Abstract
A novel mechanism for enhancement of adherence of Staphylococcus aureus to host components is described. A secreted protein, Eap (extracellular adherence protein), was purified from the supernatant of S. aureus Newman and found to be able to bind to at least seven plasma proteins, e.g., fibronectin, the α-chain of fibrinogen, and prothrombin, and to the surface of S. aureus. Eap bound much less to cells of Staphylococcus epidermidis, Streptococcus mutans, or Escherichia coli. The protein can form oligomeric forms and is able to cause agglutination of S. aureus. Binding of S. aureus to fibroblasts and epithelial cells was significantly enhanced by addition of Eap, presumably due to its affinity both for plasma proteins on the cells and for the bacteria.
Staphylococcus aureus, a human pathogen, produces a large number of proteins that specifically bind to molecules from plasma or from the human extracellular matrix. These interactions have been proposed to contribute to the colonization of the host tissues. S. aureus has been shown to bind to fibronectin (Fn) (5, 25), collagen (19), fibrinogen (Fg) (2, 3, 13), vitronectin (11, 20), and elastin (18). Many bacterial proteins that are associated with the cell wall exhibit a common amino acid sequence, an LPXTG motif, which anchors the molecule to the cell wall peptidoglycan (23, 24).
S. aureus interacts with Fg in several ways by producing different Fg binding proteins (2, 3, 13). The main mediator of the adherence of S. aureus to Fg and fibrin is a cell surface-associated protein called clumping factor (Clf) (13). The binding of S. aureus to Fg has been shown to be important in the development of endocarditis (15) and presumably in the attachment of bacteria to implanted biomaterials. In addition to Clf, S. aureus produces three extracellular proteins that can bind to Fg (2, 3).
(i) Coagulase. This protein (87 kDa from strain Newman) has the capacity to bind to both prothrombin (Pt) and Fg (21). The coagulase-Pt complex has the ability to turn Fg into fibrin threads by a mechanism different from natural clotting (7). Coagulase has been shown to be a virulence factor in pulmonary infection (22).
(ii) Efb. The second secreted protein (15.6 kDa) that binds to Fg, Efb (for extracellular Fg binding protein [17]), previously designated Fib (1), is produced constitutively. The role of Efb as a virulence factor has been demonstrated elsewhere (16), and the protective action of antibodies against Efb in an experimental infection strengthens the belief that Efb contributes to virulence (12). The physiological function of Efb is still unclear, but it was shown elsewhere that Efb-Fg complex precipitated when these two molecules were mixed in a 1:1 molar ratio, and it is unlikely that Efb is directly involved in bacterial adherence (16).
(iii) Sixty-kilodalton protein. A 60-kDa protein that interacts with both Fg and Pt has been partly characterized elsewhere (2) and is further characterized in this study. We have shown that this extracellular 60-kDa protein has a wide binding repertoire; it has affinity for at least seven plasma proteins, among which are the α-chain of Fg, Fn, and Pt. The protein also has an ability to bind to cells of S. aureus, to form oligomers, and to agglutinate S. aureus. This protein is presumably the same as a protein subsequently described by McGavin and coworkers (9, 14) and designated Map, for major histocompatibility complex class II analogous protein. However, based on the broad binding activity of this protein and its role as an adherence enhancement protein, demonstrated in this study, the term Eap (for extracellular adherence protein) will be used here.
MATERIALS AND METHODS
Bacterial strain and culture condition.
S. aureus Newman was radiolabeled by dilution of an overnight culture 1:50 in Luria-Bertani (LB) medium. The culture was grown for 5 h at 37°C in the presence of 50 μCi of [3H]thymidine (specific activity, 80 mCi/mmol). The cells were washed with phosphate-buffered saline (PBS) and then resuspended in PBS with 0.05% Tween 20 (PBST) to an optical density at 600 nm of 1.0. Specific labeling was 600 to 1,000 CFU per cpm.
Purification of Eap and coagulase.
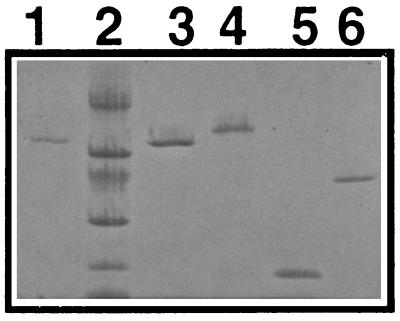
One liter of S. aureus Newman was grown for 19 h at 37°C in LB medium. The culture was centrifuged, and Fg binding proteins from the supernatant were isolated by affinity chromatography on Fg-Sepharose (Pharmacia, Uppsala, Sweden) as described before (3). Proteins were eluted with 0.7% acetic acid, dialyzed against 40 mM phosphate buffer (pH 6.5) (buffer A), and subjected to fast protein liquid chromatography (FPLC) on a Mono S column (Pharmacia), with a gradient of 0 to 100% buffer B (1 M NaCl in buffer A). Three peaks of proteins were eluted. The first one eluted at a salt concentration of 0.15 to 0.25 M NaCl (coagulase [Fig. 1, lane 4]), the second eluted at 0.35 to 0.45 M NaCl (Efb, previously named Fib [Fig. 1, lane 5]), and the third peak eluted at a concentration of 0.5 to 0.7 M NaCl (Eap [Fig. 1, lane 3]). Proteins in these three peaks were tested for coagulase activity with rabbit plasma, and only the first one was active.
FIG. 1.
SDS-PAGE of purified proteins. Lanes: 1, Eap purified from Eap-Sepharose; 2, molecular mass markers (14, 20, 30, 43, 67, and 94 kDa from bottom); 3, Eap purified by FPLC; 4, coagulase purified by FPLC; 5, Efb purified by FPLC; 6, recombinant His-Clf.
Preparation of bacterial cell surface protein extract.
To release cell surface-associated proteins, a LiCl extract of S. aureus cell surface protein was prepared as described before (11). Briefly, pelleted bacteria from 1 liter of LB medium culture were resuspended in 100 ml of 1 M LiCl, and the mixture was incubated at 37°C for 2 h with gentle agitation. After centrifugation (5,000 × g for 15 min at 4°C) to sediment the bacteria, the supernatant was dialyzed (membrane tubing with molecular weight cutoff of 3,500; Spectrum Medical Industries, Inc., Los Angeles, Calif.) against PBS overnight. The LiCl extract was used for purification of Fg binding proteins with Fg-Sepharose as described above for the supernatant.
Quantification of protein A.
The amounts of protein A from the supernatant and from the LiCl extract were estimated by determining the protein concentrations of protein A, purified with a 3-ml immunoglobulin G-Sepharose column (Pharmacia).
Enrichment of plasma proteins binding to Eap.
Eap-Sepharose was prepared by coupling 5 mg of Eap to 2 g of CNBr-activated Sepharose 4B (Pharmacia) by the procedure recommended by the manufacturer. A bed volume of 1 ml of the Eap-Sepharose was used for 0.5 ml of human plasma diluted with PBS. The column was washed with 10 bed volumes of PBS, and bound plasma proteins were eluted by adding PBS with 1 M NaCl (i.e., total NaCl concentration was 1.15 M, since PBS contains 0.15 M NaCl). The eluted proteins were dialyzed against PBS.
Binding of Fg, Pt, Fn, and collagen to coagulase and Eap.
Wells in a microtiter plate (Falcon; Becton Dickinson and Co., Paramus, N.J.) were coated overnight at room temperature (RT) with 100 μl of Eap or coagulase in PBS at a concentration range from 0.6 to 20 μg/ml. The wells were then blocked by incubation with 2% bovine serum albumin (Sigma Chemical, St. Louis, Mo.) for 1 h at 37°C. A constant amount of Fg, Pt, Fn, or collagen type II (Cn) (Southern Biotechnology, Inc., Birmingham, Ala.) (0.1 μg) in 100 μl of PBST was added to the wells followed by 2 h of incubation at RT. Bound Fg, Pt, Fn, or Cn was subsequently detected by horseradish peroxidase-conjugated rabbit immunoglobulins against Fg, Pt, Fn, or Cn diluted 1:1,000 in PBST (DAKO, Glostrup, Denmark). The plate was washed with PBST after each incubation, developed with 1,2-phenylenediamine (DAKO), and read at 492 nm.
SDS-PAGE and Western immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was run with the Phast system (Pharmacia). Protein from the second (Efb) and third (Eap) peaks from the Mono S column was run on an 8 to 25% gradient Phast gel and transferred to a nitrocellulose filter which was blocked by 1% Tween for 20 min at RT. The filter was incubated with 25 ng of 125I-labeled human Pt per ml in PBST for 2 h at RT. After being washed with PBST, the membrane was dried and examined by autoradiography (−70°C, about 48 h) with X-Omat AR film (Eastman Kodak). Radiolabeling was done with Iodobeads (Pierce, Rockford, Ill.).
Fg, human serum albumin, lysozyme, and Cn were also run on PAGE and transferred to a nitrocellulose filter. The filter was probed with Eap (10 μg/ml) for 1 h at RT, and the binding of Eap was detected with rat anti-Eap antibodies diluted 1,000-fold. Antiserum against Eap was obtained by immunizations of rats with 20 μg of Eap with Freund’s adjuvant on three occasions with a 2-week interval.
Amino acid sequence determination.
About 50 μg of Eap in 100 μl of PBS was used to determine the amino acid composition, the N-terminal sequence, and the C-terminal sequence. The Protein and Peptide Unit at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden, performed this.
Adherence of S. aureus to immobilized S. aureus proteins.
Microtiter plates were coated overnight at RT with 100 μl of either Eap, a portion of Clf, coagulase, or protein A at a concentration range from 1.9 to 30 μg/ml. Protein A was obtained from Pharmacia. Clf used here is a His6 fusion protein comprising residues 221 to 550 of the ClfA protein and was purified as described in reference 17 (Fig. 1, lane 6). The plasmid expressing Clf was kindly supplied to us by T. J. Foster (Dublin, Ireland) (13). After coating, the wells were blocked by addition of 100 μl of 2% bovine serum albumin in PBS for 1 h at 37°C. After washing three times with PBST, 100 μl of a 3H-labeled S. aureus suspension (5 × 107 cells in PBST) was added, and after 2 h of incubation at 37°C, the wells were washed three times with PBST, and bound bacteria were released by addition of two 50-μl suspensions of 3% SDS for twice 30 min at RT. The amount of bound bacteria was measured by scintillation counting. The binding ability of other bacterial species (Escherichia coli TG-1, Staphylococcus epidermidis, and Streptococcus mutans) for immobilized Eap was also tested in a similar way.
Adherence of soluble Eap to bacteria.
The binding of soluble Eap to S. aureus was analyzed. One hundred microliters of 106 CFU in PBST was added to 1.5-ml microcentrifuge tubes containing different amounts of 125I-labeled Eap ranging from 4.4 to 2,500 ng. The binding was done by shaking at room temperature for 60 min, and the cells were collected by centrifugation at 8,000 × g for 10 min. The supernatants were discarded, and the pellets were resuspended in 1.2 ml of PBST and then transferred to new microcentrifuge tubes. After another centrifugation, the pellets were washed once more, and the radioactivity associated with the pellets was measured in a gamma counter (LKB Wallac 1282; Compugamma, Turku, Finland). To determine the inhibition of radiolabeled Eap binding by cold Eap or coagulase, the same procedure was followed, with 1 μg of 125I-labeled Eap, but in the presence of cold Eap or coagulase (60, 30, 15, 7.5, or 0 μg).
Eap-Eap binding.
Three microliters of unlabeled Eap, coagulase, or Clf (200, 100, 50, 25, and 12.5 μg/ml) was spotted onto different regions of a nitrocellulose filter. The membrane was then blocked by 1% Tween for 30 min at RT and incubated with 5 ml of 100-ng/ml 125I-labeled Eap for 3 h at RT. The nitrocellulose filter was washed three times with PBST, the spots of the filter containing the different concentrations of proteins were cut out, and the radioactivities of the nitrocellulose pieces were measured in a gamma counter.
Isolation of Eap by Eap-Sepharose chromatography.
Eap was coupled to CNBr-activated Sepharose as described above. Before use, and between experiments, the column was extensively washed and treated with elution buffer to remove any loosely bound Eap. The column was equilibrated with PBS, and 0.5 liter of supernatant from a 19-h S. aureus culture was applied. The column was then washed with PBST, and the absorbed material was eluted with 1 M NaCl in PBS. The protein isolated from the Eap-Sepharose column was analyzed by SDS-PAGE with the Phast system (Fig. 1, lane 1). One hundred picomoles of a protein with the molecular mass of 60 kDa, which had been purified by Eap-Sepharose chromatography, was absorbed for 2 days into a 3- by 3-mm polyvinylidene difluoride membrane presoaked in methanol. The membrane was then washed with 20% methanol, dried, and subjected to N-terminal sequence analysis.
Agglutination of bacteria.
Bacteria (S. aureus, S. epidermidis, or S. mutans) were washed in PBS, and 100 μl containing 5 × 107 bacterial cells was placed on glass plates together with Eap at final concentrations shown in Table 2. Agglutination was readily visible within 30 min at RT and was scored as not detectable, weak, or strong.
TABLE 2.
Agglutination of bacteria in the presence of various amounts of Eapa
| Amt of protein added (μg) | S. aureus plus Eap | S. aureus plus coagulase | S. epidermidis plus Eap | S. mutans plus Eap |
|---|---|---|---|---|
| 20 | ++ | − | + | − |
| 2 | ++ | − | − | − |
| 0.2 | + | − | − | − |
| 0 | − | − | − | − |
++, strong agglutination; +, weak agglutination; −, no agglutination.
Binding of S. aureus to fibroblasts and epithelial cells.
Fibroblasts and epithelial cells were grown on eight-well glass slides (Chamber Slide system; Nunc, Naperville, Ill.) until confluent in Eagle’s medium supplemented with 10% fetal calf serum (Gibco BRL). Eap was added to the confluent cells at final concentrations shown in Fig. 7 and incubated for 30 min at RT. The wells were then washed in growth medium to remove excess Eap. Cells of S. aureus Newman, washed in PBS, were then added to a concentration of 2.5 × 105 bacteria per well. After 60 min of incubation at RT, the nonadherent bacteria were washed away. The plastic barriers between the wells were removed, the cells were dried and fixed with methanol, and the bacteria were Gram stained. Determination of bacterial adherence was done microscopically. The number of bacteria in a field of vision in the microscope was determined. Forty fields of vision were counted for each concentration of Eap, and the mean number of bacteria was calculated. Only bacteria adherent to fibroblasts or epithelial cells were counted, in case of occasional nonconfluence of fibroblasts.
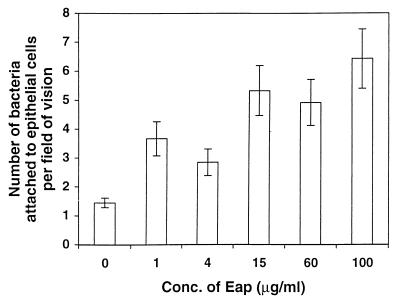
FIG. 7.
Effect of Eap on adherence of S. aureus to epithelial cells. Confluent layers of epithelial cells were preincubated with Eap at indicated concentrations for 30 min. After removal of excess Eap, bacteria were added and incubated for 60 min. After washing, cells were fixed and Gram stained, and the number of bacteria in each field of vision was determined. Forty fields were counted for each concentration of Eap. Mean values with standard errors are shown. The average number of epithelial cells per field was 10 to 12.
RESULTS
Isolation and identification of the three extracellular Fg binding proteins.
Three extracellular Fg binding proteins from S. aureus Newman were purified by Fg affinity chromatography followed by ion exchange. The FPLC profile revealed three major peaks that eluted at concentrations of 0.15 to 0.25, 0.35 to 0.45, and 0.50 to 0.70 M NaCl. The coagulase test, SDS-PAGE, and the NH2-terminal sequence analysis (IVTKDYSG in accordance with reference 21) revealed that the first peak contained the previously described 87-kDa Fg binding coagulase (2, 21) (Fig. 1, lane 4). The second peak corresponds to a 15.6-kDa protein designated Efb (17), earlier named Fib (1, 2) (Fig. 1, lane 5), and the third peak contained a 60-kDa protein (Fig. 1, lane 3). Efb and the 60-kDa protein have no coagulase activity. The 60-kDa protein is called Eap for extracellular adherence protein.
Binding of plasma and matrix proteins to Eap.
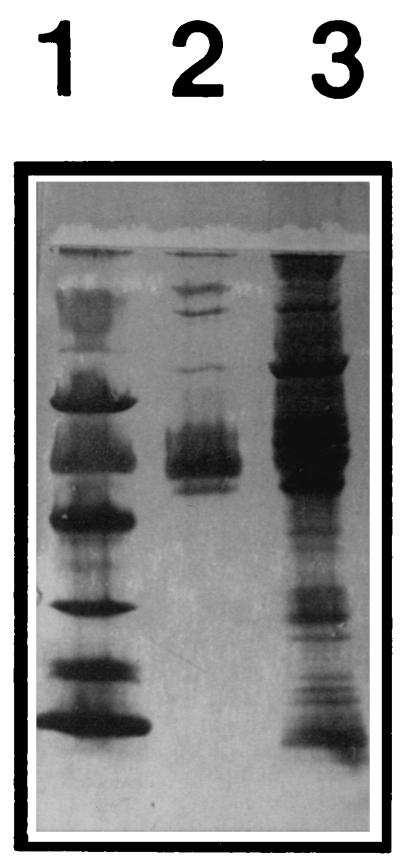
Coagulase has been reported to bind to both Fg and Pt. Similarly, Eap was previously found to bind to Fg and Pt (2). Its ability to bind to additional plasma proteins was investigated by coupling the protein to CNBr-activated Sepharose. Human plasma was pumped through, and plasma proteins that bound to the immobilized Eap were analyzed by SDS-PAGE. Figure 2 shows that at least seven proteins were enriched by this procedure. Although the identity of all these proteins has not been investigated here, the finding clearly implies a broad binding capacity of Eap. Moreover, Fn and Fg were detected by Western immunoblotting (data not shown).
FIG. 2.
Human plasma proteins before and after Eap affinity enrichment. Human plasma was run through a Sepharose column onto which Eap was coupled. Bound proteins were, after washing, eluted with PBS with 1 M NaCl, dialyzed, and subjected to PAGE and silver staining. Lane 1, molecular mass markers (14, 20, 30, 43, 67, and 94 kDa from bottom to top); lane 2, proteins enriched by Eap; lane 3, human plasma proteins before Eap-Sepharose enrichment.
A capture enzyme-linked immunosorbent assay (ELISA) was used to determine the binding ability of Fg, Fn, Pt, and Cn for Eap. Eap was immobilized at indicated concentrations in microtiter wells; a constant amount of Fg, Pt, Fn, or Cn was applied; and as shown in Fig. 3A, these proteins could all be captured by Eap except for Cn. A similar experiment was performed with coagulase. Only Fg and Pt could bind to coagulase (data not shown).
FIG. 3.
(A) Capture ELISA of Fg, Pt, Fn, and Cn. Microtiter wells were coated at the indicated concentration with Eap. Fg, Pt, Fn, or Cn (0.1 μg) was added, and binding was measured with horseradish peroxidase-conjugated antibodies against Fg (circles), Pt (squares), Fn (triangles), or Cn (diamonds). OD492, optical density at 492 nm. (B) Western affinity blot. Left, Coomassie blue-stained gel. Right, corresponding affinity blot probed with Eap followed by antibodies against Eap. Lanes in both panels: 1, Fg, α, β, and γ chains from top to bottom; 2, human serum albumin; 3, lysozyme; 4, Cn.
A Western affinity blot with iodinated Pt as probe confirmed the result from the capture ELISA that Eap can bind Pt whereas Efb cannot (data not shown).
In a Western blot experiment, Fg, human serum albumin, lysozyme, and Cn were run on PAGE gels. These proteins were probed with Eap followed by anti-Eap antibodies. Figure 3B shows that the α-chain of Fg was detected but that none of the other proteins was detected by Eap.
Localization of Eap.
To assess the localization of Eap, it was purified from both the supernatant and the LiCl extract of the cell surface of both 4- and 19-h cultures. The yields obtained indicated that approximately 70% of Eap, from both logarithmic- and stationary-phase cultures, was located in the culture supernatant, demonstrating that this is mainly a secreted protein (Table 1).
TABLE 1.
Amount of Eap isolated from 4- and 24-h cultures from the supernatant and from LiCl extracts of the bacteriaa
| Culture | Fraction | Yield (mg/liter) | % |
|---|---|---|---|
| 4 h | LiCl extractb | 0.42 | 29 |
| Supernatantb | 1.05 | 71 | |
| 24 h | LiCl extractb | 3.75 | 37 |
| Supernatantb | 6.47 | 63 | |
| Protein A | LiCl extractc | 5 | >98 |
| Supernatantc | <0.1 | <2 |
Protein A was used as a control for protein extraction.
Eap was purified by Fg affinity chromatography followed by ion exchange.
Protein A was purified by immunoglobulin G affinity chromatography.
S. aureus adherence to Eap.
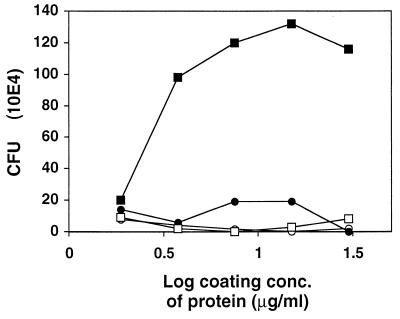
The attachment of radiolabeled S. aureus to immobilized Fg in microtiter wells was measured in the presence of Eap or Clf. The presence of Clf decreased bacterial adherence to Fg by up to 50%. In contrast, the presence of Eap resulted in a slight enhancement (data not shown), implying that Eap and Clf recognize different domains of Fg. The enhancement of S. aureus adherence led us to investigate the direct interaction between S. aureus and Eap. Figure 4 shows that an increased amount of immobilized Eap led to increased bacterial adherence. Binding of S. aureus to other S. aureus proteins (coagulase, protein A, and Clf) was not observed. At a saturating Eap concentration (10 μg/ml), about 2.5% of added bacteria bound. Under the conditions used, this is about the same level of adherence as that to immobilized Fg (data not shown).
FIG. 4.
Ability of S. aureus Newman to bind to immobilized Eap. Microtiter wells were coated at the indicated concentrations with Eap (closed squares), protein A (open squares), Clf (open circles), or coagulase (closed circles). A constant amount (5 × 107 CFU) of radiolabeled bacteria was added to the immobilized proteins. Nonadherent bacteria were removed after 2 h, and the amount of bound bacteria was determined.
Strain Newman of S. aureus was used in Fig. 4. The binding capacities of S. aureus 8325-4 and S. epidermidis, S. mutans, and E. coli were also tested. Strain Newman bound much better than did strain 8325-4 to immobilized Eap. Attachment of S. epidermidis, S. mutans, and E. coli to Eap appeared to be poor (data not shown).
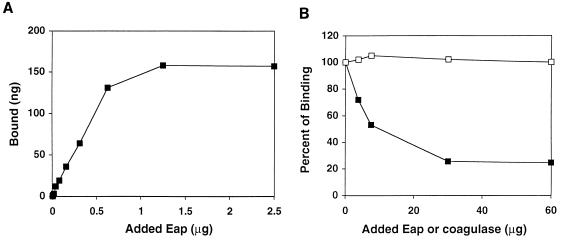
Interaction between strain Newman and Eap was also demonstrated by the binding of soluble, radiolabeled Eap to the bacterial cells (Fig. 5A). The binding of radiolabeled Eap to S. aureus was competitively inhibited by unlabeled Eap but not by coagulase (Fig. 5B). Unlabeled Eap reduced the binding of labeled protein by up to 75%. In contrast, the presence of coagulase did not interfere with the interaction between Eap and S. aureus.
FIG. 5.
(A) Binding of 125I-labeled Eap to S. aureus Newman. To a constant number of bacterial cells (106 CFU) were added different amounts of radiolabeled Eap. Binding was allowed for 1 h, the cells were washed, and bound radioactivity was determined. (B) Competitive inhibition of Eap binding to S. aureus. Cold Eap (closed squares) or coagulase (open squares) was added at the indicated concentrations to cells of S. aureus (106 CFU), to which was simultaneously added 125I-labeled Eap (1 μg). Binding of Eap was determined as described for Fig. 4.
Oligomerization of Eap.
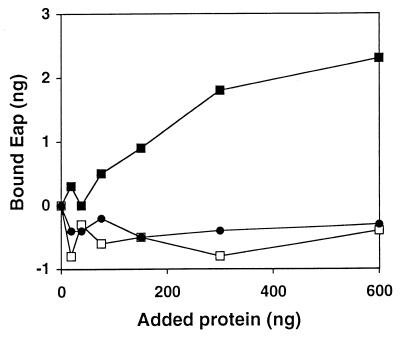
The high efficiency of Eap binding to cells of S. aureus led us to suspect that aggregation or oligomerization of Eap takes place. Eap, Clf, and coagulase were spotted at various concentrations onto a nitrocellulose filter, which was then probed with iodinated Eap. The amount of bound Eap was determined, and Fig. 6 shows that labeled Eap bound only to immobilized Eap, not to Clf or coagulase.
FIG. 6.
Binding of radiolabeled Eap to protein on nitrocellulose membranes. Different amounts of Eap (closed squares), coagulase (open squares), and Clf (closed circles) were absorbed onto separate spots on a nitrocellulose membrane. The filter was soaked in 125I-labeled Eap and then washed. Radioactivity in the individual spots was determined. Background value is subtracted (a piece of the filter with no spotted protein), hence the negative binding.
Eap-Eap interaction was also demonstrated by affinity chromatography of Eap with an Eap-Sepharose column. Eap was coupled to Sepharose, and a LiCl extract or supernatant of an S. aureus culture from the postexponential phase was pumped through the column. A 60-kDa protein was eluted from the Eap-Sepharose and showed a high degree of purity on SDS-PAGE gels (Fig. 1, lane 1). The protein could be recovered both from the LiCl extract and from the supernatant. This protein was found to bind to Fg, Pt, and Fn in a capture ELISA (as in Fig. 3A, but not shown here). The elution profile from the FPLC Mono S column was the same as that for Eap (0.5 to 0.7 M NaCl). The NH2-terminal sequence (AAKPLDKSSS) was the same as that in Eap from strain Newman (AAKPLDKS). To eliminate the possibility that this protein was a result of leakage of Eap from the column, the Eap-Sepharose column was repeatedly treated with elution buffer (PBS with 1 M NaCl) and PBS. No leakage of Eap was detected. Therefore, we conclude that this protein can be purified with an Eap-Sepharose column, presumably due to an ability of Eap to form oligomers.
Agglutination of S. aureus.
Due to its ability to form oligomers and to bind to cells of S. aureus, Eap was expected to cause bacterial aggregation. To washed cells of strain Newman, Eap was therefore added at various concentrations. Agglutination was readily visible within 30 min, and the strength was scored as a −, +, or ++. Table 2 shows that the agglutination was dose dependent and species specific. Five other strains of S. aureus also showed similar aggregation (data not shown). Protease treatment of the cells before addition of Eap eliminated aggregation (data not shown).
Adherence of S. aureus to fibroblasts and epithelial cells.
S. aureus Newman was added to fibroblasts or epithelial cells which had been preincubated with various amounts of Eap. After adherence of bacteria, the cells were methanol fixed and Gram stained and the number of adherent bacteria was determined. Figure 7 shows that the presence of Eap significantly (P < 0.01, comparing 0 and 1 μg of Eap per ml) stimulated the adherence of S. aureus to epithelial cells. A similar stimulation was found also for adherence to fibroblasts (P < 0.001, comparing 0 and 1 μg of Eap per ml) (data not shown).
Relation between Eap and Map.
Analysis of the NH2-terminal sequence of Eap (AAKPLDKS) showed that an S. aureus protein called Map (9, 14) (AAKQIDKSSS) might be related to Eap. The amino acid contents in these two proteins are similar. When the percentage of each amino acid was plotted against those of the others for Eap and Map, a regression coefficient of 0.97 was found, further implying a relationship between these two proteins. However, the C-terminal amino acid sequence of Eap (C-terminal sequence, KNKXS) is not to be found in the published sequence of Map (9).
DISCUSSION
We have previously identified an extracellular 60-kDa protein with a capacity to bind to both Pt and Fg (2). This study was now extended, and it was shown that this protein can bind also to Fn, several different plasma proteins, itself, and cells of S. aureus. It should be noted that the protein, although very sticky, has a targeted specificity, since not every protein in plasma or in a supernatant or LiCl extract of S. aureus was bound by it when affinity chromatographed. Human serum albumin, Cn, and lysozyme did not bind either in a capture ELISA or in a Western affinity blot.
Eap and another S. aureus protein named Map (9, 14) from strain FDA 574 have similar N-terminal sequences, and the amino acid compositions are also very similar, although not identical. Exact identity between these proteins is unlikely due to different C-terminal amino acid sequences. However, nucleotide sequence information (26) for a p70 protein (6) from S. aureus Wood 46 shows that this p70 protein has both C and N termini which are identical to those of Eap. Eap and Map were isolated from different strains of S. aureus, Newman and FDA 574, respectively. Partial amino acid sequence information is also available for Map from strain Newman (10), showing a C terminus similar to those of Eap and p70. The amino acid sequence identity between Map from FDA 574 and p70 is 84% for the first 540 amino acids but only 27% for the last 110 amino acids. Thus, Eap, p70, and Map seem to be members of the same family of proteins.
A novel mechanism of bacterial surface anchoring was recently described for Listeria monocytogenes (4). A C-terminal portion of internalin with a GW motif is required for association of internalin with L. monocytogenes. Association between internalin and the bacterium also takes place when added externally and when added to some other gram-positive species (4). The binding of Eap to S. aureus resembles the situation with internalin in that externally added Eap can associate with the bacterium. Another example of a bacterial protein binding to the surface of the bacterium producing it is binding of pneumococcal autolysin to teichoic acid (8).
The ability of soluble iodinated Eap to bind to S. aureus was matched by the finding that S. aureus binds to immobilized Eap, a binding as efficient as binding to immobilized Fg. The interaction between bacteria and Eap seems to be more pronounced for S. aureus, since S. epidermidis, S. mutans, and E. coli did not bind to a detectable level whereas another strain of S. aureus could also bind. Furthermore, binding of Eap to S. aureus could be competitively inhibited by Eap but not by coagulase.
To demonstrate the assumed oligomerization of Eap, Eap was coupled to Sepharose, and an S. aureus Newman supernatant was pumped through. Eap was then recovered from the Eap-Sepharose. Repetitive washes and mock elutions excluded the possibility of Eap leakage from the Eap-Sepharose. Oligomerization was confirmed in the experiment showing Eap-Eap interaction on nitrocellulose. It should again be stressed that the broad binding specificity of Eap for matrix and plasma proteins does not mean that every protein would bind to immobilized Eap; otherwise, in the affinity chromatography on Eap-Sepharose, a large number of proteins would have been enriched.
The ability to oligomerize and to bind to the bacterial cell surface implies that Eap would be able to aggregate bacteria. This was, in fact, also found, and the ability was found to be concentration dependent. The concentration required of Eap to promote aggregation is the concentration found in supernatants of overnight cultures. Eap could be the explanation for spontaneous aggregation often seen in S. aureus cultures.
This is the first report of an extracellular bacterial protein possessing both a broad binding specificity for matrix and plasma proteins and an ability to recognize the bacterial cell surface. We propose that this protein serves as a bridging molecule between host components and the bacterium, thereby acting as a stimulator of bacterial adherence by first priming the surface for adherence. This is supported by the stimulatory effect of Eap on the binding of S. aureus to fibroblasts and epithelial cells. Such an adherence mechanism would complement the adherence exerted by surface-located bacterial proteins such as Clf, Fn binding protein, collagen binding protein, vitronectin binding protein, etc. The ability of Eap to aggregate would enhance such an adherence-stimulating function. The proposed adherence enhancement mechanism offers an additional feature in that it is concentration dependent. In a situation with a high density of bacteria at the site of infection, and a high local concentration of Eap, adherence enhancement would be relatively more efficient and play a more important role than at a lower bacterial density. The kinetics of adherence would thereby be influenced by a density-sensing mechanism.
In conclusion, we have purified and characterized an extracellular protein from S. aureus, Eap, which enhances adherence of S. aureus to host components.
ACKNOWLEDGMENTS
This work was supported by a grant from the Swedish Medical Research Council (K98-16X-12218-02B) and from Biostapro AB.
David Wade is acknowledged for fruitful discussions and comments.
REFERENCES
- 1.Bodén M, Flock J-I. Cloning and characterization of a gene for a 19 kDa fibrinogen binding protein from Staphylococcus aureus. Mol Microbiol. 1994;12:599–606. doi: 10.1111/j.1365-2958.1994.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 2.Bodén M, Flock J-I. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb Pathog. 1992;12:289–298. doi: 10.1016/0882-4010(92)90047-r. [DOI] [PubMed] [Google Scholar]
- 3.Bodén M, Flock J-I. Fibrinogen-binding protein/clumping factor from Staphylococcus aureus. Infect Immun. 1989;57:2358–2363. doi: 10.1128/iai.57.8.2358-2363.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun L, Dramsi S, Dehoux P, Bierne H, Lindahl G, Cossart P. InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol Microbiol. 1997;25:285–294. doi: 10.1046/j.1365-2958.1997.4621825.x. [DOI] [PubMed] [Google Scholar]
- 5.Flock J-I, Fröman G, Jonsson K, Guss B, Signäs C, Nilsson B, Raucci G, Höök M, Wadström T, Lindberg M. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 1987;6:2351–2357. doi: 10.1002/j.1460-2075.1987.tb02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujigaki Y, Yousif Y, Morioka T, Batsford S, Vogt A, Hishida A, Miyasaka M. Glomerular injury induced by cationic 70-kD staphylococcal protein; specific immune response is not involved in early phase in rats. J Pathol. 1998;184:436–445. doi: 10.1002/(SICI)1096-9896(199804)184:4<436::AID-PATH1225>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Hendrix H, Lindhout T, Mertens K, Engels W, Hemker H C. Activation of human prothrombin by stoichiometric levels of staphylocoagulase. J Biol Chem. 1983;258:3637–3644. [PubMed] [Google Scholar]
- 8.Holtje J V, Tomasz A. Specific recognition of choline residues in the cell wall teichoic acid by the N-acetyl-L-alanine amidase of Pneumococcus. J Biol Chem. 1975;250:6072–6076. [PubMed] [Google Scholar]
- 9.Jönsson K, McDevitt D, McGavin M H, Patti J M, Höök M. Staphylococcus aureus expresses a major histocompatibility complex class II analog. J Biol Chem. 1995;270:21457–21460. doi: 10.1074/jbc.270.37.21457. [DOI] [PubMed] [Google Scholar]
- 10.Kreikemeyer, B., D. McDevitt, V. Kapur, and M. Hook. 1998. The MHC class II analog protein (Map) expressed by S. aureus: prevalence of the Map gene, expression of size variants and characterization of a second gene class. National Center for Biotechnology Information database, accession no. AJ223806.
- 11.Liang O D, Ascencio F, Fransson L A, Wadström T. Binding of heparan sulfate to Staphylococcus aureus. Infect Immun. 1992;60:899–906. doi: 10.1128/iai.60.3.899-906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mamo W, Bodén M, Flock J-I. Vaccination with Staphylococcus aureus fibrinogen binding proteins (FgBPs) reduces colonization of S. aureus in a mouse mastitis model. FEMS Immunol Med Microbiol. 1994;10:47–54. doi: 10.1111/j.1574-695X.1994.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 13.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 14.McGavin M H, Krajewska-Pietrasik D, Rydén C, Höök M. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect Immun. 1993;61:2479–2485. doi: 10.1128/iai.61.6.2479-2485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, Francois P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palma M, Nozohoor S, Schenning T, Heimdahl A, Flock J-I. Lack of the extracellular 19-kilodalton fibrinogen-binding protein from Staphylococcus aureus decreases virulence in experimental wound infection. Infect Immun. 1996;64:5284–5289. doi: 10.1128/iai.64.12.5284-5289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palma M, Wade D, Flock M, Flock J-I. Multiple binding sites in the interaction between fibrinogen and an extracellular fibrinogen binding protein from Staphylococcus aureus. J Biol Chem. 1998;273:13177–13181. doi: 10.1074/jbc.273.21.13177. [DOI] [PubMed] [Google Scholar]
- 18.Park P W, Rosenbloom J, Abrams W R, Rosenbloom J, Mecham R P. Molecular cloning and expression of the gene for elastin binding protein (ebpS) in Staphylococcus aureus. J Biol Chem. 1996;271:15803–15809. doi: 10.1074/jbc.271.26.15803. [DOI] [PubMed] [Google Scholar]
- 19.Patti J M, Jönsson K, Guss B, Switalski L, Wiberg K, Lindberg M, Höök M. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem. 1989;267:4766–4772. [PubMed] [Google Scholar]
- 20.Paulsson M, Liang O, Ascencio F, Wadström T. Vitronectin binding surface proteins of Staphylococcus aureus. Zentbl Bakteriol. 1992;277:54–64. doi: 10.1016/s0934-8840(11)80871-6. [DOI] [PubMed] [Google Scholar]
- 21.Phonimdaeng P, O’Reilly M, Nowlan P, Bramley A J, Foster T J. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol. 1990;4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 22.Sawai T, Tomono K, Yanagihara K, Yamamoto Y, Kaku M, Hirakata Y, Koga H, Tashiro T, Kohno S. Role of coagulase in a murine model of hematogenous pulmonary infection induced by intravenous injection of Staphylococcus aureus enmeshed in agar beads. Infect Immun. 1997;65:466–471. doi: 10.1128/iai.65.2.466-471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 25.Signäs C, Raucci G, Jönsson K, Lindgren P-E, Anantharamaiah G M, Magnus H, Martin L. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus and its use in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yousif, Y., R. Draeger, M. Schiltz, H. Peter, and M. Schleisier. 1997. Nucleotide sequence of a S. aureus gene encoding outer surface binding 70 kD protein. National Center for Biotechnology Information database, accession no. Y10419.