of which G-13(nt) contacts side chains of Q336 and R340 of σ2.4.
The σ2 domain interacts with the −10 element of the non-template strand in essentially the same manner as that previously observed in RNAP structures with housekeeping σA subunits (45,47,48). The invariant W-dyad (W332/W333, corresponding to W433/W434 in E. coli or W256/W257 in T. aquaticus) maintains the ds/ss DNA junction at the upstream edge of the transcription bubble, where W332 forms a stack with the base of T−12(nt). Bases of both T−11(nt) (corresponding to the conserved A−11(nt) of a canonical −10 element) and T−7(nt) are flipped out and captured by a pocket on σ2 formed by K322, Y324 and Y329, and a pocket made of L227 of σ1.2, and Asn282, R284, Leu285, Phe326 and Ser327 of σ2 respectively (Figure 5B). The ‘discriminator’ (DSR) region of non-template strand (−6 to −4) interacts with residues of σ1.2. The ‘core recognition element’ (CRE) (−3 to +2) interacts with residues of β-subunit, of which R169, I356, R362, L449 and V458 form a pocket to accommodate G+2(nt). The template strand of −10 element is bent by ∼90° at position −11, wraps around the σ3 globular domain, and passes through a cleft between σ2 and σ3 to place the downstream template strand ssDNA into the main primary channel. However, the density of the template ssDNA in the transcription bubble is not well-defined.
Map quality of the ScRNAPσHrdB−Zurd2−zitB13 was better among the two RPitc complexes (Supplementary Figure S3D). The ScRNAP in the ScRNAPσHrdB−Zurd2−zitB13 is similar to that in the ScRNAPσHrdB−Zurd3−zitB6, whereas the density of the template strand ssDNA in the transcription bubble is better resolved (Supplementary Figure S17). The 5-nt RNA base-pairing with nucleotides from −4 to + 1 of template strand DNA is in a post-translocation state with the 5’ end reaching the σ3.2 loop. The fork-loop 2 of β subunit maintains the ds/ss DNA junction at the downstream edge of the transcription bubble by separating the dsDNA (+3 to +16) inside the DNA binding clamp between the β′ clamp, β′ jaw and β lobe.
Zur actives transcription by contacting αCTD
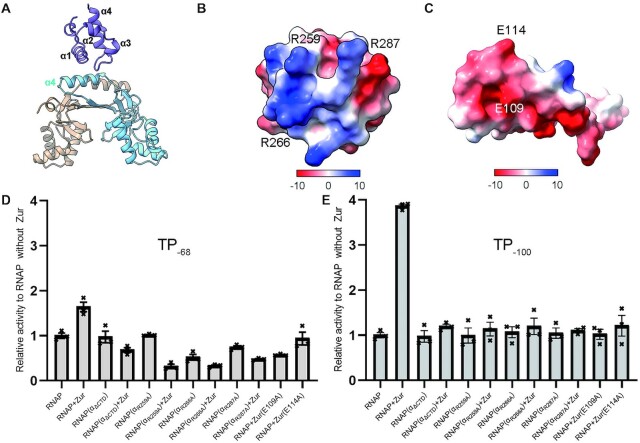
ScRNAPσHrdB−Zurd1−zitB6 complex is trapped in an active conformation in which ScZur makes interaction with ScRNAP αCTD, suggesting it is a ScZur-dependent transcription activation complex (TAC) (Figure 4). The α4 helix of C-terminal dimerization domain of one ScZur monomer packs against the α1 helix and the loop region between α3 and α4 helices of ScRNAP αCTD, burying an interfacial area of ∼200 Å2 (Figure 6). The involvement of ScRNAP αCTD in ScZur-dependent transcription activation is supported by Mango-based transcription assay with ScRNAPσHrdB assemble from a truncated α subunit without CTD region (αΔCTD) that spans residues 1–227. As shown in Figure 6D and E, the αCTD is dispensable for basal transcription of zitB but is required for Zur-dependent activation. At the interface, R259, R266 and R287 of αCTD cluster to form a positively charged patch (Figure 6B) that is complementary to the negatively charged patch formed by E109 and E114 of the Zur (Figure 6C). Mutational studies and in vitro transcriptional assays confirmed the importance of these residues for ScZur-dependent transcription activation (Figure 6D and E). Intriguingly, ScZur repressed transcription of RNAP assembled from αΔCTD and α with a single mutant in transcription assays of TP−68 (Figure 6D). Zur E109A mutant also repressed transcription of TP−68. The reason of these negative effects is unclear and requires further investigation. However, negative effects are not observed in transcription assays of TP−100 (Figure 6E). In the prototypical class II TAP-TAC (TAP, homolog of E. coli CAP in T. thermophilus, PDB ID: 5I2D), the αCTD also uses the α1 helix and the loop region between α3 and α4 helices to interact with the activating region 4 (AR4) of one TAP monomer, burying a larger surface area of ∼300 Å2 (11) (Supplementary Figure S18B). However, the distribution of surface charge in this region of T. thermophilus αCTD is mostly negative and different from the corresponding region that is positively charged in S. coelicolor αCTD.
Figure 6.
Interactions between ScZur and ScRNAP αCTD. (A) ScZur interacts with the α1 helix and the loop region between α3 and α4 helices of ScRNAP αCTD by its α4 helix of one monomer. (B) The positively charged interface of ScRNAP αCTD patch composed of R259, R266 and R287. (C) The negatively charged interface of ScZur composed of E109 and E114. (D) Transcription assays of TP−68. ScZur repressed transcription of RNAP assembled from αΔCTD and α with single mutant. Zur E109A mutant also repressed transcription. The reason of these negative effects requires further investigation. (E) Transcription assays of TP−100. ScZur shows no activation on ScRNAPσHrdB assemble from a truncated α subunit without CTD (αΔCTD). Mutating interfacial residues impaired transcription activation. Figures of structures are prepared by Chimera X. Transcription data are presented as mean ± SEM from three independent assays.
DISCUSSION
Although S. coelicolor is a model organism for the study of antibiotic-producing and morphological development of actinobacteria, no structural information is known about the promoter recognition mechanisms employed by its RNAP. Here, we provide the first structures of ScRNAP-σHrdB with a promoter. These structures show overall similarity to previously reported structures of RNAPs with σA factors. σHrdB occupies the same positions, makes the same interactions with RNAP, and uses the same strategy to recognize, bind and unwind promoter DNA. The structures demonstrate protein-protein and protein–DNA interactions involved in σHrdB-dependent transcription initiation in S. coelicolor. The actinobacteria-specific insertion β’i1 of S. coelicolor RNAP folds into two long anti-parallel α-helices as observed in RNAP structures M. tuberculosis and M. smegmatis (42,43). The conserved structural feature of the actinobacteria β’i1 indicates a functional role requiring further investigations (Supplementary Figure S16). Housekeeping σ factors such as E. coli σ70 and its orthologs possess an NCR of variable length between regions 1.2 and 2.1. The NCR of housekeeping σ factors is a target for transcription regulation (49,50). S. coelicolor σHrdB contains a significantly shorter NCR region compared to that of E. coli σ70, suggesting a different regulatory strategy is adopted by σHrdB.
Numerous positive regulators have been revealed in streptomycetes, whereas how they activate gene transcription remains obscure. Here, we show that ScZur activates zitB transcription by binding to the Zur-box positioned 8 nt upstream from the −35 element and contacting ScRNAP αCTD using the α4 helix involved in dimerization (Figure 6). The importance of interface residues is supported by Mango-based transcription assays with corresponding mutants. Sequence alignment of ScZur with the counterparts from E. coli, M. tuberculosis and X. campestris reveals that the E109 and E114 residues are not conserved (Supplementary Figure S9), suggesting the protein interaction observed here may be strain-specific (19,20,22). RNAP αCTD is a central target of transcription regulation (10,11). It is connected to αNTD by a long, flexible linker and therefore can occupy different positions relative to the remainder of RNAP. In E. coli, the RNAP αCTD recognizes AT-rich UP elements located upstream of −35 elements (from −60 to −40) in some promoters to enhance transcription (51). Various transcription factors activate RNAP by interacting with αCTD. Structures of prototypical class I and class II TAC have been reported. Obvious differences in the interfaces between αCTD and transcription activator were revealed by comparing our structure and the previously reported structures (Supplementary Figure S18). In the prototypical class I CAP-TAC (PDB ID: 6B6H), the αCTD binds both the CAP activator and the minor groove in the −40 to −45 region (10) (Supplementary Figure S18A). The 287 determinant of α3 helix and the C-terminal loop make contact with the activation region 1 (AR1) of one CAP monomer, burying a small interfacial area of ∼80 Å2. The αCTD also interacts with the 596 determinant of σ4 binding at the major groove in the −35 element mainly with its 261 determinant. In the prototypical class II TAP-TAC (TAP, homolog of CAP PDB ID: 5I2D), the αCTD uses same structural elements as observed in our structure to interact with one TAP monomer, whereas the electrostatic nature is different from that of S. coelicolor αCTD (Supplementary Figure S18B) (11). Among the three ScRNAP αCTD residues interacting with ScZur, R259 is highly conserved while R266 and R287 are only conserved in αCTD of gram-positive bacteria (Supplementary Figure S19). The recently reported B. subtilis Spx-TAC reveals a novel interface between αCTD and Spx (52). The αCTD packs against the activator using the α1 helix and the preceding loop, and buries a remarkably larger surface area of ∼700 Å2 (Supplementary Figure S18C).
We tried to assemble the ScRNAPσHrdB–Zur–DNA from both zitB6 and zitB13 promoters. The interaction between αCTD and Zur is only observed in the ScRNAPσHrdB–Zurd1−zitB6 structure. Mango-based in vitro assays reveal that ScZur can activate transcription from zitB6 promoter instead of zitB13, indicating that ScZur affects steps prior to the formation of the 13-bp transcription bubble (Figure 4D). In vivo assays demonstrate that zitB gene is gradually activated by ScZur (24). The zitB regulatory region up to −60 only allows gene activation of ∼1.3 folds while the upstream region up to −228 nt enables gene activation of ∼6 folds. We identified two additional palindromes (−67 to −93 nt, and −117 to −133) in the zitB promoter (Supplementary Figure S1). According to this information, three DNA templates were designed and used for the in vitro run-off transcription reactions. Consistent with the in vivo results, transcription of the template containing the zitB regulatory region up to −68 was enhanced ∼1.6-fold by the Zur protein while the enhancement for the other two templates up to −100 and −140 was more than 4-fold (Figure 1B-D), indicating that full activation of zitB depends on Zur binding at palindrome II and/or I. In the case of Fur-activated genes, the Fur-box sequence is located −100 to −200 bp upstream of the TSS (53). An activator binding to a DNA region located this far distance from TSS has been proposed to activate transcription by a looping mechanism (24). An upward DNA bend toward RNAP is observed in the ScZur region in the ScRNAPσHrdB−Zurd1−zitB6 structure (Supplementary Figure S20). However, it is unclear whether the DNA bending is prior to or upon binding of ScZur.
We provide the first structural insight into DNA-binding characteristics of ScZur (Figure 2). Structural comparisons of apo and holo ScZur proteins reveal binding of Zn atoms in the regulatory sites drives a conformational change activating Zur proteins (22). We observed that DNA triggers a conformational change of the DNA-binding domains. The distance between the recognition helices increased from 27 to 34 Å so that the helices are better aligned with consecutive major groove regions (Figure 3A), enabling the formation of protein-nucleic acid interactions. DNA-induced movements of the DNA-binding domains are observed in Magnetospirillum gryphiswaldense Fur–DNA complex structures, where the distance between the N-terminals of recognition helices decreased from 39 Å to 32 Å (measured from the Ca atoms of I52 residues) (54). In contrast, the DNA-induced conformational change is not observed in the structures of diphtheria toxin repressor (DtxR) activated by transition metal ions (55,56). The TOD structure of ScZur-promoter complex explains the extended footprint in previous assays (24). The structure and biochemical results demonstrate that three ScZur dimers cooperatively and sequentially binding to the target DNA region with the first and third ScZur dimer docking on one side and the second ScZur dimer in the middle docking on the opposite side, surrounding the palindromic 7–1−7 Zur-box.
In summary, our structural and biochemical results demonstrate that ScZur proteins cooperatively bind to target DNA as a trimer of dimers and activates transcriptions by contacting ScRNAP αCTD. These findings provide a mechanistic framework for understanding Zur-dependent transcription activation and may also shed light on transcription activation of other Fur-family regulators.
DATA AVAILABILITY
Atomic coordinates of 7 structures have been deposited in PDB with accession numbers 7VO9 (Zur-DNA DOD), 7VO0 (Zur-DNA TOD), 7VPD (ScRNAPσHrdB-Zurd1-zitB6), 7X76 (ScRNAPσHrdB-Zurd2-zitB6), 7X75 (ScRNAPσHrdB-Zurd3-zitB6), 7VPZ (ScRNAPσHrdB-Zurd1-zitB13) and 7X74 (ScRNAPσHrdB-Zurd2-zitB13). 12 cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession number EMD-32049 (Zur-DNA DOD), EMD-32048 (Zur-DNA TOD), EMDB-32063 (ScRNAPσHrdB-Zurd1-zitB6), EMDB-32033 (ScRNAPσHrdB-Zurd2-zitB6), EMDB-33032 (ScRNAPσHrdB-Zurd3-zitB6), EMDB-32077 (ScRNAPσHrdB-Zurd1-zitB13) and EMDB-33031 (ScRNAPσHrdB-Zurd2-zitB13), EMDB-33034 (Zurd1-CTD-DNA from ScRNAPσHrdB–Zurd1-zitB6), EMDB-33035 (Zurd2-DNA from ScRNAPσHrdB-Zurd2-zitB6), EMDB-33036 (Zurd3-DNA from ScRNAPσHrdB-Zurd3-zitB6), EMDB-33037 (Zurd1-DNA from ScRNAPσHrdB-Zurd1-zitB13) and EMDB-33038 (Zurd1-DNA from ScRNAPσHrdB-Zurd2-zitB13).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jing Liu, Yao Han and Xinqiu Guo at the Instrument Analysis Center (IAC) of Shanghai Jiao Tong University, and Fangfang Wang of the Electron Microscopy System at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, China for providing assistance in data collection.
Contributor Information
Xu Yang, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Yiqun Wang, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Guiyang Liu, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Zixin Deng, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Shuangjun Lin, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China.
Jianting Zheng, State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China; Joint International Research Laboratory of Metabolic & Developmental Sciences, Shanghai Jiao Tong University, Shanghai 200240, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2019YFA0905400]; National Natural Science Foundation of China [32070040]. Funding for open access charge: National Key R&D Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ward A.C., Allenby N.E.. Genome mining for the search and discovery of bioactive compounds: the streptomyces paradigm. FEMS Microbiol. Lett. 2018; 365:fny240. [DOI] [PubMed] [Google Scholar]
- 2. Casas-Pastor D., Muller R.R., Jaenicke S., Brinkrolf K., Becker A., Buttner M.J., Gross C.A., Mascher T., Goesmann A., Fritz G.. Expansion and re-classification of the extracytoplasmic function (ECF) sigma factor family. Nucleic Acids Res. 2021; 49:986–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murakami K.S. Structural biology of bacterial RNA polymerase. Biomolecules. 2015; 5:848–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smidova K., Zikova A., Pospisil J., Schwarz M., Bobek J., Vohradsky J.. DNA mapping and kinetic modeling of the HrdB regulon in streptomyces coelicolor. Nucleic Acids Res. 2019; 47:621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim J.E., Choi J.S., Kim J.S., Cho Y.H., Roe J.H.. Lysine acetylation of the housekeeping sigma factor enhances the activity of the RNA polymerase holoenzyme. Nucleic Acids Res. 2020; 48:2401–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kerr L., Hoskisson P.A.. Reconciling DNA replication and transcription in a hyphal organism: visualizing transcription complexes in live streptomyces coelicolor. Microbiology. 2019; 165:1086–1094. [DOI] [PubMed] [Google Scholar]
- 7. Tabib-Salazar A., Liu B., Doughty P., Lewis R.A., Ghosh S., Parsy M.L., Simpson P.J., O’Dwyer K., Matthews S.J., Paget M.S.. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res. 2013; 41:5679–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 9. Lawson C.L., Swigon D., Murakami K.S., Darst S.A., Berman H.M., Ebright R.H.. Catabolite activator protein: DNA binding and transcription activation. Curr Opin. Struct. Biol. 2004; 14:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu B., Hong C., Huang R.K., Yu Z., Steitz T.A.. Structural basis of bacterial transcription activation. Science. 2017; 358:947–951. [DOI] [PubMed] [Google Scholar]
- 11. Feng Y., Zhang Y., Ebright R.H.. Structural basis of transcription activation. Science. 2016; 352:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi W., Jiang Y., Deng Y., Dong Z., Liu B.. Visualization of two architectures in class-II CAP-dependent transcription activation. PLoS Biol. 2020; 18:e3000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang C.L., Philips S.J., Wu X.X., Chen K., Shi J., Shen L.Q., Xu J.C., Feng Y., O’Halloran T.V., Zhang Y.. CueR activates transcription through a DNA distortion mechanism. Nat. Chem. Biol. 2021; 17:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y., Liu C., Zhou W., Shi W., Chen M., Zhang B., Schatz D.G., Hu Y., Liu B.. Structural visualization of transcription activated by a multidrug-sensing MerR family regulator. Nat. Commun. 2021; 12:2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D., Darst S.A., Campbell E.A., Paget M.S.. Structural, functional, and genetic analyses of the actinobacterial transcription factor rbpA. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:7171–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis E., Chen J., Leon K., Darst S.A., Campbell E.A.. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by carD. Nucleic Acids Res. 2015; 43:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kandari D., Joshi H., Bhatnagar R.. Zur: zinc-sensing transcriptional regulator in a diverse set of bacterial species. Pathogens. 2021; 10:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mikhaylina A., Ksibe A.Z., Scanlan D.J., Blindauer C.A.. Bacterial zinc uptake regulator proteins and their regulons. Biochem. Soc. Trans. 2018; 46:983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilston B.A., Wang S., Marcus M.D., Canalizo-Hernandez M.A., Swindell E.P., Xue Y., Mondragon A., O’Halloran T.V.. Structural and mechanistic basis of zinc regulation across the e. coli zur regulon. PLoS Biol. 2014; 12:e1001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lucarelli D., Russo S., Garman E., Milano A., Meyer-Klaucke W., Pohl E.. Crystal structure and function of the zinc uptake regulator FurB from mycobacterium tuberculosis. J. Biol. Chem. 2007; 282:9914–9922. [DOI] [PubMed] [Google Scholar]
- 21. Shin J.H., Jung H.J., An Y.J., Cho Y.B., Cha S.S., Roe J.H.. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in zur. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5045–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu F., Su Z., Chen P., Tian X., Wu L., Tang D.J., Li P., Deng H., Ding P., Fu Q.et al.. Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator. Nucleic Acids Res. 2021; 49:6511–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang D.L., Tang D.J., Liao Q., Li H.C., Chen Q., He Y.Q., Feng J.X., Jiang B.L., Lu G.T., Chen B.et al.. The zur of xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res. 2008; 36:4295–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi S.H., Lee K.L., Shin J.H., Cho Y.B., Cha S.S., Roe J.H.. Zinc-dependent regulation of zinc import and export genes by zur. Nat. Commun. 2017; 8:15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kouba T., Pospisil J., Hnilicova J., Sanderova H., Barvik I., Krasny L.. The core and holoenzyme forms of RNA polymerase from mycobacterium smegmatis. J. Bacteriol. 2019; 201:e00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi J., Wen A., Jin S., Gao B., Huang Y., Feng Y.. Transcription activation by a sliding clamp. Nat. Commun. 2021; 12:1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trachman R.J. 3rd, Autour A., Jeng S.C.Y., Abdolahzadeh A., Andreoni A., Cojocaru R., Garipov R., Dolgosheina E.V., Knutson J.R., Ryckelynck M.et al.. Structure and functional reselection of the Mango-III fluorogenic RNA aptamer. Nat. Chem. Biol. 2019; 15:472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fernandez-Leiro R., Scheres S.H.W.. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol. 2017; 73:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 30. Wang J. Experimental charge density from electron microscopic maps. Protein Sci. 2017; 26:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L.et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF chimerax: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J.et al.. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019; 75:861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B.et al.. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018; 27:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shi J., Gao X., Tian T., Yu Z., Gao B., Wen A., You L., Chang S., Zhang X., Zhang Y.et al.. Structural basis of Q-dependent transcription antitermination. Nat. Commun. 2019; 10:2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Autour A., S C.Y.J., A D.C., Abdolahzadeh A., Galli A., Panchapakesan S.S.S., Rueda D., Ryckelynck M., Unrau P.J. Fluorogenic RNA mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018; 9:656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bishop E.P., Rohs R., Parker S.C., West S.M., Liu P., Mann R.S., Honig B., Tullius T.D.. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chem. Biol. 2011; 6:1314–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B.. The role of DNA shape in protein–DNA recognition. Nature. 2009; 461:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sagendorf J.M., Markarian N., Berman H.M., Rohs R.. DNAproDB: an expanded database and web-based tool for structural analysis of DNA-protein complexes. Nucleic Acids Res. 2020; 48:D277–D287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Punjani A., Fleet D.J.. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021; 213:107702. [DOI] [PubMed] [Google Scholar]
- 42. Lilic M., Chen J., Boyaci H., Braffman N., Hubin E.A., Herrmann J., Muller R., Mooney R., Landick R., Darst S.A.et al.. The antibiotic sorangicin a inhibits promoter DNA unwinding in a mycobacterium tuberculosis rifampicin-resistant RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:30423–30432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hubin E.A., Lilic M., Darst S.A., Campbell E.A.. Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 2017; 8:16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lane W.J., Darst S.A.. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010; 395:671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuo Y., Steitz T.A.. Crystal structures of the e. coli transcription initiation complexes with a complete bubble. Mol. Cell. 2015; 58:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kang J.G., Hahn M.Y., Ishihama A., Roe J.H.. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from streptomyces coelicolor A3(2). Nucleic Acids Res. 1997; 25:2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S.A.. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife. 2015; 4:e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S., Darst S.A., Campbell E.A.. Structure and function of the mycobacterial transcription initiation complex with the essential regulator rbpA. Elife. 2017; 6:e22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bae B., Chen J., Davis E., Leon K., Darst S.A., Campbell E.A.. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife. 2015; 4:e08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shin Y., Qayyum M.Z., Pupov D., Esyunina D., Kulbachinskiy A., Murakami K.S.. Structural basis of ribosomal RNA transcription regulation. Nat. Commun. 2021; 12:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi J., Li F., Wen A., Yu L., Wang L., Wang F., Jin Y., Jin S., Feng Y., Lin W.. Structural basis of transcription activation by the global regulator spx. Nucleic Acids Res. 2021; 49:10756–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Teixido L., Carrasco B., Alonso J.C., Barbe J., Campoy S.. Fur activates the expression of salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS One. 2011; 6:e19711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng Z.Q., Wang Q., Liu Z., Zhang M.F., Machado A.C.D., Chiu T.P., Feng C., Zhang Q., Yu L., Qi L.et al.. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 2015; 6:7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. D’Aquino J.A., Tetenbaum-Novatt J., White A., Berkovitch F., Ringe D. Mechanism of metal ion activation of the diphtheria toxin repressor dtxR. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:18408–18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. White A., Ding X., vanderSpek J.C., Murphy J.R., Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998; 394:502–506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of 7 structures have been deposited in PDB with accession numbers 7VO9 (Zur-DNA DOD), 7VO0 (Zur-DNA TOD), 7VPD (ScRNAPσHrdB-Zurd1-zitB6), 7X76 (ScRNAPσHrdB-Zurd2-zitB6), 7X75 (ScRNAPσHrdB-Zurd3-zitB6), 7VPZ (ScRNAPσHrdB-Zurd1-zitB13) and 7X74 (ScRNAPσHrdB-Zurd2-zitB13). 12 cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession number EMD-32049 (Zur-DNA DOD), EMD-32048 (Zur-DNA TOD), EMDB-32063 (ScRNAPσHrdB-Zurd1-zitB6), EMDB-32033 (ScRNAPσHrdB-Zurd2-zitB6), EMDB-33032 (ScRNAPσHrdB-Zurd3-zitB6), EMDB-32077 (ScRNAPσHrdB-Zurd1-zitB13) and EMDB-33031 (ScRNAPσHrdB-Zurd2-zitB13), EMDB-33034 (Zurd1-CTD-DNA from ScRNAPσHrdB–Zurd1-zitB6), EMDB-33035 (Zurd2-DNA from ScRNAPσHrdB-Zurd2-zitB6), EMDB-33036 (Zurd3-DNA from ScRNAPσHrdB-Zurd3-zitB6), EMDB-33037 (Zurd1-DNA from ScRNAPσHrdB-Zurd1-zitB13) and EMDB-33038 (Zurd1-DNA from ScRNAPσHrdB-Zurd2-zitB13).