Abstract
Background
Lysyl oxidase-like 2 (LOXL2) plays a role in tumor microenvironment formation and metastasis of hepatocellular carcinoma (HCC), which has a high mortality burden. Liver cancer stem cells (LCSCs) are related with the major malignant phenotypes of HCC. The function of LOXL2 in regulation of LCSCs remains unknown.
Methods
CD133+HepG2 and CD133+Hep3B cells were sorted by fluorescence-activated cell sorting (FACS) from two human hepatoblastoma cell lines. Spheroid formation, apoptosis, cell cycle, as well as transwell assays were performed upon LOXL2 knockdown in CD133+HepG2 and CD133+Hep3B cells. Protein and mRNA levels were quantified by Western blotting, immunofluorescence and reverse transcription-PCR (RT-PCR).
Results
Knockdown of LOXL2 decreased spheroid formation, migration and invasion (P<0.05), also induced apoptosis (P<0.05) and cell cycle arrest (P<0.05) in CD133+HepG2 and CD133+Hep3B cells. Knockdown of LOXL2 effectively inhibited expression of the anti-apoptosis proteins baculoviral inhibitor of apoptosis protein (IAP) repeat-containing 3 (BIRC3) and murine double minute 2 (MDM2) (P<0.01), as well as autophagy marker microtubule-associated protein 1 light chain 3 B (LC3B) and autophagy gene ATG5 in CD133+HepG2 and CD133+Hep3B cells (P<0.01).
Conclusions
The results revealed that LOXL2 inhibition could reduce the proliferation and expansion of LCSCs, making LOXL2 inhibitors an attractive and novel therapeutic strategy of HCC.
Keywords: Hepatocellular carcinoma (HCC), lysyl oxidase-like 2 (LOXL2), liver cancer stem cells (LCSCs), apoptosis
Introduction
Hepatocellular carcinoma (HCC) is very common and has a high mortality burden (1,2). Hepatitis B and C, metabolic dysfunction associated fatty liver disease and alcohol intake are the main risk factors of HCC (2,3). Hepatocarcinogenesis is accompanied by resistance to cellular death signals and a failure to clear damaged precancerous hepatocytes by the immune system (4,5). The poor prognosis and response to therapy of HCC is in part due to the presence of cancer stem cells (CSCs), an immune-privileged cell population that can evade immune surveillance more effectively than non-CSCs (6,7). Liver cancer stem cells (LCSCs) are a subset of hepatocarcinoma cells with pluripotent and self-renew properties which are related to the major malignant phenotypes of HCC (8-10). The expression of CSC-related genes such as NANOG, SOX2 and OCT4 can be promoted by oncogenes and/or overexpression of oncoproteins such as the HBV envelope protein PreS1, thereby driving the malignant biological behaviors of LCSCs (11). LCSCs can be characterized through cell surface markers, such as CD133, which is the original marker of hematopoietic stem cells, neural stem cells and liver stem/progenitor cells as a transmembrane cell surface single chain glycoprotein (12,13). The expression of CD133 in tumor cells is related to the stem cell-like characteristics, which is an important indicator of malignant progression, patient survival and recurrence (14,15).
Lysyl oxidase-like 2 (LOXL2) belongs to the lysyl oxidase (LOX) family, members of which are implicated in diverse pathophysiology including developmental regulation, cell motility, cellular senescence, and tumor suppression or promotion (16,17). The roles played by LOXL2 in fibrosis, tumorigenesis, and metastasis have been extensively reported, and it has also been recognized as a therapeutic target (16-18). Inhibition of LOXL2 slowed cancer progression of breast cancer, squamous cell carcinoma and renal cell carcinoma (19-21). LOXL2 can induce epithelial to mesenchymal transition (EMT), its overexpression negatively affects the clinicopathological features of different tumor types (22). Moreover, the expression of LOXL2 in CD133+ CSCs of glioblastoma multiforme was >9-fold higher compared to CD133- differentiated glioblastoma cells, suggesting its potential as a target for CSCs (23).
In HCC, LOXL2 is upregulated and plays a key role in HCC metastasis by mediating EMT and extracellular matrix remodeling, subsequently enhancing HCC invasion (22,24). LOXL2 is also crucial in tumor microenvironment formation and metastatic niche in HCC (24,25). However, the functions and regulatory mechanism of LOXL2 in LCSCs remains unknown.
Cell death is subdivided into apoptosis, autophagy death and necrosis (26). The activation of anti-apoptosis proteins benefits tumor cells from escaping apoptotic cell death (4,27). Baculoviral inhibitor of apoptosis protein (IAP) repeat-containing 3 (BIRC3; also known as cIAP2), one of the human IAP family, is often overexpressed in HCC tissues and indicates a poor prognosis of HCC patients (28,29). BIRC3 also induces EMT, proliferation, migration of hepatoblastoma cells in vitro (29). Moreover, murine double minute 2 (MDM2) is a negative-regulatory factor of p53, which is a central player in apoptosis (30,31). MDM2 is also an up-regulator of XIAP, another member of the IAP family, which mediated MDM2-p53 signaling pathway, promoting tumorigenesis and favoring resistance of neoplastic cells to irradiation-induced apoptosis (32,33).
In this study, we sorted the CD133+HepG2 and CD133+Hep3B cells from two human hepatoblastoma cell lines. The cellular function of the two cell lines, including proliferation, migration, invasion as well as apoptosis and cell cycle assay, were demonstrated upon LOXL2 gene knockdown. To understand whether LOXL2 benefited LCSCs from escaping apoptotic cell death, we investigated the change of gene expression of anti-apoptosis proteins BIRC3 and MDM2 in CD133+HepG2 and CD133+Hep3B cells upon LOXL2 knockdown. Moreover, to explore whether LOXL2 was related with autophagic cell death, we investigated the expression change of autophagy marker microtubule-associated protein 1 light chain 3 B (LC3B) and autophagy gene ATG5 in CD133+HepG2 and CD133+Hep3B cells upon LOXL2 gene knockdown. We present the following article in accordance with the MDAR reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-298/rc).
Methods
Cell lines
The human hepatoblastoma cell lines HepG2 (accession number: TCHu 72) and Hep3B (accession number: SCSP-5045) were purchased from Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; PAN, Germany) and 1% penicillin and streptomycin (Gibco) at 37.0 ℃ in 5% CO2. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The scheme of the current study was approved by the Institutional Review Board and Ethics Committee of the First Affiliated Hospital of Dalian Medical University. This study does not involve human participants, specimen, field samples or animal experiment.
Isolation of LCSCs through fluorescence-activated cell sorting (FACS)
HepG2 or Hep3B cells were resuspended in phosphate-buffered saline (PBS), then incubated with FcR blocking reagent (Miltenyi Biotec, Bergisch Gladbac, Germany) and phycoerythrin (PE)-conjugated CD133 antibodies (Miltenyi Biotec) at 4 ℃ for 20 min before sorting by FACS on a BD Influx (BD Biosciences, Franklin Lakes, NJ, USA) (12). About 60% of CD133+ cells presenting strongly positive were sorted. Isotype-matched mouse IgG was used to exclude non-specifically-stained cells. CD133+HepG2 and CD133+Hep3B cells were cultured in DMEM/F12 (HyClone, Logan, UT, USA) with 20 ng/mL recombinant epidermal growth factor (EGF; PeproTech, Rocky Hill, NJ, USA), 20 ng/mL basic fibroblast growth factor (bFGF; PeproTech) and 2% B27 supplement (Gibco) at 37.0 ℃ in 5% CO2. The CD133 positive proportion of cells was detected by flow cytometry.
In vitro LOXL2 gene silencing
CD133+HepG2 cells and CD133+Hep3B cells were first transfected with shRNA lentiviral vector (Target No. 77650-1; GeneChem, Shanghai, China) against LOXL2 (LOXL2-LV) or negative control (NC-LV). The vectors were encoded with green fluorescent protein (GFP). Cells were transduced with 25 multiplicity of infection (MOI) lentiviral particles, and HiTransG P (GeneChem, Shanghai, China) was used to promote transfer efficiency. After 48 to 72 hours, the effect of cell infection was observed under fluorescence microscope to make sure that more than 80% of cells show GFP. Cells were transduced for 72 hours before treated with 2 µg/mL puromycin for 2 weeks.
After stable knockout of LOXL2 by transfected with LOXL2 shRNA lentivirus, the CD133+Hep3B cell line presented with growth retardation, even died. CD133+Hep3B cell line was more susceptible to LOXL2 gene knockdown. Thus, we shifted to transient transfection of CD133+Hep3B cell line by siRNA in the follow-up study after spheroid formation assay. Pre-validated Silencer® select siRNAs (Thermo Fisher Scientific) against LOXL2 [LOXL2-siRNA, sequence (5'-3'): sense CAAGAUUCCGGAAAGCGUAtt; antisense UACGCUUUCCGGAAUCUUGag] or negative control (NC-siRNA, 4390843) were transfected respectively into 5×105 CD133+Hep3B cells at a dose of 25 pmol in six-well plates using the Lipofectamine RNAiMAXTM transfection reagent (Invitrogen, Waltham, MA, USA) for 24 h. Transfection efficiency was confirmed by reverse transcription-PCR (RT-PCR) and Western blotting.
RT-PCR
Total RNA was extracted from CD133+HepG2 cells and CD133+Hep3B cells. Then, cDNA was synthetized by reverse transcription with the Evo M-MLV RT Kit for qPCR (AG Bio, China). PCR was performed by Applied Biosystems 7500 PCR System (Thermo Fisher Scientific) with SYBR® Green Premix Pro Taq HS qPCR Kit (AG Bio). GAPDH was taken as the internal control. The amplification conditions were as follows: pre-denaturation (95 ℃, 30 s), denaturation (95 ℃, 5 s), annealing and extension (60 ℃, 30 s), all for a total 40 cycles. The experimental Ct values were normalized against GAPDH. 2−ΔΔCT method was used to analyze the data.
Western blotting
Total protein lysate was extracted on ice using radioimmunoprecipitation assay (RIPA) buffer, phenylmethanesulfonyl fluoride, phosphatase inhibitor, and protease inhibitor (KeyGEN, Nanjing, China). After separated in SDS-PAGE, proteins were transferred to polyvinylidene difluoride (PVDF) membrane. Then, the PVDF membrane was blocked with 5% buffer bovine serum albumin in TBST and exposed to primary antibody at 4 ℃ overnight. Next, the PVDF membrane was washed in TBST (10 min ×3 times) and exposed to secondary antibody (1:5,000; Abbkine, China) at room temperature for 1–1.5 hours (34). The band was incubated in BeyoECL Plus (Beyotime, China) and detected by chemiluminescence. The primary antibodies include anti-LOXL2 antibody (ab179810, Abcam), anti-BIRC3 antibody (ab32059, Abcam), anti-MDM2 antibody (ab259265, Abcam), anti-LC3B antibody (ab192890, Abcam), anti-ATG5 antibody (ab108327, Abcam).
Spheroid formation
Cells were collected and washed with PBS. After ACCUTASE (Sigma-Aldrichl, USA) digestion, single cells (1×104) were seeded in ultra-low adhesion 6-well plate (Crystalgen, Commack, NY, USA) containing serum-free culture (SFC) medium, which composed of DMEM/F12 (Hyclone), 20 ng/mL EGF (PeproTech), 20 ng/mL bFGF (PeproTech), and 2% B27 supplement (Life Technologies). Add 1 mL of fresh SFC into the cells twice a week. After culturing for 10–15 days, the numbers of tumor spheroids (>50 µm) were determined.
Flow cytometric apoptosis analysis
Cells in logarithmic growth phase were collected and digested into single ones by ACCUTASE (Sigma-Aldrichl). A total of 1×105–1×106 single cells were washed with cold PBS and 1× binding buffer, respectively. Next, cells were resuspended in 100 µL binding buffer with 5 µL Annexin V-FITC and 10 µL PI staining solution, and incubated in the dark for 15 min at room temperature. Then, apoptosis was analyzed with a FACS Calibur instrument.
Flow cytometry cell cycle analysis
Cells were collected and digested into single ones by ACCUTASE (Sigma-Aldrichl). Then, the cells were washed with cold PBS, and fixed with 70% ethanol at 4 ℃ overnight. After being washed, cells were incubated with 2 mL staining solution (propidium iodide) in the dark for 30 min at room temperature. Then, cell cycle was analyzed with a FACSCalibur instrument (BD Biosciences).
Transwell assay for migration and invasion
Cells were starved for 24 hours, and suspended with RPMI 1640 medium. A total of 5×104 cells in 200 µL RPMI 1640 medium were added into the upper chambers of 8 µm pore size transwells. Seven hundred µL medium with 10% FBS were added into the lower ones. For the invasion assay, the bottom of the transwell chamber was laid with diluted Matrigel (BD Biosciences). The cells in transwell were cultured at 37 ℃ in 5% CO2 for 24 hours. Next, the transwell was treated with 4% paraformaldehyde for 30 min, and 0.1% crystal violet for 25 min. After PBS washing, cells in the upper chambers were wiped off, and cells at the bottom of the chamber were observed under the microscope, and five fields were randomly selected for counting (×100 objective).
Immunofluorescence assay
Cells were fixed in 4% paraformaldehyde for 30 min and permeabilized in 0.25% Triton X-100 for 15 min. After blocked by 5% goat serum at room temperature for 45 min, cells were exposed to primary antibodies (1:500; Abcam) mentioned above at 4 ℃ overnight. Next, cells were exposed to fluorescein (FITC)-conjugated goat anti-rabbit antibody (1:250; Abbkine, China) in the dark for 1 hour at room temperature. Fluorescence images were acquired by a fluorescence microscope (Olympus, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed in Prism 8 software (GraphPad Software, La Jolla, CA, USA). Data of continuous variables are shown as mean ± standard deviation (SD). The data were analyzed upon three independent experiments. The difference between two independent groups was analyzed with two-sided unpaired Student’s t-test. P value <0.05 is regarded as statistically significant.
Results
Cell sorting of CD133-positive subsets in HepG2 and Hep3B hepatoma cell lines
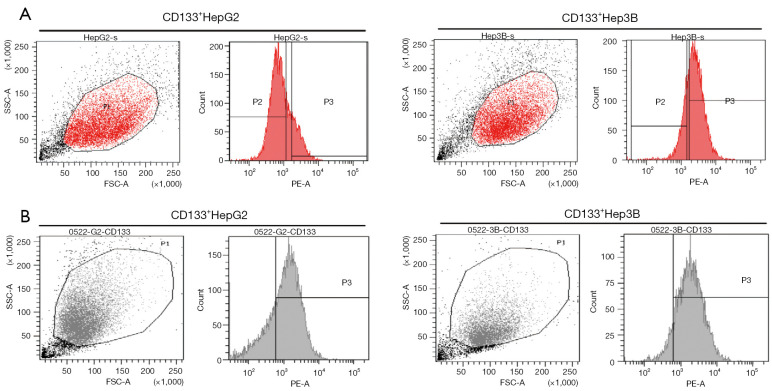
The CD133-positive subsets in HepG2 and Hep3B cell lines were sorted by flow cytometry, the proportion of subsets were 19.8%, 72.4%, respectively (Figure 1A). About 60% of CD133+ cells presenting strongly positive were sorted. The CD133+HepG2 and CD133+Hep3B cells obtained by sorting were cultured with DMEM/F12, EGF, FGF, and B27. Then, we detected the CD133 positivity of CD133+HepG2 and CD133+Hep3B cell lines by flow cytometry, which were 74%, 90.5%, respectively (Figure 1B).
Figure 1.
Cell sorting and the CD133 positivity after cell culture of CD133-positive subsets in HepG2 and Hep3B hepatoma cell lines. (A) The CD133-positive subsets in HepG2 and Hep3B cell lines were sorted by flow cytometry, the proportion of the subsets were 19.8%, 72.4%, respectively. About 60% of CD133+ cells presenting strongly positive were sorted. The sorted cells were cultured with DMEM/F12, EGF, bFGF, and B27. (B) The CD133 positivity of CD133+HepG2 and CD133+Hep3B cell lines which detected by flow cytometry were 74%, 90.5%, respectively. EGF, recombinant epidermal growth factor; bFGF, basic fibroblast growth factor.
Knockdown of LOXL2 impairs LCSCs spheroid formation
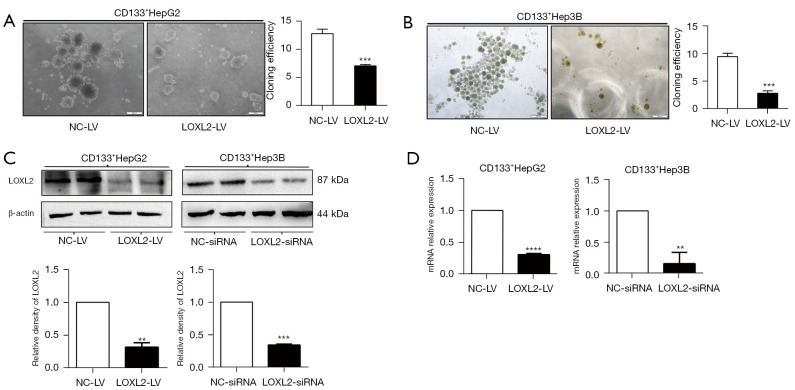
The ability of colony formation depends on the replication immortality of tumor cells, which is one of the hallmarks of tumor (35). We firstly explore whether knockdown of LOXL2 gene could impair tumor spheroid formation of LCSCs. LOXL2 or control shRNA lentiviral vectors were transferred into CD133+HepG2 and CD133+Hep3B cell lines. For CD133+HepG2 cells, spheroid formation was significantly decreased upon silencing LOXL2 (Figure 2A). When switching to CD133+Hep3B cells, a more significant inhibition effect was observed (Figure 2B). CD133+Hep3B cell line was more susceptible to LOXL2 gene knockdown. After stable knockout of LOXL2 by transfected with LOXL2 shRNA lentivirus, the CD133+Hep3B cell line presented with growth retardation, even died. Thus, we shifted to transient transfection of CD133+Hep3B cell line by siRNA to knockdown LOXL2 in the following study. Results of Western blot and PCR showed that LOXL2 was effectively knocked down in the two cell lines (Figure 2C,2D), which were transfected with shRNA lentiviral vector (LOXL2-LV/NC-LV) or siRNAs (LOXL2-siRNA/NC-siRNA) respectively. However, these results indicate that LOXL2 knockdown impairs LCSCs spheroid formation.
Figure 2.
Knockdown of LOXL2 impairs the spheroid formation of LCSCs. CD133+ (A) HepG2 and (B) Hep3B cells for sphere-forming assay were transfected with shRNA LOXL2-LV or NC-LV to observe cell survival upon LOXL2 knockdown. After culturing for 10–15 days, the number of tumor spheroids (>50 µm) was counted. LOXL2 knockdown impaired the cell sphere-forming, especially the CD133+Hep3B cells presenting with growth retardation. CD133+Hep3B cells in the following study were shifted to transiently transfection with LOXL2-siRNA or NC-siRNA. (C) Western blotting and (D) PCR were performed to determine transfection efficiency of the two ways. Relative intensity values for the proteins were obtained using Image J software. The data were analyzed upon three independent experiments and shown as mean ± SD. **, P<0.01; ***, P<0.001; ****, P<0.0001. NC-LV, lentiviral vector against negative control; LOXL2-LV, lentiviral vector against LOXL2; LOXL2, lysyl oxidase-like 2; LOXL2-siRNA, siRNAs against LOXL2; NC-siRNA, siRNAs against negative control; LCSCs, liver cancer stem cells; SD, standard deviation.
Knockdown of LOXL2 inhibits LCSCs migration, and invasion
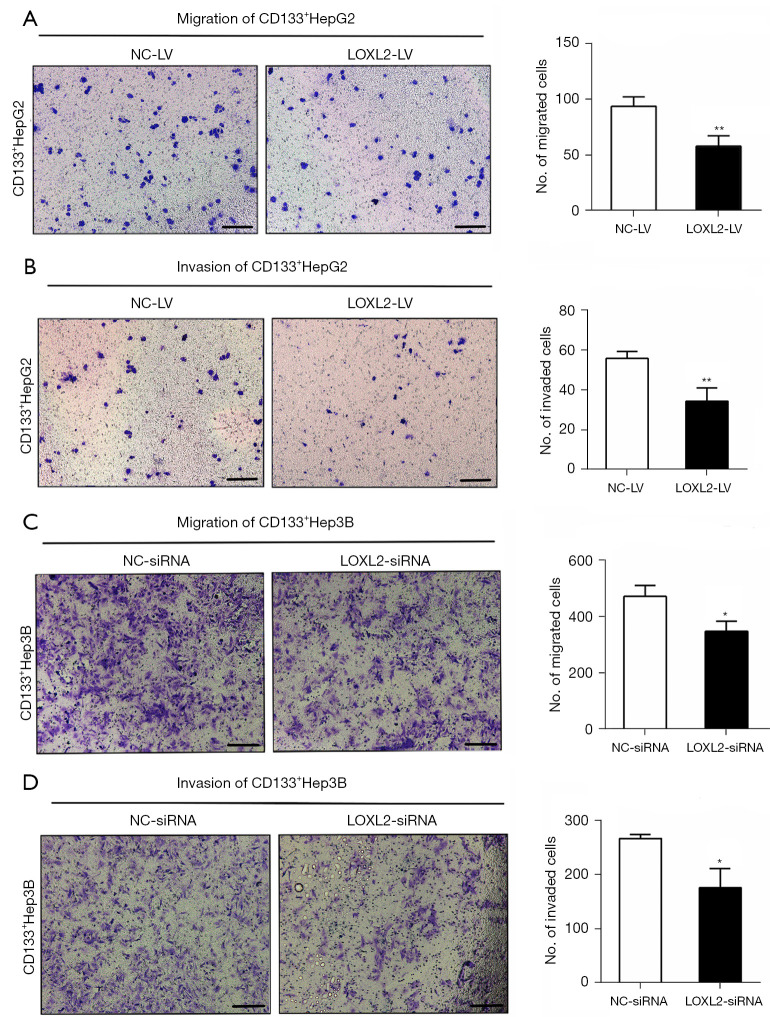
As LOXL2 gene plays a key role in HCC metastasis, subsequently enhancing HCC invasion, we asked whether silencing LOXL2 could inhibit LCSCs expansion. The transwell migration and invasion assays were performed in CD133+HepG2 (Figure 3A,3B) and CD133+Hep3B cells (Figure 3C,3D) upon LOXL2 knockdown. Result showed that the migration and invasion effect of both two cell lines were decreased upon LOXL2 knockdown compared to the controls. It indicates that LOXL2 knockdown inhibits LCSCs migration, and invasion.
Figure 3.
Knockdown of LOXL2 inhibits LCSCs migration and invasion. The transwell (A) migration and (B) invasion assays of CD133+HepG2 cells were performed upon LOXL2 knockdown by transfected with shRNA lentiviral vector (LOXL2-LV/NC-LV). After culturing for 24 hours in the transwells, cells were fixed and stained by 0.1% crystal violet. The average number of migrating and invading cells are showed in graphs. The transwell (C) migration and (D) invasion assays of CD133+Hep3B cells were performed upon LOXL2 knockdown by transfected with siRNAs (LOXL2-siRNA/NC-siRNA). Scale bar: 50 µm. The data were analyzed upon three independent experiments and shown as mean ± SD. *, P<0.05; **, P<0.01. NC-LV, lentiviral vector against negative control; LOXL2-LV, lentiviral vector against LOXL2; LOXL2, lysyl oxidase-like 2; LOXL2-siRNA, siRNAs against LOXL2; NC-siRNA, siRNAs against negative control; LCSCs, liver cancer stem cells; SD, standard deviation.
Knockdown of LOXL2 induces apoptosis and cell cycle arrest in LCSCs
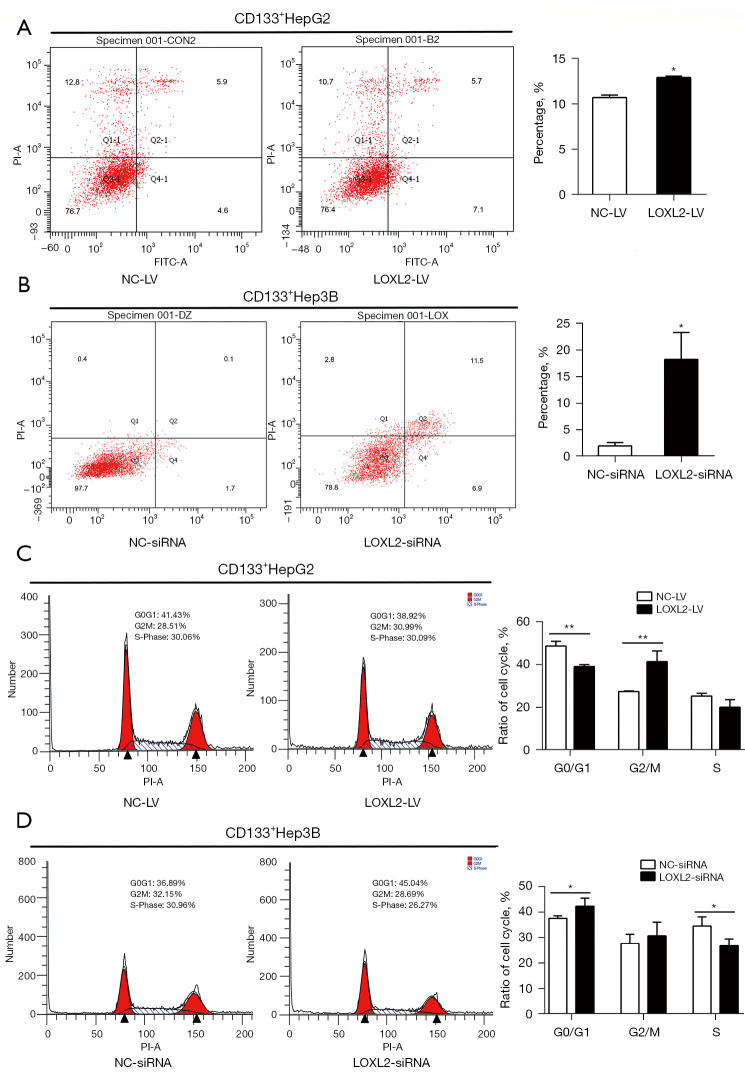
Anti-apoptosis is one of the signs of malignant tumors. We assessed apoptosis of CD133+HepG2 and CD133+Hep3B cells by flow cytometry using Annexin V-FITC and PI. The results showed that knockdown of LOXL2 significantly increased the proportion of cell apoptosis in both CD133+HepG2 (Figure 4A) and CD133+Hep3B cells (Figure 4B). Furthermore, we explored whether knockdown of LOXL2 could also affect cell cycle progression in the two cell lines. We found that the proportion of CD133+HepG2 cell decreased in G0/G1 phase, increased in G2/M phase, and was unaltered in S phase upon LOXL2 knockdown (Figure 4C). Correspondingly, the proportion of CD133+Hep3B cells increased in G0/G1 phase, decreased in S phase, and did not change in G2/M phase (Figure 4D). Knockdown of LOXL2 induced CD133+HepG2 cell cycle arrest in G2/M phase, and CD133+Hep3B cells arrest in G0/G1 phase. The difference may due to the CD133+HepG2 and CD133+Hep3B cells for cell cycle analysis were knocked down LOXL2 by different methods. However, these results indicated that LOXL2 knockdown induced the apoptotic phenotype and regulated the cell cycle of LCSCs.
Figure 4.
Knockdown of LOXL2 induces apoptosis and cell cycle arrest in LCSCs. CD133+HepG2 and CD133+Hep3B cells for flow cytometry analysis were transfected with shRNA lentiviral vector (LOXL2-LV/NC-LV) or siRNAs (LOXL2-siRNA/NC-siRNA), respectively, to knockdown LOXL2. Cells for apoptosis were gated by Annexin V-FITC and PI staining. Knockdown of LOXL2 induced the apoptosis of CD133+ (A) HepG2 and (B) Hep3B cells. LOXL2 knockdown also induced cell cycle arrested in CD133+ (C) HepG2 and (D) Hep3B cells. Data represented as % of total cells. The data were analyzed upon three independent experiments and presented as mean ± SD. *, P<0.05; **, P<0.01. NC-LV, lentiviral vector against negative control; LOXL2-LV, lentiviral vector against LOXL2; LOXL2, lysyl oxidase-like 2; LOXL2-siRNA, siRNAs against LOXL2; NC-siRNA, siRNAs against negative control; LCSCs, liver cancer stem cells; SD, standard deviation.
Knockdown of LOXL2 inhibits the expression of the anti-apoptosis proteins BIRC3 and MDM2 in LCSCs
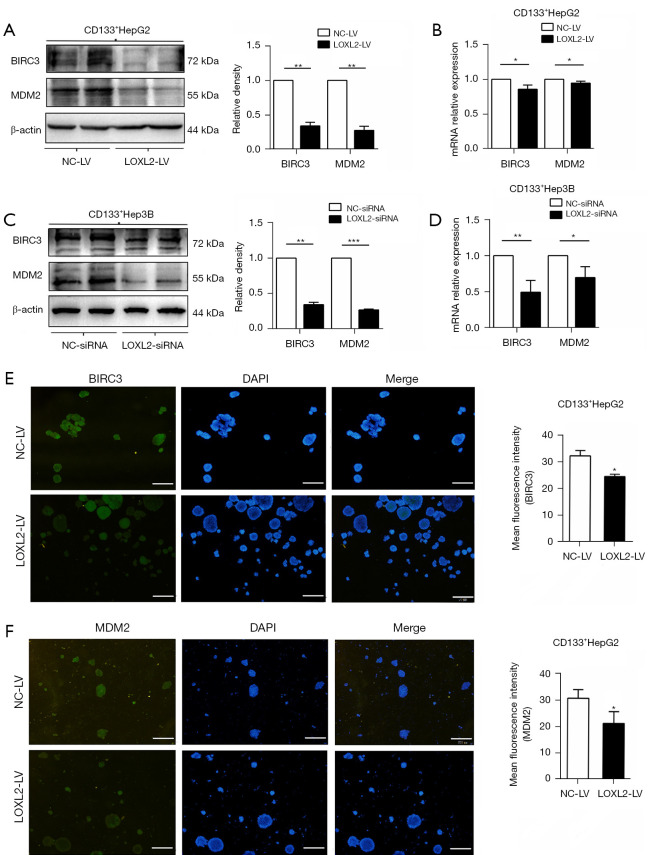
The activation of anti-apoptosis proteins benefits tumor cells from escaping apoptotic cell death (4,27). To understand whether LOXL2 favored the survival of LCSCs by anti-apoptosis, we explored the expression of BIRC3 and MDM2 in CD133+HepG2 and CD133+Hep3B cells upon LOXL2 knockdown. The results showed that LOXL2 knockdown significantly decreased protein and mRNA expression of BIRC3 and MDM2 in the two cell lines compared to controls (Figure 5A-5D). Immunofluorescence analysis of CD133+HepG2 cells was consistent with the western blotting and RT-PCR results, with reduced level of BIRC3 and MDM2 upon LOXL2 knockdown (Figure 5E,5F). These results indicated that one of the molecular mechanisms of apoptosis induced by LOXL2 knockdown in LCSCs may lie in the downregulation of anti-apoptosis protein BIRC3 and MDM2.
Figure 5.
Knockdown of LOXL2 downregulates BIRC3 and MDM2 in LCSCs. Western blotting and PCR of CD133+ (A,B) HepG2 and (C,D) Hep3B cells were performed upon LOXL2 knockdown to determine the expression of BIRC3 and MDM2 by transfected with shRNA lentiviral vector (LOXL2-LV/NC-LV) or siRNAs (LOXL2-siRNA/NC-siRNA) respectively. Relative intensity values for the proteins were obtained using Image J software. (E,F) Immunofluorescence staining was performed for BIRC3 and MDM2 in CD133+HepG2 cells upon LOXL2 knockdown. Scale bar: 200 µM. Mean fluorescence intensity values were obtained using Image J software. The data were analyzed upon three independent experiments. *, P<0.05; **, P<0.01; ***, P<0.001. BIRC3, baculoviral inhibitor of apoptosis protein repeat-containing 3; MDM2, murine double minute 2; NC-LV, lentiviral vector against negative control; LOXL2-LV, lentiviral vector against LOXL2; LOXL2, lysyl oxidase-like 2; LOXL2-siRNA, siRNAs against LOXL2; NC-siRNA, siRNAs against negative control; LCSCs, liver cancer stem cells.
Knockdown of LOXL2 inhibits the expression of autophagy marker LC3B and autophagy gene ATG5 in LCSCs
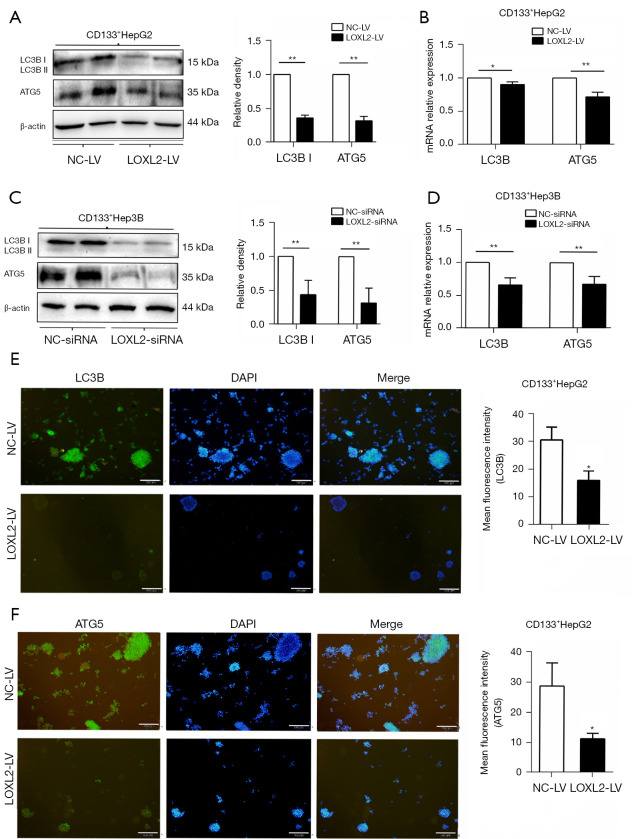
As to CD133+Hep3B cells presented with growth retardation or even died after stable knockout of LOXL2, we wondered whether LOXL2 took a part in regulation of autophagic cell death of LCSCs except anti-apoptosis. So, we further explored the expression of autophagy marker LC3B and autophagy gene ATG5 in CD133+HepG2 and CD133+Hep3B cells upon LOXL2 gene knockdown. The results showed that LC3B and ATG5 expression at protein and mRNA level were both significantly decreased in the two cell lines upon LOXL2 knockdown (Figure 6A-6D). Also, it was found using immunofluorescence that inhibition of LOXL2 reduced the expression of LC3B and ATG5 in CD133+HepG2 cells (Figure 6E,6F). These results indicated that LOXL2 knockdown inhibited the expression of autophagy marker LC3B and autophagy gene ATG5 in LCSCs.
Figure 6.
Knockdown of LOXL2 inhibits the expression of autophagy marker LC3B and autophagy gene ATG5 in LCSCs. Western blotting and PCR of CD133+ (A,B) HepG2 and (C,D) Hep3B cells were performed upon LOXL2 knockdown to determine the expression of LC3B and ATG5 by transfected with shRNA lentiviral vector (LOXL2-LV/NC-LV) or siRNAs (LOXL2-siRNA/NC-siRNA) respectively. Relative intensity values for the proteins were obtained using Image J software. (E,F) Immunofluorescence staining was performed for LC3B and ATG5 in CD133+HepG2 cells upon LOXL2 knockdown. Scale bars 200 µM. Mean fluorescence intensity values were obtained using Image J software. The data were analyzed upon three independent experiments. *, P<0.05; **, P<0.01. LC3B, microtubule-associated protein 1 light chain 3 B; NC-LV, lentiviral vector against negative control; LOXL2-LV, lentiviral vector against LOXL2; LOXL2, lysyl oxidase-like 2; LOXL2-siRNA, siRNAs against LOXL2; NC-siRNA, siRNAs against negative control; LCSCs, liver cancer stem cells.
Discussion
LCSCs, a subset of hepatocarcinoma cells with CSC-like properties, are important therapeutic target in HCC (8,9,13). Here we show that knockdown of LOXL2 inhibits the proliferation, invasion and migration of CD133+HepG2 and CD133+Hep3B cells, and induces cell apoptosis and cell cycle arrest. This is consistent with the conclusions of Wong et al., they demonstrated that LOXL2 facilitates HCC metastasis by inducing bone marrow-derived cells recruitment into the metastatic site (24). These results indicated that LOXL2 inhibition could reduce the proliferation and expansion of LCSCs. Targeting LCSCs by LOXL2 inhibitors may become an attractive and novel therapeutic strategy in HCC.
We tried to explore the potential mechanisms of LOXL2 inhibiting LCSCs function. In the current study, the growth of CD133+HepG2 and CD133+Hep3B cells significantly slowed down after stable knockout of LOXL2. The CD133+Hep3B cell was even stagnant or dead. Apparently, knockdown of LOXL2 may lead to LCSCs death via some mechanism. We first investigated whether knockdown of LOXL2 blunted the anti-apoptosis mechanism of LCSCs. We found that knockdown of LOXL2 effectively downregulated the anti-apoptosis proteins BIRC3 and MDM2 expression in CD133+HepG2 and CD133+Hep3B cells. This is consistent with our previous finding by microarray that BIRC3 and MDM2 are possible LOXL2 downstream genes in SMMC-7721 human hepatoblastoma cell lines (36). We speculate that LOXL2 might benefit LCSCs from escaping apoptotic cell death by regulating BIRC3 and MDM2. Knockdown of LOXL2 could induce LCSCs apoptosis through downregulation of BIRC3 and MDM2, which may become a promising strategy of targeting LCSCs.
Another mechanisms of LOXL2 inhibiting LCSCs function is that LOXL2 might play a role in regulation of LCSCs autophagy. We investigated the expression change of autophagy marker LC3B and autophagy gene ATG5 upon LOXL2 gene knockdown to understand whether LOXL2 was related with autophagic cell death. Our results show that knockdown of LOXL2 downregulated the expression of LC3B and ATG5 in CD133+HepG2 and CD133+Hep3B cells. This phenomenon could be explained by the previous reports that autophagy favors LCSCs resistance to hypoxia and nutrient deficiency in the tumor microenvironment of HCC (37-39). Autophagy positively regulates LCSCs by suppressing p53 (40). Thus, we speculate that LOXL2 might take a part in regulation of LCSCs autophagy.
Evidences show that both LOXL2 and BIRC3 induce EMT in HCC (22,29), BIRC3 is upregulated in the active autophagy response and that inhibition of autophagy suppresses the expression of BIRC2 and BIRC3 (41,42). We speculate LOXL2 maybe upregulate BIRC3 through an autophagy-mediated pathway. MDM2 is a negative regulator of the tumor suppressor p53, the MDM2-p53 axis participates in the transformation of normal hepatocytes into hepatoma cells (31). Autophagy also regulates p53 activity. Mitophagy, a specific type of autophagy, positively regulates LCSCs by suppressing p53, which would otherwise be activated by mitochondrial PINK1 to downregulate the expression of the CSC-related gene NANOG (40). We speculate that there may be a possible LOXL2/MDM2/autophagy/p53 pathway, which might mediate apoptosis and autophagy to favor the survival of LCSCs in the face of intracellular and extracellular stress.
However, the specific mechanism of LOXL2 in LCSCs is not well established. The relationship between LOXL2, apoptosis and autophagy needs to be further explored. Moreover, BIRC3, MDM2 and other possible LOXL2 downstream genes need further investigations, such as p53 which is known associated with MDM2 and autophagy.
In summary, knockdown of LOXL2 inhibits the proliferation, invasion and migration of LCSCs, and induces cell apoptosis and cell cycle arrest. Knockdown of LOXL2 effectively inhibited the expression of anti-apoptosis proteins BIRC3 and MDM2, as well as autophagy marker LC3B and autophagy gene ATG5 in LCSCs. LOXL2 inhibition could reduce the proliferation and expansion of LCSCs, making LOXL2 inhibitors an attractive and novel therapeutic strategy of HCC.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to acknowledge support from the Laboratory of Integrated Traditional Chinese and Western Medicine of the First Affiliated Hospital of Dalian Medical University.
Funding: This work was supported by National Natural Science Foundation of China (No. 81673728).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The scheme of the current study was approved by the Institutional Review Board and Ethics Committee of the First Affiliated Hospital of Dalian Medical University. This study does not involve human participants, specimen, field samples or animal experiment.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-298/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-298/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-298/coif). The authors have no conflicts of interest to declare.
References
- 1.Fenton SE, Burns MC, Kalyan A. Epidemiology, mutational landscape and staging of hepatocellular carcinoma. Chin Clin Oncol 2021;10:2. 10.21037/cco-20-162 [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. 10.1038/nrdp.2016.18 [DOI] [PubMed] [Google Scholar]
- 3.Raees A, Kamran M, Özkan H, et al. Updates on the Diagnosis and Management of Hepatocellular Carcinoma. Euroasian J Hepatogastroenterol 2021;11:32-40. 10.5005/jp-journals-10018-1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marquardt JU, Edlich F. Predisposition to Apoptosis in Hepatocellular Carcinoma: From Mechanistic Insights to Therapeutic Strategies. Front Oncol 2019;9:1421. 10.3389/fonc.2019.01421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn JC, Qureshi TA, Singal AG, et al. Deep learning in hepatocellular carcinoma: Current status and future perspectives. World J Hepatol 2021;13:2039-51. 10.4254/wjh.v13.i12.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsuchiya H, Shiota G. Immune evasion by cancer stem cells. Regen Ther 2021;17:20-33. 10.1016/j.reth.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sistigu A, Musella M, Galassi C, et al. Tuning Cancer Fate: Tumor Microenvironment's Role in Cancer Stem Cell Quiescence and Reawakening. Front Immunol 2020;11:2166. 10.3389/fimmu.2020.02166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nio K, Yamashita T, Kaneko S. The evolving concept of liver cancer stem cells. Mol Cancer 2017;16:4. 10.1186/s12943-016-0572-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan LH, Luk ST, Ma S. Turning hepatic cancer stem cells inside out--a deeper understanding through multiple perspectives. Mol Cells 2015;38:202-9. 10.14348/molcells.2015.2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv D, Chen L, Du L, et al. Emerging Regulatory Mechanisms Involved in Liver Cancer Stem Cell Properties in Hepatocellular Carcinoma. Front Cell Dev Biol 2021;9:691410. 10.3389/fcell.2021.691410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, Zhu Y. Targeting liver cancer stem cells for the treatment of hepatocellular carcinoma. Therap Adv Gastroenterol 2019;12:1756284818821560. 10.1177/1756284818821560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu YM, Li XF, Liu H, et al. Ultrasound-targeted microbubble destruction-mediated downregulation of CD133 inhibits epithelial-mesenchymal transition, stemness and migratory ability of liver cancer stem cells. Oncol Rep 2015;34:2977-86. 10.3892/or.2015.4270 [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Borlak J. Advances in Liver Cancer Stem Cell Isolation and their Characterization. Stem Cell Rev Rep 2021;17:1215-38. 10.1007/s12015-020-10114-6 [DOI] [PubMed] [Google Scholar]
- 14.Marquardt S, Solanki M, Spitschak A, et al. Emerging functional markers for cancer stem cell-based therapies: Understanding signaling networks for targeting metastasis. Semin Cancer Biol 2018;53:90-109. 10.1016/j.semcancer.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 15.Ciuffreda L, Falcone I, Incani UC, et al. PTEN expression and function in adult cancer stem cells and prospects for therapeutic targeting. Adv Biol Regul 2014;56:66-80. 10.1016/j.jbior.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 16.Wen B, Xu LY, Li EM. LOXL2 in cancer: regulation, downstream effectors and novel roles. Biochim Biophys Acta Rev Cancer 2020;1874:188435. 10.1016/j.bbcan.2020.188435 [DOI] [PubMed] [Google Scholar]
- 17.Ye M, Song Y, Pan S, et al. Evolving roles of lysyl oxidase family in tumorigenesis and cancer therapy. Pharmacol Ther 2020;215:107633. 10.1016/j.pharmthera.2020.107633 [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Yang A, Jia J, et al. Lysyl Oxidase (LOX) Family Members: Rationale and Their Potential as Therapeutic Targets for Liver Fibrosis. Hepatology 2020;72:729-41. 10.1002/hep.31236 [DOI] [PubMed] [Google Scholar]
- 19.Ferreira S, Saraiva N, Rijo P, et al. LOXL2 Inhibitors and Breast Cancer Progression. Antioxidants (Basel) 2021;10:312. 10.3390/antiox10020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin A, Salvador F, Moreno-Bueno G, et al. Lysyl oxidase-like 2 represses Notch1 expression in the skin to promote squamous cell carcinoma progression. EMBO J 2015;34:1090-109. 10.15252/embj.201489975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong X, Yu JJ. Silencing of lysyl oxidase-like 2 inhibits the migration, invasion and epithelial-to-mesenchymal transition of renal cell carcinoma cells through the Src/FAK signaling pathway. Int J Oncol 2019;54:1676-90. 10.3892/ijo.2019.4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuevas EP, Eraso P, Mazón MJ, et al. LOXL2 drives epithelial-mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Sci Rep 2017;7:44988. 10.1038/srep44988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko V, Arnotskaya N, Pak O, et al. Molecular determinants of the interaction between glioblastoma CD133+ cancer stem cells and the extracellular matrix. Int Rev Neurobiol 2020;151:155-69. 10.1016/bs.irn.2020.03.005 [DOI] [PubMed] [Google Scholar]
- 24.Wong CC, Tse AP, Huang YP, et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology 2014;60:1645-58. 10.1002/hep.27320 [DOI] [PubMed] [Google Scholar]
- 25.Wu L, Zhu Y. The function and mechanisms of action of LOXL2 in cancer (Review). Int J Mol Med 2015;36:1200-4. 10.3892/ijmm.2015.2337 [DOI] [PubMed] [Google Scholar]
- 26.Booth LA, Tavallai S, Hamed HA, et al. The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell Signal 2014;26:549-55. 10.1016/j.cellsig.2013.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Càceres J, Fabregat I. Apoptosis in liver carcinogenesis and chemotherapy. Hepat Oncol 2015;2:381-97. 10.2217/hep.15.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frazzi R. BIRC3 and BIRC5: multi-faceted inhibitors in cancer. Cell Biosci 2021;11:8. 10.1186/s13578-020-00521-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu PY, Hu B, Ma XL, et al. New insight into BIRC3: A novel prognostic indicator and a potential therapeutic target for liver cancer. J Cell Biochem 2019;120:6035-45. 10.1002/jcb.27890 [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Yu H, Hu W. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai) 2014;46:180-9. 10.1093/abbs/gmt147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao H, Chen X, Wang Z, et al. The role of MDM2-p53 axis dysfunction in the hepatocellular carcinoma transformation. Cell Death Discov 2020;6:53. 10.1038/s41420-020-0287-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu L, Zhu N, Zhang H, et al. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell 2009;15:363-75. 10.1016/j.ccr.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu H, Costa M. XIAP's Profile in Human Cancer. Biomolecules 2020;10:1493. 10.3390/biom10111493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tie L, Xiao H, Wu DL, et al. A brief guide to good practices in pharmacological experiments: Western blotting. Acta Pharmacol Sin 2021;42:1015-7. 10.1038/s41401-020-00539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Wang X, Zhang Z, et al. ACOT11 promotes cell proliferation, migration and invasion in lung adenocarcinoma. Transl Lung Cancer Res 2020;9:1885-903. 10.21037/tlcr-19-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu LH, Zhang Y, Zhu Y, et al. Mechanism Analyses for Elucidating the Role of LOXL2 knockdown in Hepatocellular Carcinoma. J Agr Sci Tech 2015;5:370-9. [Google Scholar]
- 37.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, et al. Autophagy in malignant transformation and cancer progression. EMBO J 2015;34:856-80. 10.15252/embj.201490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song YJ, Zhang SS, Guo XL, et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett 2013;339:70-81. 10.1016/j.canlet.2013.07.021 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Hu SB, Wang LY, et al. Autophagy-dependent generation of Axin2+ cancer stem-like cells promotes hepatocarcinogenesis in liver cirrhosis. Oncogene 2017;36:6725-37. 10.1038/onc.2017.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu K, Lee J, Kim JY, et al. Mitophagy Controls the Activities of Tumor Suppressor p53 to Regulate Hepatic Cancer Stem Cells. Mol Cell 2017;68:281-92.e5. 10.1016/j.molcel.2017.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He W, Wang Q, Xu J, et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2- and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy 2012;8:1811-21. 10.4161/auto.22145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comb WC, Cogswell P, Sitcheran R, et al. IKK-dependent, NF-κB-independent control of autophagic gene expression. Oncogene 2011;30:1727-32. 10.1038/onc.2010.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as