Abstract
The roles of micropeptides in cell cycle regulation and cancer development remain largely unknown. Here we found that a micropeptide STMP1 (small transmembrane protein 1) was up-regulated in multiple malignancies including hepatocellular carcinoma (HCC), and its high level was associated with short recurrence-free survival of HCC patients. Gain- and loss-of-function analyses revealed that STMP1 accelerated cell proliferation and clonogenicity in vitro and tumor growth in vivo, and silencing STMP1 blocked G1/S transition. Mechanistically, STMP1 promoted the mRNA and protein levels of CCNE2, CDK2, and E2F1. STMP1 was localized in the inner membrane of mitochondria and interacted with mitochondrial complex IV and then enhanced its activity. Moreover, treatment with the mitochondrial complex IV inhibitor tetrathiomolybdate dramatically abrogated the promoting effect of STMP1 on cell proliferation and the expression of cyclin E2, CDK2, and E2F1. These results suggest that STMP1 may promote G1/S transition and cell proliferation by enhancing mitochondrial complex IV activity, which highlights STMP1 as a new regulator of the cell cycle and a potential target for anti-cancer therapy.
Keywords: micropeptide, STMP1, G1/S transition, mitochondria, HCC
Graphical abstract

The roles of micropeptide STMP1 in cancer development remain unknown. Sang et al. show that STMP1 is up-regulated in tumor tissue, accelerates G1/S transition and tumor growth by up-regulating CCNE2, CDK2, and E2F1 in a mitochondrial complex IV-dependent manner. Their findings suggest STMP1 as a potential target for anti-cancer therapy.
Introduction
More and more small open reading frames have been identified in a wide variety of genome transcripts, some of which have been proved to encode micropeptides with less than 100 amino acids in length.1 Growing evidence suggests that micropeptides play important roles in physiological processes, including development,2 differentiation,3 stress response,4 metabolism,5 and cell survival.6 Whether dysfunction of micropeptides contributes to the development of disease, like cancer, remains largely unknown.
Loss of cell cycle control, especially at the G1/S transition, results in uncontrolled cell proliferation and consequent tumor development.7,8 The G1/S transition is tightly controlled by the pRb-E2F1 pathway, which consists of proliferation-stimulatory proteins (e.g., CDK4/6/2, cyclin D/E, E2F) and proliferation-inhibitory proteins (e.g., Rb, p15, p16, p21, p27).9 Deregulation of the pRb-E2F1 pathway is observed in multiple malignancies and targeting the promoter of G1/S transition has been considered as a promising strategy for cancer therapy.8,10 To date, only two micropeptides have been identified to affect cell cycle progression, and their deregulation results in abnormal cell proliferation. In breast cancer, up-regulation of micropeptide CASIMO1 promotes G0/G1 transition and cell proliferation by interacting with squalene epoxidase and subsequent modulation of lipid droplet homeostasis.11 Micropeptide PINT87aa, which is down-regulated in glioblastoma, interacts with polymerase-associated factor complex to suppress oncogenic transcriptional elongation, thus blocking G1/S transition and cell proliferation.6 Obviously, extensive investigations are required to find more micropeptides that play key roles in G1/S transition.
Hepatocellular carcinoma (HCC) is a common malignancy with rapid growth, early metastasis, and high mortality.12,13 The role of micropeptide in the development of HCC has not been reported yet. In an attempt to identify micropeptide that is associated with cell cycle and HCC, we screened for the micropeptides that were co-expressed with cell cycle regulators and deregulated in HCC. STMP1 (small transmembrane protein 1) appeared as an attractive candidate. STMP1 (also known as C7orf73, PL-5283, and Mm47) was first identified from bovine heart mitochondria by tandem mass spectrometry of protein ions.14 In zebrafish larvae, knockdown of STMP1 resulted in severe dysfunction of ventilatory activities upon exposure to hypoxia.15 Knockout of STMP1 in mouse primary macrophages impaired Nlrp3 inflammasome responses, leading to reduced activation of caspase-1 and compromised interleukin-1b secretion.16 However, whether STMP1 exists in human cells and regulates human physiological and pathological processes remains unknown.
In this study, we found that STMP1 was up-regulated in human HCC tissues, and higher STMP1 level was associated with shorter recurrence-free survival of patients. Furthermore, STMP1 accelerated G1/S transition, cell proliferation, and clonogenicity of HCC cells in vitro, and promoted tumor growth in vivo. Mechanism analyses revealed that STMP1 increased the levels of G1/S promoters (cyclin E2, CDK2, and E2F1) by enhancing mitochondrial complex IV activity. Our findings suggest the fundamental roles of STMP1 in cell cycle control and tumor development, and provide attractive molecular target for HCC therapy.
Results
STMP1 is up-regulated in HCC tissues and promotes tumor cell growth in vitro and in vivo
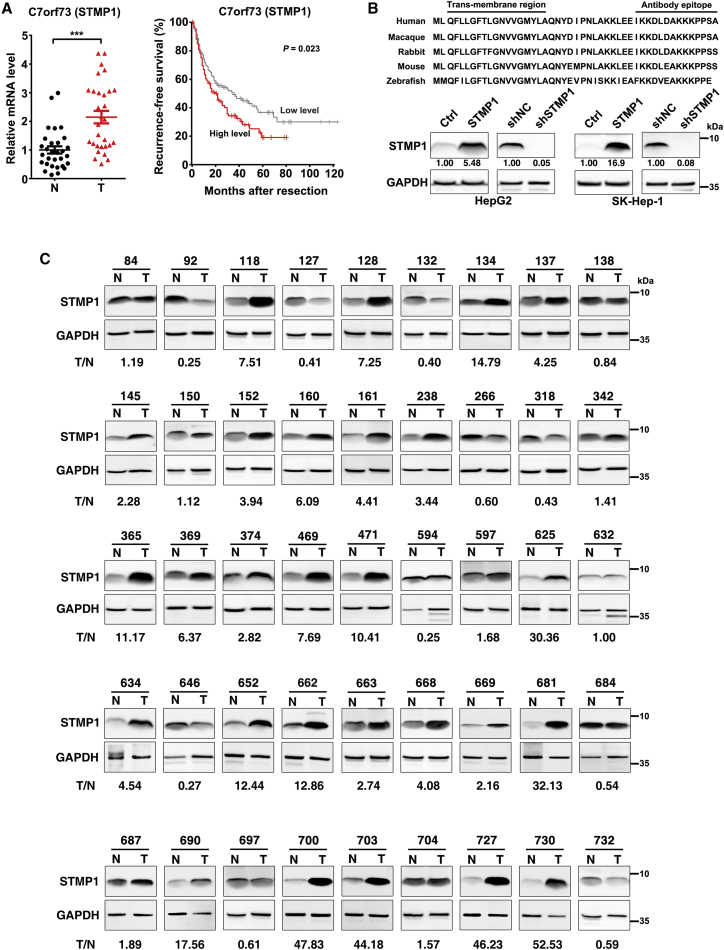
To identify novel micropeptides that regulate cell cycle and HCC development, we performed a bioinformatics analysis based on 753 reviewed putative coding genes from UniProt datasets and found three candidate genes that satisfied the following criteria (Figure S1A): (1) length of the predicted peptide less than 100 amino acids, (2) high conservation across various vertebrate species, and (3) significant expression alteration in HCC. Subsequent Gene Ontology analysis revealed that C7orf73 (STMP1) was highly co-expressed with the cell cycle regulators (Figure S1B), and it was therefore selected for further exploration. Consistently, the RNA level of STMP1 was significantly increased in the HCC tissues of our study cohort (Figure 1A, left panel). Analysis on the transcriptome data from Gene Expression Profiling Interactive Analysis (GEPIA) also revealed that STMP1 RNA was frequently up-regulated across a variety of cancer types (Figure S1C), and its high level was associated with short recurrence-free survival of HCC patients (Figure 1A, right panel). Moreover, based on The Cancer Genome Atlas (TCGA) data, the STMP1 level was much higher in HCC with higher TNM stage (Figure S1D), and in HCC with poor histological differentiation (Figure S1E).
Figure 1.
Up-regulation of STMP1 is a frequent event in HCC tissues and is associated with a worse prognosis for HCC patients
(A) Up-regulation of STMP1 was associated with short recurrence-free survival of HCC patients. Left, the mRNA level of STMP1 was assessed in 30 paired HCC (T) and adjacent nontumor liver tissues (N) by qPCR assay. U6 was used as an internal control. The mean value of adjacent noncancerous liver tissues was set as relative level 1. Right, a Kaplan-Meier survival analysis for HCC patients according to STMP1 level obtained from the GEPIA website. The median STMP1 level of 364 HCC tissues was set as the cutoff point for separating the STMP1 high-level cohort (n = 182) and STMP1 low-level cohort (n = 182). (B) Detection of endogenous and exogenous STMP1 protein in tumor cells. Top, alignment for the amino acid sequences of STMP1 across vertebrate species. The transmembrane region and antibody epitope region are indicated. Bottom, the STMP1 transcript encodes an ∼5 kDa micropeptide. HepG2 and SK-Hep-1 cells were infected with the indicated lentiviruses for 72 h before western blotting. GAPDH was used as an internal control. (C) The STMP1 protein level was significantly increased in HCC tissues. The levels of STMP1 were examined by western blotting in 45 paired HCC and adjacent nontumor liver tissues. The intensity for each band, representing protein level, was densitometrically quantified. The STMP1 level was normalized by the GAPDH level in each sample. The value under each pair of samples (T/N) indicates the fold change of STMP1 level in HCC, relative to that in adjacent nontumor tissue. The data are presented as mean ± SD (A); p values were assessed by paired Student’s t test (A, left), or log rank test (A, right) (∗∗∗p < 0.001).
Based on the information from NCBI, human STMP1 transcript contains 2461 bp (Figure S2) and is predicted to encode a 47-aa micropeptide which is highly conserved throughout vertebrate evolution (Figure 1B, upper panel). To validate the coding capacity of STMP1 in human cells, a STMP1-Flag fusion construct (Figure S3A) was delivered into the human hepatoma cells. A band with expected size at 5 kDa was detected in the cells expressing STMP1-Flag fusion protein by anti-Flag antibody (Figure S3B), and two unique peptide fragments in the STMP1 peptide were further detected by mass spectrometry following immunoprecipitation using anti-Flag gel (Figure S4), indicating that the STMP1 transcript encodes a bona fide micropeptide.
To detect the endogenous STMP1, a customized polyclonal antibody against the C-terminal of STMP1 (anti-STMP1) was produced. As shown, the STMP1-Flag fusion protein was detected by anti-STMP1 antibody (Figure S3B). Moreover, anti-STMP1 detected a significant decrease in the signal of STMP1-Flag fusion protein when cells were transfected with siSTMP1-CDS, a small interfering RNA (siRNA) targeting the coding sequence of STMP1 (Figures S3B and S3C). Consistently, anti-STMP1 also detected a significant reduction in the signal of cellular endogenous STMP1 when cells were infected with shSTMP1 that targeted 3′UTR of STMP1 (Figure 1B, lower panel). Using anti-STMP1 antibody, we found that compared with their noncancerous liver tissues, the protein level of STMP1 was markedly up-regulated in HCC tissues and 27 out of 45 (60%) HCCs displayed more than 2-fold increase in the STMP1 protein level (Figures 1C and S5), which was consistent with the alterations in STMP1 RNA level (Figure 1A, left panel). These results confirm the specificity of the anti-STMP1 antibody and the existence of the micropeptide STMP1 in human cells, and suggest that elevated STMP1 expression is a frequent event in human HCC and may be involved in hepatocarcinogenesis.
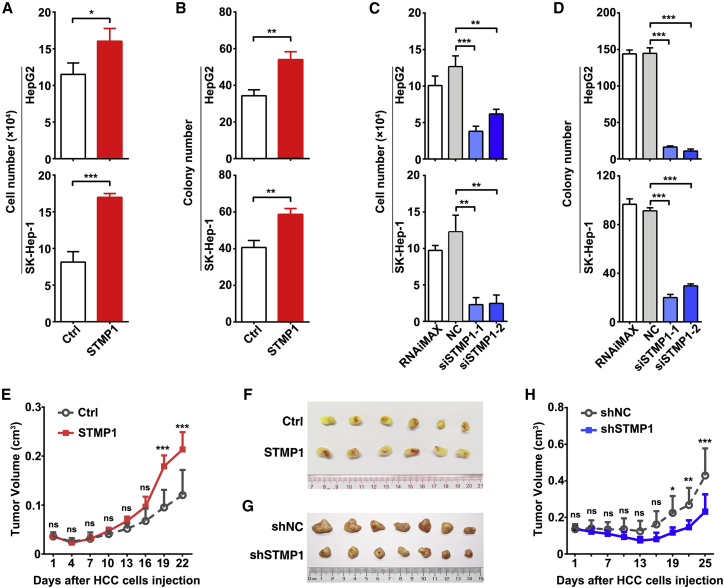
We next evaluated whether STMP1 regulated cell growth, and found that overexpression of STMP1 increased cell number and promoted colony formation of hepatoma cells (Figures 2A, 2B, S6A, and S6B), whereas silencing of STMP1 inhibited cell growth and clonogenicity (Figures 2C, 2D, S6C, and S6D). Moreover, cell growth and colony formation of hepatoma cells were promoted by overexpression of 5′UTR-ORFwt containing the 5′UTR region and wild-type ORF of human STMP1, but were not affected by overexpression of 5′UTR-ORFmut in which the start codon ATG was mutated to ATT (Figures S3A and S7), indicating that the proliferation-stimulatory effect of STMP1 is mediated by its protein but not RNA. Further investigations using an in vivo mouse xenograft model showed significant increase in the size of xenografts from STMP1-overexpressing hepatoma cells (Figures 2E and 2F), and obvious decrease in the size of xenografts derived from STMP1-silencing hepatoma cells (Figures 2G and 2H). These results suggest that STMP1 may function as an oncogenic micropeptide to promote cell growth in vitro and in vivo.
Figure 2.
STMP1 promotes tumor cell growth in vitro and in vivo
(A and B) Overexpression of STMP1 promoted cell growth (A) and clonogenicity (B). HepG2 or SK-Hep-1 cells were infected with the indicated lentiviruses for 72 h, then performed the cell counting and colony formation assays. (C and D) Silencing of STMP1 inhibited cell growth (C) and clonogenicity (D). RNAiMAX, cells treated with Lipofectamine RNAiMAX without RNA duplexes. NC, cells transfected with negative control RNA duplex. siSTMP1-1 and siSTMP1-2, cells transfected with siRNA targeting different sequences of STMP1. (E and F) STMP1 overexpression promoted xenograft growth in vivo. (G and H) Silencing of STMP1 suppressed tumor growth in vivo. For (E–H), SK-Hep-1 stable cell lines were subcutaneously injected into NCG mice (n = 6 or 7 mice/group). Representative photographs of excised tumors (F, G) and the curve of tumor growth (E, H) are shown. The data from at least three independent experiments are presented as mean ± SD (A–D); p values were assessed by unpaired Student’s t test (A–D), or two-way ANOVA (E, H) (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant).
STMP1 promotes G1/S transition by up-regulating CCNE2, CDK2, and E2F1
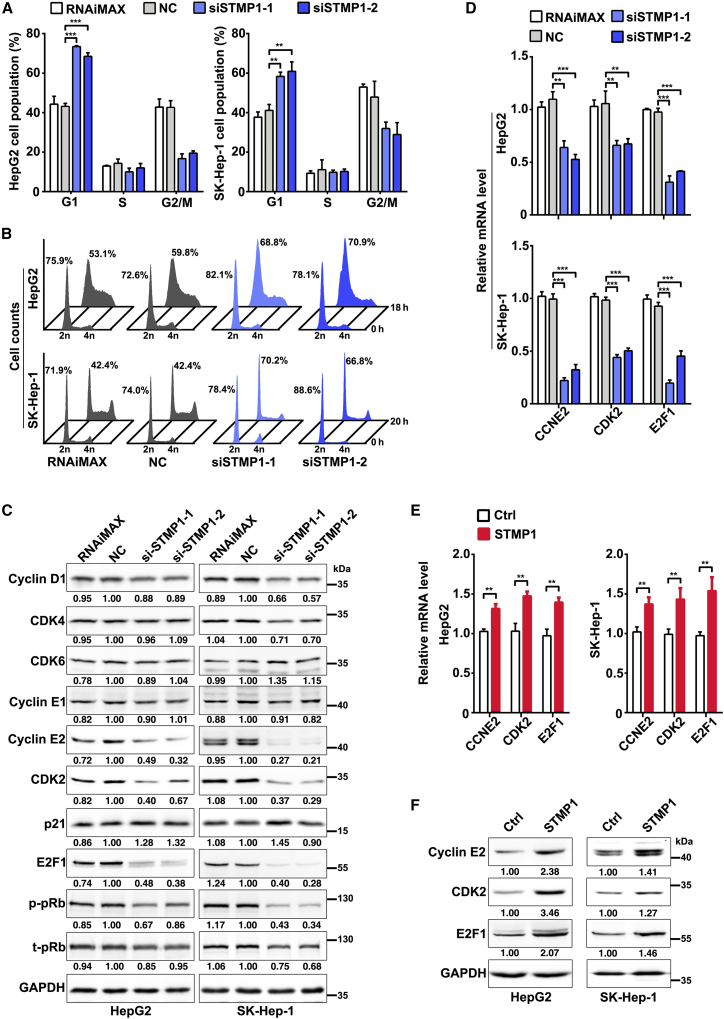
We then explored whether STMP1 facilitated tumor cell growth by accelerating cell cycle progression. As shown, silencing STMP1 caused marked accumulation of the G1 population in nocodazole-synchronized models (Figure 3A). Consistently, serum starvation-stimulation assays revealed that compared with NC transfectants, the transition of siSTMP1 transfectants from G1-phase to S-phase was significantly delayed (Figure 3B), suggesting that STMP1 may promote G1/S transition.
Figure 3.
Silencing STMP1 inhibits G1/S transition and decreases CCNE2, CDK2, and E2F1 levels
(A) Inhibition of STMP1 induced a substantial increase in the G1 population. NC-transfected or siSTMP1-transfected cells were synchronized with nocodazole for 20 h before FACS analysis. (B) Inhibition of STMP1 blocked S-phase entry upon serum stimulation. NC- or siSTMP1-transfectants were serum-deprived for 48 h, followed by serum re-addition and then cell harvest at the indicated timepoints. The time point when serum was re-added was set as 0 h. (C) Knockdown of STMP1 reduced the protein levels of cyclin E2, CDK2, E2F1, and phosphorylated pRB (p-pRb). (D) Knockdown of STMP1 reduced the mRNA levels of CCNE2, CDK2 and E2F1. (E) Overexpression of STMP1 increased the mRNA levels of CCNE2, CDK2, and E2F1. HepG2 or SK-Hep-1 cells were transfected with the indicated plasmids for 72 h prior to qPCR. (F) Overexpression of STMP1 increased the protein levels of cyclin E2, CDK2, and E2F1. For (C) and (F), the level of target protein relative to GAPDH level is indicated under each band. The data from at least three independent experiments are presented as mean ± SD (A, D, E); p values were assessed by unpaired Student’s t test (∗∗p < 0.01; ∗∗∗p < 0.001).
We next analyzed whether STMP1 affected the expression levels of key components in the cyclin/CDK-pRb-E2F pathway, which is the master controller of G1/S transition. We found that STMP1 knockdown caused a prominent decrease in the protein levels of cyclin E2, CDK2, E2F1, and phosphorylated pRB, but had no significant effect on other molecules examined (Figures 3C and S8A). Furthermore, silencing STMP1 also reduced the mRNA levels of CCNE2, CDK2, and E2F1 (Figure 3D). Consistently, overexpressing STMP1 increased the mRNA and protein levels of CCNE2, CDK2, and E2F1 (Figures 3E, 3F, and S8B). These results indicate that STMP1 may promote G1/S transition by up-regulating CCNE2, CDK2, and E2F1, and in turn facilitate cell proliferation.
STMP1 interacts with mitochondrial complex IV and enhances mitochondrial respiration
Identification of the subcellular localization of a micropeptides can enable insight into its putative biological function. We found that STMP1 was co-localized with mitochondrial-targeted DsRed (MT-DsRed) or mitochondrial protein TOMM20 (translocase of outer mitochondrial membrane 20) (Figures 4A and S9, upper panel), but not endoplasmic reticulum (ER)-targeted DsRed (ER-DsRed) or ER protein BiP (Figures 4B and S9, lower panel). The endogenously, naturally expressed STMP1 peptide from the mitochondria of hepatoma cells was also identified by mass spectrometry (Figure S10), and endogenous STMP1 was co-fractionated with the mitochondrial protein MTCO2 (mitochondrially encoded cytochrome c oxidase II) (Figure 4C), suggesting localization of STMP1 in mitochondria. Notably, the proteinase K digestion pattern of STMP1 is similar to UQCRC2 (ubiquinol-cytochrome c reductase core protein 2), an inner mitochondrial membrane (IMM) protein, implying that STMP1 is localized in the IMM of mitochondria (Figure 4D). We next explored whether STMP1 may regulate the mitochondrial function. Interestingly, the basal and maximal oxygen consumption rates and ATP production were increased by overexpressing STMP1 (Figure 4E), but were decreased by silencing STMP1 in hepatoma cells (Figure 4F). However, STMP1 knockdown had no discernible effect on mitochondrial reactive oxygen species (ROS) level, mitochondrial membrane potential, and mitochondrial DNA content (Figure S11). These data indicate that STMP1 may enhance mitochondrial respiration.
Figure 4.
STMP1 is localized to mitochondria and enhances mitochondrial respiration
(A, B, and C) STMP1 was located in mitochondria. For (A) and (B), SK-Hep-1 cells that expressed STMP1-Flag were transfected with MT-DsRed (a marker of mitochondria) (A) or ER-DsRed (a marker of ER) (B) plasmids for 36 h. Shown is immunofluorescence staining for Flag (green) and the nucleus (blue). Scale bar, 10 μm. For (C), immunoblotting was performed for cellular endogenous STMP1, MTCO2, and GAPDH in the cytosol and mitochondrial fractions of SK-Hep-1 cells. (D) STMP1 was localized to the IMM. Mitochondria isolated from HepG2 or SK-Hep-1 cells were subjected to proteinase K digestion in isotonic, hypotonic, or membrane perorated (1% Trion X-100) buffer, then the pellet collected by centrifugation before western blotting. TOMM20, AK2, and UQCRC2 were used as the markers for the outer mitochondrial membrane, the intermembrane space, and the IMM, respectively. (E and F) STMP1 enhanced mitochondrial respiration. Hepatoma cells were infected with the indicated lentiviruses for 72 h before detection for oxygen consumption rate (OCR). The basal and maximal respiration and ATP production of mitochondria were calculated based on the OCR detected by Seahorse XFe24 analyzer. The data from at least three independent experiments are presented as mean ± SD (E and F); p values were assessed by unpaired Student′s t test (∗p < 0.05; ∗∗∗p < 0.001).
The mitochondrial electron-transfer chain is composed of four complexes. We therefore explored whether STMP1 interacted with these complexes. The blue native-PAGE (BN-PAGE) analysis showed that STMP1 was detected in the low-molecular-weight (<232 kDa) complex (Figures 5A and 5B, left panels), which is similar to the molecular weight of mitochondrial complex IV. Consistently, MTCO2, a subunit of mitochondrial complex IV, was detected at the same electrophoretic migration position as STMP1 (Figures 5A and 5B, right panels), suggesting that STMP1 may associate with mitochondrial complex IV. Subsequent immunoprecipitation assays revealed that cellular MTCO2 was detected in the STMP1-precipitated complex (Figure 5C), further confirming the interaction between STMP1 and complex IV. In addition, overexpressing STMP1 enhanced the activity of complex IV (Figure 5D), whereas silencing STMP1 reduced it (Figure 5E). However, overexpressing or silencing STMP1 did not affect the levels of MTCO2 (Figures 5A and 5B), indicating that STMP1 may not influence the assembly of mitochondrial complex IV. These results imply that STMP1 may regulate mitochondrial respiration by interacting with mitochondrial complex IV and then increasing its activity.
Figure 5.
STMP1 interacts with mitochondrial complex IV and enhances its activity
(A and B) STMP1 interacted with mitochondrial complex IV. SK-Hep-1 cells were infected with the indicated lentiviruses for 72 h, then were passed and cultured for 4 days before harvesting. The mitochondria were isolated from the cells, suspended in solution buffer, then the mitochondrial respiratory chain complexes were separated by BN-PAGE. MTCO2 was used to indicate the mitochondrial complex IV. Arrowheads indicate the protein band of STMP1 or MTCO2. (C) HEK293T cells were transfected with the MT-3×Flag or STMP1-Flag plasmid for 48 h, followed by immunoprecipitation using anti-Flag gel and then western blotting using anti-Flag or anti-MTCO2 antibody. (D and E) STMP1 enhanced the activity of mitochondrial complex IV. HepG2 and SK-Hep-1 cells were infected with the indicated lentiviruses for 72 h before detection for respiratory complex IV activity. The complex IV activity was determined by calculating the absorbance decrease rate at 550 nm. Change in OD 550 nm observed in cell lysate over time (left panel) and complex IV activity was represented as a change in optical density per minute and shown as mOD/minute per 250 μg cell lysates (right panel). The data from at least three independent experiments are presented as mean ± SD (D, right; E, right); p values were assessed by unpaired Student′s t test (∗p < 0.05; ∗∗p < 0.01).
STMP1 promotes CCNE2, CDK2, and E2F1 expression by increasing mitochondrial complex IV activity
Complex IV of the mitochondrial respiratory chain is a copper-dependent enzyme. Tetrathiomolybdate ((NH4)2MoS4) is a copper chelator that can inhibit the activity of mitochondrial complex IV. As shown, hepatoma cells treated with (NH4)2MoS4 showed significant decrease in the complex IV activity (Figure S12) and in the mRNA and protein levels of CCNE2, CDK2, and E2F1 (Figures 6A, 6B, and S13A). Consistently, (NH4)2MoS4 treatment caused a marked arrest of hepatoma cells at the G1-phase (Figure 6C). Furthermore, the roles of STMP1 in enhancing the expression of CCNE2/CDK2/E2F1 (Figures 6D, 6E, and S13B) and in promoting the growth of hepatoma cells (Figure 6F) were abrogated by (NH4)2MoS4 treatment. These data suggest that STMP1 may promote CCNE2, CDK2, and E2F1 expression and cell proliferation by increasing mitochondrial complex IV activity.
Figure 6.
STMP1 increases the levels of CCNE2, CDK2, and E2F1 by enhancing mitochondrial complex IV activity
(A and B) (NH4)2MoS4-treatment reduced the levels of CCNE2, CDK2, and E2F1. HepG2 and SK-Hep-1 cells were treated with 100 μM of (NH4)2MoS4 for 24 h and then subjected to qPCR (A) or western blotting (B). (C) (NH4)2MoS4-treatment blocked G1/S transition. HepG2 and SK-Hep-1 cells were serum-deprived for 48 h and then maintained in culture medium with 10% FBS and 100 μM of (NH4)2MoS4 for 24 h before FACS analysis. (D and E) (NH4)2MoS4-treatment abrogated the STMP1-induced increases in CCNE2, CDK2, and E2F1 levels. HepG2 and SK-Hep-1 cells were transfected with the indicated plasmid for 72 h, then treated with 100 μM of (NH4)2MoS4 for 24 h before qPCR assay (D) or western blotting (E). (F) Promoting effect of STMP1 on cell proliferation was abolished by the (NH4)2MoS4 treatment. Hepatoma cells were infected with the indicated lentivirus for 72 h, then were reseeded. After 24 h, the cells were treated with 100 μM of (NH4)2MoS4 for 24 h, then were cultured in drug-free medium for another 24 h before cell counting. For (B) and (E), the level of target protein relative to GAPDH level is indicated under each band. The data from at least three independent experiments are presented as mean ± SD (A, C, D, F); p values were assessed by unpaired Student’s t test (∗∗p < 0.01; ∗∗∗p < 0.001; ns, not significant).
Taken together, our findings suggest that STMP1, a mitochondria-localized micropeptide, enhances the activity of mitochondrial complex IV and then increases the expression levels of CCNE2, CDK2, and E2F1, which in turn facilitates G1/S transition, consequently resulting in cell proliferation and tumor growth.
Discussion
It is well known that cell cycle progression is controlled by elaborate regulatory networks. Deregulation of cell cycle, especially deficiency in G1/S transition, is a key step during cancer development.9 A large number of proteins and non-coding RNAs have been characterized as cell cycle regulators. As a class of newly identified small molecules, the biological function of micropeptides in the cell cycle control is not clear. In this study, we discovered an oncogenic micropeptide, STMP1, which is frequently up-regulated in HCC tissues and accelerates G1/S transition and tumor growth by up-regulating CCNE2, CDK2, and E2F1 in a mitochondrial complex IV-dependent manner.
Cyclin E2, CDK2, and E2F1 are the key drivers of G1/S transition and cell division. The cyclin E-CDK2 complex accelerates the phosphorylation of pRb and subsequent release of E2F, resulting in transcription of S-phase genes and consequent G1/S transition and cell proliferation.17 Therefore, the expression levels of CCNE2, CDK2, and E2F1 are tightly controlled to guard against runaway proliferation. The expression of CCNE2 and CDK2 is regulated by transcription factors such as c-Myc,18 and by microRNAs (miRNAs) like miR-15019 and miR-34a.20 The level of E2F1 is controlled by transcription factors, like c-Myc18 and KLF6,21 by the ubiquitin-proteasome system including SCF-cyclin F22 and POH1,23 by miRNAs, such as miR-34a24 and miR-20b-5p,25 and by long non-coding RNAs (lncRNAs), such as lnc-APUE.26 However, the roles of micropeptides in the pRb-E2F1 pathway remain unknown. Herein, we showed that micropeptide STMP1 up-regulated the expression of proteins both upstream (cyclin E2 and CDK2) and downstream (E2F1) of Rb, suggesting STMP1 as a new promoter of G1/S transition. These findings extend our understanding on the regulatory network of the cell cycle control.
Based on observations from human specimens as well as in vitro and in vivo models, we identified the promoting function of STMP1 in cell proliferation. STMP1 was frequently up-regulated in different cancer types, including HCC, and higher STMP1 level was associated with worse prognosis, higher TNM stage, and poor histological differentiation of HCC patients. Both gain- and loss-of-function studies showed that STMP1 promoted cell proliferation and clonogenicity of HCC cells in vitro and tumor growth in vivo. These findings highlight the fundamental role of STMP1 in regulating cancer growth and the potential to target STMP1 as an anti-cancer strategy.
Otto Warburg described the dependence of cancer cells on aerobic glycolysis for their growth and proliferation,27 which led many to the assumption that cancer cells had mitochondrial defects and oxidative phosphorylation (OXPHOS) was down-regulated. However, recent studies have shown that mitochondrial bioenergetics, biosynthesis, and signaling are crucial for tumorigenesis,28 and up-regulation of OXPHOS is observed in a variety of cancers,29 potentially rendering them sensitive to OXPHOS inhibition. Several OXPHOS inhibitors, like metformin and arsenic trioxide, have been proven as potential anti-cancer agents.28,29 Here, we revealed that STMP1 enhanced the activity of mitochondrial complex IV, mitochondrial respiration, and ATP production, thus increasing tumor cell proliferation. Our data imply that inhibition of OXPHOS may act as a target for anti-proliferation therapy of cancer.
The crosstalk between mitochondria and other organelles has a pivotal role in tumorigenesis and cancer development.30,31 It has been shown that mitochondria regulate the function of nucleus through mitochondria-to-nucleus retrograde signaling.32 Several reports have shown that dysfunction of mitochondrial complexes may affect cell cycle progression and the expression of cell cycle regulators; however, the role of mitochondrial complex IV in cell cycle control remains unreported. In this study, we showed that micropeptide STMP1 interacted with complex IV and enhanced its activity, then increased the expression of cyclin E2, CDK2, and E2F1. Inhibition of complex IV activity by (NH4)2MoS4 decreased cyclin E2/CDK2/E2F1 levels and caused G1 arrest. Furthermore, the effect of STMP1 in increasing cyclin E2/CDK2/E2F1 levels and cell proliferation was abrogated by (NH4)2MoS4. These data reveal that the STMP1-enhanced complex IV activity may promote the expression of nuclear genes, cyclin E2, CDK2, and E2F1, through retrograde regulation. The retrograde response can be triggered by a decrease in the level of ATP, increased signaling by ROS, or the release of Ca2+ from mitochondria.32 Herein, STMP1 enhanced mitochondrial respiration and ATP production, but had no significant effect on mitochondrial ROS level, mitochondrial membrane potential, and mitochondrial DNA content, indicating that STMP1 may activate the mitochondria-to-nucleus retrograde signaling by increasing ATP levels. The increased AMP levels arising from a decrease in mitochondrial ATP synthesis may activate AMPK signaling pathway, which has been shown to lead to G1 arrest and the downregulation of cell cycle-related genes, like cyclin E1, CDK2, and E2F133, 34, 35; however, its underlying mechanism is still not clear. Consistently, silencing of STMP1 increased the level of phosphorylated AMPKα (pAMPKα-Thr172) (Figure S14), implying that STMP1 may activate the retrograde signaling by inactivating AMPK pathway. Our findings may offer new insight into the regulation of nuclear function by mitochondria, and open the possibility for future therapeutic intervention.
Collectively, we disclose a STMP1-mitochondrial complex IV-CCNE2/CDK2/E2F1 regulatory cascade and its role in G1/S transition and tumor growth. Our findings characterize the new function of micropeptides and the complex network of cell cycle control, and implicate STMP1 as a potential target for therapeutic application.
Materials and methods
More details are provided in Supplemental materials and methods.
Tissue specimens and cell lines
Paired human HCC and adjacent nontumor liver tissues were collected from patients who underwent radical tumor resection in Sun Yat-sen University Cancer Center, Guangzhou, China. Both tumor and nontumor tissues were confirmed histologically. No local or systemic treatment had been conducted before surgery. After surgical resection, no other anti-cancer therapy was administered before relapse. Tissues were immediately snap frozen in liquid nitrogen until use. Informed consent was obtained from each patient, and the study was approved by the Institutional Research Ethics Committee.
The cells used in this study included human hepatoma cell lines (HepG2, SK-Hep-1) and transformed human embryonic kidney cell line (HEK293T). All cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Gibco). For animal experiments, the stable cell lines were established by infecting SK-Hep-1 cells with lentiviruses that expressed the target sequences, including subline stably expressing STMP1 (SK-STMP1) and its control line (SK-Ctrl), and subline with stable silencing of STMP1 (SK-shSTMP1) and its control line (SK-shNC).
Analysis of gene expression
The expression level of target genes was analyzed by real-time qPCR or western blotting.
STMP1 antibody production
A custom polyclonal antibody against the C-terminal of STMP1 was generated by GenScript (Nanjing, China). Briefly, the C-terminal peptide (IKKDLDAKKKPPSA) of STMP1 was chemically synthesized, cross-linked to keyhole limpet hemocyanin, then used to immunize rabbits. Sera were collected and affinity-purified against the peptide immunogen. The specificity of our customed antibody was validated by western blotting, using cells with STMP1 overexpression or STMP1 silencing.
RNA oligoribonucleotides and vectors
The following siRNAs were used: siSTMP1-1, siSTMP1-2, and siSTMP1-CDS targeting human STMP1 transcripts (NM_001130929.2), the negative control RNA duplex (NC) for siRNA was non-homologous to any human genome sequence. All siRNA duplexes were purchased from GenePharma (Shanghai, China).
The vectors pCDH-Ctrl, pCDH-STMP1, pCDH-5′UTR-ORFwt, pCDH-5′UTR-ORFmut, pCDH-shNC, pCDH-shSTMP1, pCDH-MT-3×Flag, pCDH-STMP1-Flag, pCDH-Puro-STMP1-Flag, MT-DsRed, and ER-DsRed were generated as described in Supplemental materials and methods.
All oligonucleotide sequences are listed in Table S1.
Lentivirus production and infection
For lentivirus production, HEK293T cells were cotransfected with the lentivirus expression vector and the Lenti-X HTX Packaging System (Clontech, Palo Alto, CA). The lentiviral supernatant was harvested and stored in aliquots at −80°C until use. Target cells were grown to 40% confluence and then infected with lentiviral supernatant supplemented with 10 μg/mL polybrene (Sigma-Aldrich, St. Louis, MO) for 72 h.
Cell transfection
Cells were transfected with RNA oligoribonucleotides using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). The final concentrations of RNA duplex were 25 nM. Transfection with plasmid DNA alone or together with RNA duplex was performed using Lipofectamine 2000 (Invitrogen).
Cell counting assay
Cell counting assay was used to evaluate cell growth. For loss-of-function assays, the siRNA-transfected HepG2 (2 ×104) and SK-Hep-1 (1 ×104) cells were grown in a 24-well plate and cultured for 3 days before cell counting. For gain-of-function assays, cells were infected with the indicated lentiviruses for 72 h, then HepG2 (2 ×104) and SK-Hep-1 (1 ×104) cells were reseeded in a 24-well plate for 3 days before analysis.
Colony formation assay
Forty-eight hours after transfection or 72 h after infection, aliquots of the viable cells (500 for HepG2 cells, and 150 for SK-Hep-1 cells) were placed in a 6-well plate and maintained in complete medium for 18 (HepG2) or 10 (SK-Hep-1) days unless indicated. Colonies were fixed in methanol and stained with a 0.1% crystal violet solution for 15 min before counting.
Mouse xenograft models
All mouse experiments were approved by the Institutional Animal Care and Use Committee at Sun Yat-sen University. Experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication No. 80-23, revised 1996) and according to the Institutional Ethical Guidelines for animal experiments of Sun Yat-sen University.
Five-week-old male NOD-Prkdcem26Cd52Il2rgem26Cd22/Nju (NCG) (six or seven mice for each group) mice were used. For gain-of-function study, SK-STMP1 or its control line SK-Ctrl cells (2.5×106) were suspended in 100 μL serum-free DMEM/Matrigel (1:1), and injected subcutaneously into left or right side of the posterior flank. Mice were killed 22 days after implantation. For loss-of-function study, SK-shSTMP1 and its control line SK-shNC (4×106) were resuspended in 100 μL serum-free DMEM/Matrigel (1:1) and then injected subcutaneously into either side of the posterior flank. The experiments ended 25 days after tumor cell inoculation. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and calculated with the following formula: (L × W2) × 0.5.
Cell cycle analysis
All cell cycle analyses were performed using Krishan’s reagent (0.05 mg/mL propidium iodide, 0.1% sodium citrate, 0.02 mg/mL RNase A, 0.3% NP-40, pH 8.3) and fluorescence-activated cell sorting (FACS) (Gallios, Beckman Coulter, Miami, FL). Nuclear debris and overlapping nuclei were gated out.
Immunofluorescence staining
Immunofluorescence staining assay was performed to examine the localization of STMP1.
Isolation of mitochondria
Mitochondria were isolated using a mitochondria isolation kit (89,874, Thermo Fisher Scientific). Briefly, 2 × 107 cells were washed twice with precooling 1 × PBS and lysed in mitochondrial isolation buffer, incubated on ice for 10 min, followed by differential centrifugation and further purified by sucrose gradient centrifugation.
Proteinase K protection assay
For proteinase K treatment, purified mitochondria from HepG2 or SK-Hep-1 were resuspended in iso-osmotic buffer (10 mM MOPS-KOH pH 7.2, 250 mM sucrose, 1 mM EDTA) without or with 0.04 U/mL of proteinase K (TAKARA, Palo Alto, CA), or resuspended in hypo-osmotic buffer (10 mM MOPS-KOH pH 7.2, 1 mM EDTA) or membrane perforated buffer (10 mM Tris, 1 mM EDTA, 1% Triton X-100) with 0.04 U/mL of proteinase K, incubated at 37°C for 20 min, followed by adding protease inhibitor cocktail (B14001; Bimake, Houston, TX) and PMSF (ST505; Beyotime, Shanghai, China). The pellets were collected by centrifugation at 12,000×g, 4°C for 5 min, homogenized in lysis buffer containing protease inhibitor cocktail, and then subjected to western blotting. TOMM20, AK2 (adenylate kinase 2), and UQCRC2 were used as the controls for the outer mitochondrial membrane protein, the intermembrane space protein, and the IMM protein, respectively.
Measurement of oxygen consumption rate
The oxygen consumption rate of HepG2 or SK-Hep-1 cells was examined by XF Cell Mito Stress Test (103,015-100; Seahorse Bioscience, Billerica, MA) using a Seahorse XF24 Extracellular Flux analyzer (Seahorse Bioscience). Seahorse injection ports were loaded with a final concentration of 2 μM oligomycin (port A), 1 μM carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) (port B), and 0.5 μM rotenone and antimycin A (port C), which were sequentially added to the cells.
BN-PAGE
The isolated mitochondria were solubilized with 1.5% Triton X-100 in solubilization buffer. The supernatant was mixed with BN-loading buffer and subjected to BN gel electrophoresis on a native-PAGE 4%–13% Bis-Tris gel. The proteins were transferred to PVDF membranes, followed by incubation with primary antibody and detection for proteins.
Co-immunoprecipitation
HEK293T cells transfected with the MT-3×Flag or STMP1-Flag plasmid were harvested in lysis buffer. The Flag-protein complex was immunoprecipitated using anti-Flag gel, and detected by western blotting.
Assay for mitochondrial complex IV enzyme activity
Activity of mitochondrial complexes IV was determined by complex IV enzyme activity microplate assay kit (ab109910; Abcam, Cambridge, UK) following the instructions of the manufacturer.
Bioinformatics tool and statistical analysis
The databases used for bioinformatic analysis included the following: UniPort (https://www.uniprot.org/) was used for predicting potential protein-coding gene. GEPIA (http://gepia.cancer-pku.cn) was used to analyze the levels of micropeptides in tumor and adjacent nontumor tissues, the association between the level of candidate gene and the survival of HCC patients, and the STMP1-related genes in liver cancer tissues. DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov) was used for functional annotation clustering of STMP1-related genes in liver cancer. TCGA (https://portal.gdc.cancer.gov/) (Nagy et al., 2021) database was used to analyze the relationship between the STMP1 level and the disease stage of HCC patients.
Data are presented as the mean ± SD of at least three independent experiments. The differences between groups were analyzed using Student t test when only two groups were compared, or assessed using one-way analysis of variance when more than two groups were compared. All statistical tests were two-sided; p values <0.05 were considered statistically significant. All analyses were performed using the GraphPad Prism program (GraphPad Software Inc., San Diego, CA).
Acknowledgments
This work was funded by National Key R&D Program of China (2019YFA0906001, 2019YFA0906003), National Natural Science Foundation of China (91940305, 31771554, 31970731), Science and Information Technology Bureau of Guangzhou (201904020040), Guangdong Basic and Applied Basic Research Foundation (2019A1515011657), and Fundamental Research Funds for the Central Universities (19lgpy168). We thank Jia-Ling Xu and Yi-Hang Li in School of Life Sciences, Sun Yat-sen University for technical assistance.
Author contributions
Y.S. designed the study, performed the experiments, interpreted the data, and wrote the manuscript. J.-Y.L., F.-Y.W., X.-Y.L., and Z.-Q.C. performed the experiments and interpreted the data. Y.Z. and S.-M.Z. supervised the project, designed the study, interpreted the data, and wrote the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.04.012.
Contributor Information
Shi-Mei Zhuang, Email: LSSZSM@mail.sysu.edu.cn.
Ying Zhu, Email: zhuy68@mail.sysu.edu.cn.
Supplemental information
References
- 1.Makarewich C.A., Olson E.N. Mining for micropeptides. Trends Cell Biol. 2017;27:685–696. doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chanut-Delalande H., Hashimoto Y., Pelissier-Monier A., Spokony R., Dib A., Kondo T., Bohere J., Niimi K., Latapie Y., Inagaki S., et al. Pri peptides are mediators of ecdysone for the temporal control of development. Nat. Cell Biol. 2014;16:1035–1044. doi: 10.1038/ncb3052. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y.F., Xiao M.H., Chen H.X., Meng Y., Zhao N., Yang L., Tang H., Wang J.L., Liu X., Zhu Y., et al. A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 2019;10:528. doi: 10.1038/s41419-019-1767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 5.Lee C., Zeng J., Drew B.G., Sallam T., Martin-Montalvo A., Wan J., Kim S.J., Mehta H., Hevener A.L., de Cabo R., et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015;21:443–454. doi: 10.1016/j.cmet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent L.N., Leone G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 8.Sherr C.J., Bartek J. Cell cycle–targeted cancer therapies. Annu. Rev. Cancer Biol. 2017;1:41–57. doi: 10.1146/annurev-cancerbio-040716-075628. [DOI] [Google Scholar]
- 9.Rubin S.M., Sage J., Skotheim J.M. Integrating old and new paradigms of G1/S control. Mol. Cell. 2020;80:183–192. doi: 10.1016/j.molcel.2020.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polycarpou-Schwarz M., Groß M., Mestdagh P., Schott J., Grund S.E., Hildenbrand C., Rom J., Aulmann S., Sinn H.P., Vandesompele J., et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene. 2018;37:4750–4768. doi: 10.1038/s41388-018-0281-5. [DOI] [PubMed] [Google Scholar]
- 12.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 13.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carroll J., Altman M.C., Fearnley I.M., Walker J.E. Identification of membrane proteins by tandem mass spectrometry of protein ions. Proc. Natl. Acad. Sci. U S A. 2007;104:14330–14335. doi: 10.1073/pnas.0706817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D., Xi Y., Coccimiglio M.L., Mennigen J.A., Jonz M.G., Ekker M., Trudeau V.L. Functional prediction and physiological characterization of a novel short trans-membrane protein 1 as a subunit of mitochondrial respiratory complexes. Physiol. Genom. 2012;44:1133–1140. doi: 10.1152/physiolgenomics.00079.2012. [DOI] [PubMed] [Google Scholar]
- 16.Bhatta A., Atianand M., Jiang Z., Crabtree J., Blin J., Fitzgerald K.A. A mitochondrial micropeptide is required for activation of the Nlrp3 inflammasome. J. Immunol. 2020;204:428–437. doi: 10.4049/jimmunol.1900791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson J.P., Cook J.G. Cell cycle proliferation decisions: the impact of single cell analyses. FEBS J. 2017;284:362–375. doi: 10.1111/febs.13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhawe K., Roy D. Interplay between NRF1, E2F4 and MYC transcription factors regulating common target genes contributes to cancer development and progression. Cell Oncol. (Dordr) 2018;41:465–484. doi: 10.1007/s13402-018-0395-3. [DOI] [PubMed] [Google Scholar]
- 19.Li X., Liu F., Lin B., Luo H., Liu M., Wu J., Li C., Li R., Zhang X., Zhou K., et al. miR150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int. J. Oncol. 2017;50:1097–1108. doi: 10.3892/ijo.2017.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen D., Li Y., Mei Y., Geng W., Yang J., Hong Q., Feng Z., Cai G., Zhu H., Shi S., et al. miR-34a regulates mesangial cell proliferation via the PDGFR-β/Ras-MAPK signaling pathway. Cell Mol. Life Sci. 2014;71:4027–4042. doi: 10.1007/s00018-014-1599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y., Li H., Ma X., Fan Y., Ni D., Zhang Y., Huang Q., Liu K., Li X., Wang L., et al. KLF6 suppresses metastasis of clear cell renal cell carcinoma via transcriptional repression of E2F1. Cancer Res. 2017;77:330–342. doi: 10.1158/0008-5472.can-16-0348. [DOI] [PubMed] [Google Scholar]
- 22.Burdova K., Yang H., Faedda R., Hume S., Chauhan J., Ebner D., Kessler B.M., Vendrell I., Drewry D.H., Wells C.I., et al. E2F1 proteolysis via SCF-cyclin F underlies synthetic lethality between cyclin F loss and Chk1 inhibition. EMBO J. 2019;38:e101443. doi: 10.15252/embj.2018101443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B., Ma A., Zhang L., Jin W.L., Qian Y., Xu G., Qiu B., Yang Z., Liu Y., Xia Q., et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat. Commun. 2015;6:8704. doi: 10.1038/ncomms9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han R., Chen X., Li Y., Zhang S., Li R., Lu L. MicroRNA-34a suppresses aggressiveness of hepatocellular carcinoma by modulating E2F1, E2F3, and Caspase-3. Cancer Manag. Res. 2019;11:2963–2976. doi: 10.2147/cmar.s202664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia L., Li F., Qiu J., Feng Z., Xu Z., Chen Z., Sun J. Oncogenic miR-20b-5p contributes to malignant behaviors of breast cancer stem cells by bidirectionally regulating CCND1 and E2F1. BMC Cancer. 2020;20:949. doi: 10.1186/s12885-020-07395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S.Y., Zhu Y., Li R.N., Huang J.H., You K., Yuan Y.F., Zhuang S.M. LncRNA lnc-APUE is repressed by HNF4α and promotes G1/S phase transition and tumor growth by regulating MiR-20b/E2F1 Axis. Adv. Sci. (Weinh) 2021;8:2003094. doi: 10.1002/advs.202003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 28.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin. Cancer Res. 2018;24:2482–2490. doi: 10.1158/1078-0432.ccr-17-3070. [DOI] [PubMed] [Google Scholar]
- 30.Xia M., Zhang Y., Jin K., Lu Z., Zeng Z., Xiong W. Communication between mitochondria and other organelles: a brand-new perspective on mitochondria in cancer. Cell Biosci. 2019;9:27. doi: 10.1186/s13578-019-0289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivian C.J., Brinker A.E., Graw S., Koestler D.C., Legendre C., Gooden G.C., Salhia B., Welch D.R. Mitochondrial genomic backgrounds affect nuclear DNA methylation and gene expression. Cancer Res. 2017;77:6202–6214. doi: 10.1158/0008-5472.can-17-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quiros P.M., Mottis A., Auwerx J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 33.Owusu-Ansah E., Yavari A., Mandal S., Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 34.Yang W., Park I.J., Yun H., Im D.U., Ock S., Kim J., Seo S.M., Shin H.Y., Viollet B., Kang I., et al. AMP-activated protein kinase α2 and E2F1 transcription factor mediate doxorubicin-induced cytotoxicity by forming a positive signal loop in mouse embryonic fibroblasts and non-carcinoma cells. J. Biol. Chem. 2014;289:4839–4852. doi: 10.1074/jbc.m113.496315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi H., Liu Y., Li S., Chen Y., Li L., Cao Y., Mingyao E., Shi P., Song C., Li B., et al. Activation of AMPK attenuated cardiac fibrosis by inhibiting CDK2 via p21/p27 and miR-29 family pathways in rats. Mol. Ther. Nucleic Acids. 2017;8:277–290. doi: 10.1016/j.omtn.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.