Abstract
Both membrane-bound and periplasmic nitrate reductases have been found in denitrifying bacteria. Yet the role of periplasmic nitrate reductase in denitrification has not been clearly defined. To analyze the function of the periplasmic nitrate reductase in Pseudomonas sp. strain G-179, the nap gene cluster was identified and found to be linked to genes involved in reduction of nitrite and nitric oxide and anaerobic heme biosynthesis. Mutation in the nap region rendered the cells incapable of growing under anaerobic conditions with nitrate as the alternative electron acceptor. No nitrate reduction activity was detected in the Nap− mutant, but that activity could be restored by complementation with the nap region. Unlike the membrane-bound nitrate reductase, the nitrate reduction activity in strain G-179 was not inhibited by a low concentration of azide. Nor could it use NADH as the electron donor to reduce nitrate or use chlorate as the alternative substrate. These results suggest that the periplasmic nitrate reductase in this strain plays a primary role in dissimilatory nitrate reduction.
The complete pathway for microbial denitrification has been established as NO3− → NO2− → NO → N2O → N2 (26, 29). Denitrification normally occurs under oxygen-limiting conditions. It plays a major role in completing the nitrogen cycle by converting nitrate or nitrite to nitrogen gas. In practical applications, microbial denitrification has been widely used for wastewater treatment and water purification (16). On the other hand, N2O has been shown to have detrimental effects on the stratospheric ozone layer (10). Nitrogen oxides (NOx), along with CO and hydrocarbons, can lead to an increase in the amount of tropospheric ozone. Thus, production of N2O and NO due to incomplete denitrification is of concern.
Two types of dissimilatory nitrate reductases have been found in denitrifying bacteria. One is known as the respiratory membrane-bound nitrate reductase and the other as the periplasmic nitrate reductase (13, 29). The membrane-bound enzyme has been studied in Pseudomonas aeruginosa (8), Paracoccus denitrificans (6, 11), Pseudomonas stutzeri (7), Pseudomonas fluorescens (18), and most extensively in Escherichia coli (13). The membrane-bound nitrate reductase consists of three polypeptides α, β, and γ. The large subunit (α) contains the cofactor, molybdopterin guanine dinucleotide, which is the active site of the enzyme. The enzyme is anchored to the cytoplasmic membrane by the γ subunit. All three peptides are commonly encoded by the narGHJI gene cluster. The periplasmic nitrate reductase consists of two subunits, NapA and NapB (4, 5). NapA is the large subunit, which contains the molybdenum cofactor and a [4Fe-4S] cluster. NapB is a c-type cytochrome. NapC is presumably membrane bound and functions as the electron transporter. The nap gene cluster has been characterized in a number of denitrifying bacteria, including Ralstonia eutropha (23) and P. denitrificans (5).
In addition to their differences in protein structure and gene composition, the membrane-bound and the periplasmic nitrate reductases have unique biochemical properties. The membrane-bound enzyme can reduce chlorate and use NADH as the electron donor (2, 9). Its activity is inhibited by low concentrations of azide. The periplasmic nitrate reductase is not sensitive to low concentrations of azide and cannot reduce chlorate or use NADH as the electron donor. Therefore, the two enzymatic activities can be distinguished based on these properties.
Both membrane-bound and periplasmic nitrate reductases are present in P. denitrificans (2). The periplasmic enzyme is expressed aerobically as well as anaerobically; however, the majority of the nitrate reduction activity under anaerobic conditions is contributed by the membrane-bound enzyme. It is proposed that the periplasmic nitrate reductase catalyzes the first step of aerobic denitrification, since this organism is found to be capable of aerobic denitrification. Mutation in the membrane-bound nitrate reductase does not affect growth on nitrate under anaerobic conditions, probably due to the presence of the periplasmic enzyme (3). In fact, the expression of the periplasmic nitrate reductase in this mutant is increased under anaerobic conditions. In R. eutropha, the periplasmic nitrate reductase is not required for denitrification and the NAP− mutant shows only a delay in growth after transition from aerobic to anaerobic respiration (23). It has been suggested that the periplasmic nitrate reductase plays a role in that transition. It has also been speculated that periplasmic nitrate reduction is used by organisms to dispose of excess reducing power (19, 20). To define the function of the periplasmic nitrate reductase in Pseudomonas sp. strain G-179, the nap region was characterized in this study. Both genetic and biochemical analyses indicated that the periplasmic nitrate reductase was the primary enzyme responsible for catalyzing the first step of denitrification in this organism.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Table 1 lists all the strains and plasmids used in this study. The suicide plasmid pARO180 (17) used for construction of insertion mutants was purchased from the American Type Culture Collection.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| G-179 | Wild type, Rifr | 25 |
| RTC07 | norB::Tn5, Tn5 derivative of RTC01, Rifr, Kamr | 25 |
| RTC13 | norD::Tn5, Tn5 derivative of RTC01, Rifr, Kamr | 25 |
| TW01 | ΔnapABC::Kam, Rifr, Kamr | This study |
| Plasmids | ||
| pARO180 | Apr, oriT from RP4 | 17 |
| pTJS75 | Broad-host range vector, IncP1, Tetr | 22 |
| pNAP01 | 5.5-kb KpnI-HindIII fragment with napFDABC region | This study |
| pNAP02 | 4.2-kb KpnI-BglII fragment containing napFDAB region | This study |
| pNAP03 | 3.5-kb KpnI-BamHI containing intact napFDA region | This study |
| pNAP05 | 1.9-kb BamHI-HindIII fragment downstream from napB | This study |
| pET-21a(+) | Apr | Novagen |
| pNAPA | 2.5-kb NdeI-BamHI fragment containing napA in pET-21a(+) | This study |
Growth conditions.
Pseudomonas sp. strain G-179 was grown in tryptic soy broth (TSB) at 28°C. The concentrations for both rifampin and kanamycin were 50 μg/ml when used. The concentrations for tetracycline were 3 and 12 μg/ml, respectively, for broth and solid medium. To grow wild-type strain G-179 under denitrifying conditions, potassium nitrate was added at a concentration of 1.5 g/liter. For mutant strain TW01 (NAP−), 5 mM sodium nitrite was used for anaerobic growth. To introduce plasmid DNA into the G-179 strain, triparental mating with pRK2013 as the helper was performed. Conjugation was carried out on TSB plates at 28°C. Exconjugants were selected on TSB plates with rifampin (50 μg/ml) and kanamycin (50 μg/ml) or tetracyline (12 μg/ml), depending on the plasmid used. For growth on nitrous oxide, TSB in a serum bottle was saturated with nitrous oxide before inoculation, and the gas phase was again filled with nitrous oxide after inoculation.
Determination of nitrate, nitrite, and chlorate concentrations.
Ion chromatography (Dionex Corporation, Sunnyvale, Calif.) using an IONPAC AS11-HC analytical column and an autosampler was used to measure nitrate, nitrite, and chlorate. The eluent used was NaOH at a concentration of 0.6 g/liter with a flow rate of 1.5 ml/min. Detection and quantification are based on conductivity.
Measurement of nitrate reduction activity.
Whole-cell nitrate reduction activity was measured with cells grown under anaerobic conditions and with cells grown under microaerobic conditions. To induce denitrification under microaerobic conditions, wild-type and mutant strains were grown in 1-liter flasks containing 600 ml of TSB medium supplemented with potassium nitrate (1.5 g/liter). Flasks were shaken at 125 rpm. The cells were washed with TSB medium till no nitrate or nitrite was detected with a nitrate or nitrite strip (EM Science, Gibbstown, N.J.). Cell pellets were resuspended in 10 ml of TSB. An aliquot of 2 ml of cell suspension was used for the whole-cell assay carried out in a serum bottle filled with argon gas. Disappearance of nitrate or appearance of nitrite was measured.
The assay for nitrate reductase activity with artificial electron donors was modified from methods used previously (7, 11). The reaction mixture in 1 ml contained an enzyme sample, 200 μM methyl viologen or benzyl viologen, 5 mM potassium nitrate, and 50 mM potassium phosphate buffer (pH 7.3). The reaction was started with 50 μl of freshly prepared sodium dithionite solution (16 mg/ml) in 0.8% NaHCO3. The reaction was stopped by adding 100-μl aliquots into 0.9 ml of NaOH solution (0.6 g/liter), the eluent for ion chromatography. The reaction without sodium dithionite was used as a control.
Isolation of periplasmic proteins was based on the method described for type-2 spheroplasts by Alefounder and Ferguson (1). The EDTA concentration was increased to 5 mM. The periplasmic fraction was loaded onto a native gel without dialysis. The native gel was run at 4°C, and the nitrate reductase activity was detected by incubating the gel in 2-mg/ml methyl viologen solution containing 50 mM Tris (pH 7.6) and 5 mM potassium nitrate. Methyl viologen was reduced by dithionite.
Isolation of cosmid clones.
To identify the DNA region responsible for reduction of nitrate, nitrite, and nitric oxide, the DNA fragments with the Tn5 insertion from previously studied mutant strains RTC07 and RTC13 (25) were isolated and used to probe the DNA cosmid library of the wild-type G-179 strain. Genomic DNA was isolated with the genomic DNA isolation kit from Qiagen (Santa Clarita, Calif.). To construct the DNA library, the wild-type DNA was partially digested with Sau3A and the fractions of >20 kb in size were ligated into the BamHI site of the SuperCos 1 vector from Stratagene (Menasha, Wis.). To isolate DNA fragments with Tn5 insertions, genomic DNA from mutant strains was digested with EcoRI and BamHI and ligated into pUC18. Positive clones were picked from Luria-Bertani plates containing 50 μg of kanamycin/ml. Inserts from these clones were labeled with the nonradioactive DNA Labeling Kit from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Colony hybridization was carried out with the Chemiluminescent Detection Kit provided by the same company. A total of three cosmid clones were isolated, and they all hybridized to the nirK probe of G-179. Sequencing results revealed that strains RTC07 and RTC13 had Tn5 inserted in the nor region.
Construction of insertion mutant.
The 5.5-kb KpnI-HindIII fragment containing the napEFABC region was cloned into the pARO180 vector. An internal SalI-BglII region was deleted and replaced by a kanamycin resistance cassette. The construct was introduced into the wild-type G-179 strain by conjugation. After 6 to 12 passes in TSB medium containing rifampin and kanamycin, colonies with double crossovers were selected based on poor growth on ampicillin-containing plates (500 μg/ml). Insertions were confirmed by Southern blot analysis with the 5.5-kb KpnI-HindIII fragment as a probe. The mutant strain selected for detailed study was designated TW01.
Overexpression of NapA protein.
The DNA region containing the napA gene was amplified by PCR. An NdeI site was incorporated into the translational start site in the first primer (5′-ACGTACGTACATATGACGGCAGAACTCACGCGGCGTGATGTGC-3′). A six-histidine tail before the stop codon and a BamHI site after the stop codon were introduced in the second primer (5′-TACGGAT CCTCGAGTCAGTGATGGTGGTGATGGTGGGCGACGGGAAGGATCT TGACTGC-3′). The PCR product was cloned into the NdeI and BamHI sites of the pET-21a(+) vector (Novagen, Madison, Wis.), resulting in construct pNAPA. This plasmid was introduced into Escherichia coli BL21(DE3) for overexpression. The NapA protein with a His tag was primarily found in inclusion bodies after isopropyl-β-d-thiogalactopyranoside induction. The protein was purified with Ni-nitrilotriacetic acid spin columns under denaturing conditions as described by the manufacturer (Novagen). The NapA protein was further purified by cutting the protein band from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. The identity of NapA was verified by sequencing the N terminus. Polyclonal antibody against the NapA protein was generated in rabbits by Cocalico Biologicals, Inc. (Reamstown, Pa.).
Nucleotide sequence accession number.
The sequence reported in Fig. 1 has been deposited in the GenBank database under accession no. AF083948. The sequence for 16S rDNA (genes coding for rRNA) of G-179 is under accession no. AF109171.
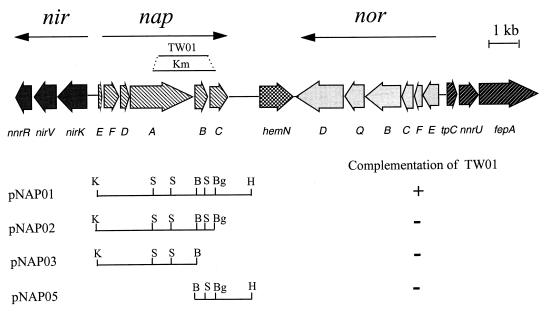
FIG. 1.
Organization of the gene clusters involved in reduction of nitrate, nitrite, and nitric oxide. K, KpnI; B, BamHI; Bg, BglII; H, HindIII; S, SalI.
RESULTS
Gene organization of the DNA region containing the nap gene cluster.
The structural gene of Cu-type nitrite reductase (nirK) has been previously isolated and characterized in Pseudomonas sp. strain G-179 (27). Sequence analysis of its downstream region in this study revealed a nap gene cluster containing genes for the periplasmic nitrate reductase (Fig. 1). The first open reading frame (ORF) in this cluster is the napE gene, which has been previously identified by comparing the nap gene cluster from P. denitrificans with the partial sequence upstream of the nirK gene (5). The second ORF shows about 30% identity with the ferredoxin-type electron transport protein NapF from E. coli (12). Following napF is an ORF that encodes a protein with 25% identity to the putative cytoplasmic protein NapD from P. denitrificans (5). NapD and NapF have been found to be important for optimal periplasmic nitrate reductase activity (21). The derived protein product from the ORF after napD is NapA, which has about 76% identity to the large subunit of periplasmic nitrate reductase from P. denitrificans. A conserved [4Fe-4S] binding motif was found near the N terminus. In fact, the proposed specificity-determining regions between these two proteins are very similar, and the likely molybdenum ligand cysteine (cysteine-203 of the NapA precursor in strain G-179) is conserved. The region containing the first 30 amino acids is very hydrophobic and could serve as the leader peptide. Hydropathy analysis indicated that the mature NapA was hydrophilic, suggesting NapA of G-179 is located in the periplasmic space, similar to the NapA protein of P. denitrificans. The NapB subunit of G-179 also appears to have a leader peptide, and Ala-32 of the NapB precursor could be the first residue of the mature protein. NapC has an N-terminal membrane-spanning region, suggesting it is a membrane-anchored cytochrome. A potential FNR box, TTGATTTTCATCAA, was located 85 bp upstream of the putative translational start site of napE, indicating the napEFDABC cluster could be regulated by an FNR-like regulator(s) under anaerobic conditions (28).
Interestingly, the napEFDABC region is directly linked to other DNA regions involved in denitrification (Fig. 1). The hemN gene, which encodes the anaerobic coproporphyrinogen III oxidase for anaerobic heme biosynthesis, is located downstream of nap. Further downstream is the norEFCBQD region involved in nitric oxide reduction. Beyond the nor region are three ORFs that, based on sequence similarities, may encode proteins involved in metal uptake or transport functions.
Mutation and complementation analysis of the nap region.
The linkage of the nap region to other genes involved in denitrification drew our attention to its possible role in denitrification. To construct mutant with a deletion in the nap region, a SalI-BglII fragment was replaced with a kanamycin resistance cassette from Tn5 (Fig. 1). The resulting mutant strain, TW01, showed no growth with nitrate as the alternative electron acceptor under denitrifying conditions. It could grow on nitrite, although there was a slight decrease in growth rate compared to that of the wild type (data not shown). No nitrate reductase activity was detected in a whole-cell assay when induced under microaerobic conditions in the presence of nitrate (Table 2). The wild-type strain accumulated nitrite in the growth medium. No nitrite accumulation, however, was observed with TW01.
TABLE 2.
Accumulation of nitrite in the growth medium and the whole-cell nitrate reduction activities of different strains grown under microaerobic conditionsa
| Strain | Nitrite accumulation (mM [±SD]) | Activity (nmol of nitrate min−1 mg−1 of protein [±SD]) |
|---|---|---|
| WT | 13 ± 2 | 26 ± 0.2 |
| TW01 | 0 | 0 |
| TW01(pNAP01) | 12 ± 3 | 30 ± 12 |
The culture medium contained 16 mM NO3−. Cells were harvested after 16 h of growth. The whole-cell assay was carried out in a 15-ml serum bottle with a 5-ml cell suspension in TSB. Anaerobic conditions were created by flushing and filling the bottle with argon gas, and the reaction was started by adding 5 mM KNO3.
For complementation study, the 5.5-kb KpnI-HindIII fragment containing the napFDABC region was cloned into pTJS75 under control of the lac promoter. The resulting construct, pNAP01, was able to restore the ability of strain TW01 to grow on nitrate as the alternative electron acceptor (Fig. 1). Even though nitrate reductase activity was found in the crude extract, the mutant strain with NapC− construct pNAP02 could not grow on nitrate. This is consistent with the suggestion that NapC functions as the electron transporter for NapAB in vivo. Other constructs without a complete nap region also failed to complement TW01. These results indicated that the nap region was required for dissimilatory nitrate reduction in strain G-179.
Biochemical characterization of the nitrate reduction system.
Requirement of the nap region for dissimilatory nitrate reduction suggested that the periplasmic nitrate reductase was the primary enzyme carrying out dissimilatory nitrate reduction. To verify this hypothesis, the nitrate reduction system of strain G-179 was characterized based on effects of electron donors, azide sensitivity, and substrate specificity. Both membrane-bound and periplasmic nitrate reductases can use reduced benzyl viologen (BV+) and methyl viologen (MV+) as electron donors (2, 9). Because BV+ is more permeable to the cell membrane than MV+ in intact cells, presence of membrane-bound nitrate reductase often results in a much higher activity with BV+. In the whole-cell assay with the wild-type strain, the nitrate reduction activities with BV+ or MV+ as the electron donor were similar (Table 3). No BV+- or MV+-dependent activity was detected in the mutant strain. When NADH was used as the electron donor in the crude extract assay, no reduction of nitrate was observed. These results suggest that the nitrate reduction activity was primarily due to the periplasmic nitrate reductase.
TABLE 3.
Effects of artificial electron donors and azide on the activity of nitrate reduction in wild-type strain G-179a
| Assay | Sp act (nmol of nitrate min−1 mg of protein−1 [±SD]) with indicated electron donor
|
|||
|---|---|---|---|---|
| MV+ | BV+ | BV+ + azide | NADH | |
| Whole cell | 337 ± 15 | 395 ± 7 | 401 ± 34 | ND |
| Crude extract | 277 ± 14 | 310 ± 20 | 303 ± 20 | nd |
Cells were grown under anaerobic conditions with nitrate as the electron acceptor. When used the concentration of azide was 40 μM. ND, not determined; nd, not detected.
At micromolar concentrations, azide inhibits the membrane-bound nitrate reductase but has no effect on the periplasmic enzyme (2, 9). The BV+-dependent nitrate reductase activity in both whole cells and crude extract was not inhibited by 40 μM azide (Table 3). No chlorate reduction was detected with either whole cells or crude extract (data not shown). All these results indicate that the nitrate reduction system in the G-179 strain had the typical biochemical properties of periplasmic nitrate reductase. Activities characteristic of membrane-bound nitrate reductase were not detected.
Native gel and immunoblot analyses.
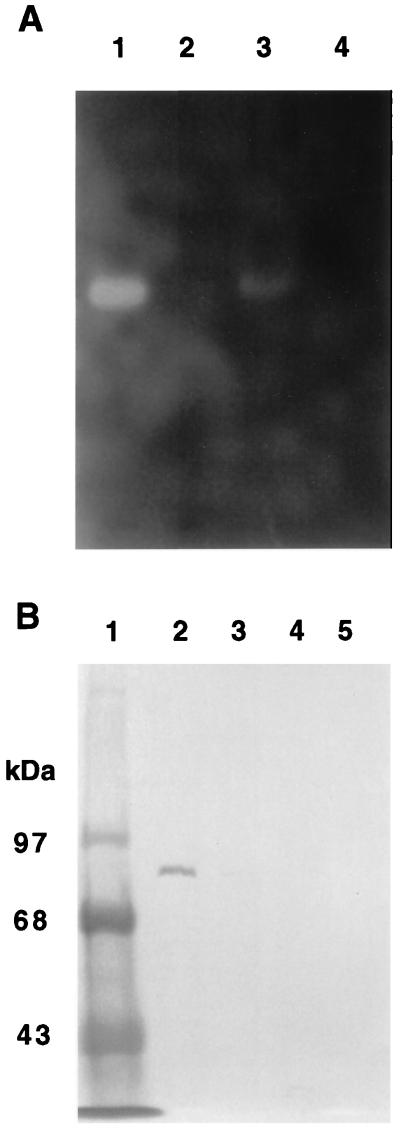
To confirm the presence of a periplasmic nitrate reductase encoded by the nap region, proteins from different subcellular fractions were isolated. Nitrate reductase activity was visualized as a bleached area against a dark blue background in a native gel with MV+ as the electron donor. No activity band was found in the cytoplasmic fraction of the wild type (data not shown). A strong activity band was, however, detected in the periplasmic fraction (Fig. 2A). In the membrane fraction, a weaker band in the same location was also observed. No activity bands were found with the mutant strain TW01 from periplasmic, cytoplasmic, or membrane fractions.
FIG. 2.
Detection of nitrate reductase from anaerobic cultures. (A) Native gel. Lane 1, wild-type periplasmic fraction; lane 2, TW01 soluble fraction; lane 3, wild-type membrane fraction; lane 4, TW01 membrane fraction. All lanes contained 80 μg of protein. (B) Western blot analysis using polyclonal antibody against the NapA protein. Lane 1, molecular size standard; lane 2, wild-type periplasmic proteins; lane 3, wild-type membrane proteins; lane 4, mutant TW01 total soluble proteins; lane 5, mutant TW01 total membrane proteins.
To further confirm the identity of the nitrate reductase, the purified NapA subunit was used to generate polyclonal antibody for immunoblot analysis. When the Western blot was developed with the NapA antibody, a positive protein band with a molecular size of about 90 kDa was detected in the wild-type periplasmic fraction (Fig. 2B). A weak band corresponding to the same molecular size was also observed in the membrane fraction. This suggests that a portion of the periplasmic nitrate reductase was associated with the membrane, probably through static or hydrophobic interactions. No positive band was found in the cytoplasmic fraction (data not shown). The 90-kDa positive band was also absent in all fractions from the mutant strain TW01. These results indicate that the nap region encodes a periplasmic nitrate reductase, consistent with its sequence similarity to other periplasmic nitrate reductases.
Phylogenetic analysis.
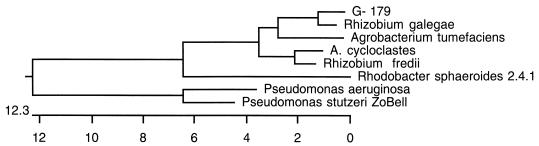
To facilitate the comparison of Pseudomonas sp. strain G-179 with other organisms which may have the same biochemical properties in nitrate reduction, the 16S rDNA sequence and fatty acid profiles were analyzed. The fatty acid profile is similar to those of Agrobacterium tumefaciens and Achromobacter cycloclastes, another unclassified denitrifier. In addition, both strain G-179 and A. cycloclastes had small amounts of cyclopropane and 10-methyl-branched fatty acids. The 16S rDNA sequence of strain G-179 showed 97% similarity to that of Rhizobium galegae, while the 16S rDNA sequence of A. cycloclastes was more similar to Rhizobium fredii. The phylogenetic tree is presented in Fig. 3. This result suggests that Pseudomonas sp. strain G-179 is likely a Rhizobium species. Attempts to isolate either small plasmids or megaplasmids from strain G-179 were not successful.
FIG. 3.
Phylogenetic analysis of Pseudomonas sp. strain G-179. The phylogenetic tree was obtained by the clustal method with DNASTAR software.
DISCUSSION
This study provides both genetic and biochemical evidence for the requirement of periplasmic nitrate reductase in the first step of denitrification in Pseudomonas sp. strain G-179. This finding brings a new perspective to our understanding of the mechanism of dissimilatory nitrate reduction in denitrifying bacteria. Here we propose that there are two systems responsible for dissimilatory nitrate reduction in denitrifiers. The first system, represented by P. denitrificans, uses the membrane-bound nitrate reductase as the primary enzyme for nitrate reduction, while the periplasmic enzyme has only secondary functions, including aerobic denitrification, transition to anaerobic respiration, or dissipating excess reducing equivalents. In the second system, represented by strain G-179, the periplasmic nitrate reductase is required for the first step of denitrification. The status and function of the membrane-bound nitrate reductase in this system are unknown. The mechanism by which strain G-179 uses the periplasmic nitrate reductase to gain energy is also unclear. It has been shown, however, that Nir− mutants RTC22 and RTC23 can grow on nitrate (25), suggesting dissimilatory reduction of nitrate in G-179 is an energy-generating process.
The function of the NapC protein is required for the in vivo activity of the periplasmic nitrate reductase (Fig. 1). Since the NapC protein is membrane bound, the NapAB subunits may have a close contact with the membrane or even associate with the membrane through static or hydrophobic interactions. In fact, one of the similarities between the membrane-bound nitrate reductase and the periplasmic enzyme is that both systems have a membrane component to transport electrons. The NarGH subunits are located in the cytoplasm and anchored to the membrane through NarI (2, 14, 15, 29). On the other hand, the dissimilatory nitrite reductase, the second enzyme in the pathway, is often located in the periplasmic space in many gram-negative denitrifiers. Thus, when a membrane-bound nitrate reductase is employed, a transport mechanism may be required for nitrate to cross the cytoplasmic membrane and for nitrite to return to the periplasmic space. If the periplasmic nitrate reductase catalyzes the nitrate reduction, the entire reaction of denitrification can be completed in the periplasmic space.
The periplasmic nitrate reductase is ubiquitous in nature, and its role may vary depending on the organism. Our observations in this report point toward a need for further examination of the roles of both periplasmic and membrane-bound nitrate reductases among different denitrifying bacteria. During the preparation of this report, the DNA sequence of the nap region from Rhodobacter sphaeroides f. sp. denitrificans was deposited in the GenBank database (accession no. AF069545). It was indicated that the nap region in this organism was also required for denitrification. If biochemical characterization substantiates that finding, it would further strengthen the hypothesis that the periplasmic nitrate reductase has a major role in this organism as well. In addition, based on our preliminary results, the nitrate reduction system in A. cycloclastes was not sensitive to a low concentration of azide and showed little activity with chlorate. This indicates that biochemical properties of the nitrate reduction system observed with strain G-179 may not be unique. From an ecological or evolutionary point of view, the distribution of denitrifying bacteria with the periplasmic enzyme as the major nitrate reductase warrants much closer examination. It has been shown that the periplasmic nitrate reductase provides one of the important mechanisms for aerobic nitrate reduction (2, 9). Oxygen-insensitive nitrate respiration may make a significant and previously unrecognized contribution to the flux from nitrate to nitrite in oxic and micro-oxic environments (9). In addition, aerobic nitrate reduction and aerobic denitrification may have potential applications in water and industrial wastewater treatment.
ACKNOWLEDGMENTS
We thank Vasantha Nagarajan and Ethel Jackson for their strong support to our research program in denitrification. We also thank Roslyn Young for sequencing 16S rDNA of G-179.
REFERENCES
- 1.Alefounder P R, Ferguson S J. The location of dissimilatory nitrite reductase and the control of dissimilatory nitrate reductase by oxygen in Paracoccus denitrificans. Biochem J. 1980;192:231–240. doi: 10.1042/bj1920231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell L C, Richardson D J, Ferguson S J. Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- 3.Bell L C, Page M D, Berks B C, Richardson D J, Ferguson S J. Insertion of transposon Tn5 into a structural gene of the membrane-bound nitrate reductase of Thiosphaera pantotropha results in anaerobic overexpression of periplasmic nitrate reductase activity. J Gen Microbiol. 1993;139:3205–3214. doi: 10.1099/00221287-139-12-3205. [DOI] [PubMed] [Google Scholar]
- 4.Berks B C, Richardson D J, Robinson C, Reilly A, Aplin R T, Ferguson S J. Purification and characterization of the periplasmic nitrate reductase from Thiosphaera pantotropha. Eur J Biochem. 1994;220:117–124. doi: 10.1111/j.1432-1033.1994.tb18605.x. [DOI] [PubMed] [Google Scholar]
- 5.Berks B C, Richardson D J, Reilly A, Willis A C, Ferguson S J. The napEDABC gene cluster encoding the periplasmic nitrate reductase system of Thiosphaera pantotropha. Biochem J. 1995;309:983–992. doi: 10.1042/bj3090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berks B C, Page M D, Richardson D J, Reilly A, Cavill A, Outen F, Ferguson S J. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol Microbiol. 1995;15:319–331. doi: 10.1111/j.1365-2958.1995.tb02246.x. [DOI] [PubMed] [Google Scholar]
- 7.Blümle S, Zumft W G. Respiratory nitrate reductase from denitrifying Pseudomonas stutzeri, purification, properties and target of proteolysis. Biochim Biophys Acta. 1991;105:102–108. [Google Scholar]
- 8.Carlson C, Ferguson L P, Ingraham J L. Properties of dissimilatory nitrate reductase purified from the denitrifier Pseudomonas aeruginosa. J Bacteriol. 1982;151:162–171. doi: 10.1128/jb.151.1.162-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter J P, Hsiao Y H, Spiro S, Richardson D J. Soil and sediment bacteria capable of aerobic nitrate respiration. Appl Environ Microbiol. 1995;61:2852–2858. doi: 10.1128/aem.61.8.2852-2858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad R. Soil microorganisms as controllers of atmospheric trace gases. Microbiol Rev. 1996;60:609–640. doi: 10.1128/mr.60.4.609-640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craske A, Ferguson S J. The respiratory nitrate reductase from Paracoccus denitrificans. Eur J Bioichem. 1986;158:429–436. doi: 10.1111/j.1432-1033.1986.tb09771.x. [DOI] [PubMed] [Google Scholar]
- 12.Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol. 1996;19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- 13.Hille R. The mononuclear molybdenum enzymes. Chem Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 14.Jones R W, Ingledew W J, Graham A, Garland P B. Topography of nitrate reductase of the cytoplasmic membrane of Escherichia coli: the nitrate-reducing site. Biochem Soc Trans. 1978;6:1287–1289. doi: 10.1042/bst0061287. [DOI] [PubMed] [Google Scholar]
- 15.Kristjansson J K, Hollocher T C. Substrate binding site for nitrate reductase of Escherichia coli is on the inner aspect of the membrane. J Bacteriol. 1979;137:1227–1233. doi: 10.1128/jb.137.3.1227-1233.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mateju V, Cizinska S, Krejci J, Janoch T. Biological water denitrification. Enzyme Microb Technol. 1992;14:172–183. [Google Scholar]
- 17.Parke D. Construction of mobilizable vectors derived from plasmids RP4, pUC18, and pUC19. Gene. 1990;93:135–137. doi: 10.1016/0378-1119(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 18.Philippot L, Clays-Josserand A, Lensi R, Trinsoutreau I, Normand P, Potier P. Purification of the dissimilative nitrate reductase of Pseudomonas fluorescens and the cloning and sequencing of its corresponding genes. Biochim Biophys Acta. 1997;1350:272–276. doi: 10.1016/s0167-4781(97)00007-9. [DOI] [PubMed] [Google Scholar]
- 19.Potter L, Cole J. The periplasmic nitrate reductase of Escherichia coli—a comparison with the Nap systems of other bacteria. Biochem Soc Trans. 1998;26:S217. doi: 10.1042/bst026s217. [DOI] [PubMed] [Google Scholar]
- 20.Reyes F, Roldán M D, Klipp W, Castillo F, Moreno-Vivián C. Isolation of periplasmic nitrate reductase genes from Rhodobacter sphaeroides DSM 158: structural and functional differences among prokaryotic nitrate reductase. Mol Microbiol. 1996;19:1307–1318. doi: 10.1111/j.1365-2958.1996.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 21.Reyes F, Gavira M, Castillo F, Moreno-Vivián C. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem J. 1998;331:897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidhauser T J, Helinski D R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985;164:446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui R A, Warnecke-Eberz U, Hengsberger A, Schneider B, Kostka S, Friedrich B. Structure and function of a periplasmic nitrate reductase in Alcaligenes eutrophus H16. J Bacteriol. 1993;175:5867–5876. doi: 10.1128/jb.175.18.5867-5876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson K, Metheringham R, Griffiths L, Cole J. Characterization of Escherichia coli K-12 mutants defective in formate-dependent nitrite reduction, essential roles for hemN and the menFDBCE operon. Arch Microbiol. 1997;168:403–411. doi: 10.1007/s002030050515. [DOI] [PubMed] [Google Scholar]
- 25.Ye R W, Averill B A, Tiedje J M. Characterization of Tn5 mutants deficient in dissimilatory nitrite reduction in Pseudomonas sp. strain G-179, which contains a copper nitrite reductase. J Bacteriol. 1992;174:6653–6658. doi: 10.1128/jb.174.20.6653-6658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye R W, Averill B A, Tiedje J M. Denitrification: production and consumption of nitric oxide. Appl Environ Microbiol. 1994;60:1053–1058. doi: 10.1128/aem.60.4.1053-1058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye R W, Fries M R, Serguei S G, Averill B A, Tiedje J M. Characterization of the structural gene encoding a copper-containing nitrite reductase and homology of this gene to DNA from other denitrifiers. Appl Environ Microbiol. 1993;59:250–254. doi: 10.1128/aem.59.1.250-254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye R W, Haas D, Ka J O, Krishnapillai V, Zimmermann A, Baird C, Tiedje J M. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires ANR, an analog of FNR. J Bacteriol. 1995;177:3606–3609. doi: 10.1128/jb.177.12.3606-3609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zumft G W. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]