Abstract
Infections with multidrug resistant (MDR) Enterococcus faecium (Efm) are a growing problem. Vancomycin resistance in enterococci has long challenged treatment, necessitating the use of linezolid or daptomycin. Subsequently, daptomycin-, linezolid-, vancomycin-resistant Efm (DLVRE) infections have emerged. Case reports and guidelines for treating DLVRE infections are limited. Here, we describe the clinical and laboratory management of an MDR Efm protracted intraabdominal (IA) infection and breakthrough DLVRE bacteremia. Serial Efm resistance was evaluated using whole genome sequencing (WGS), susceptibility testing, and synergy analysis. Prior to in vitro synergy testing, combination antimicrobial therapy with daptomycin (DAP) and ceftaroline (CPT) was employed to treat the patient’s central line-associated DLVRE bloodstream infection. In vitro antimicrobial testing revealed no synergy between daptomycin and ceftaroline; however, the patient’s bacteremia cleared following initiation of both in conjunction with catheter removal. Sequencing of the DLVRE isolates revealed multiple genomic mutations which explained both linezolid and daptomycin resistance phenotypes and confirmed the presence of a plasmid containing the vanA operon. Sequential WGS of two additional bacterial isolates from the same patient revealed protracted colonization with a single DLVRE clone and suggested the development of bacterial subpopulations. Pairing clinical isolate susceptibilities with WGS and synergy testing should be encouraged in clinical practice to better inform antimicrobial management in cases of multidrug resistance.
Abbreviations: WGS, whole genome sequencing; DAP, daptomycin; LZD, linezolid; CPT, ceftaroline; Efm, Enterococcus faecium; DLVRE, daptomycin-, linezolid-, vancomycin-resistant Enterococcus faecium; PBP5, penicillin binding protein 5; IA, intraabdominal; MIC, minimal inhibitory concentration; DNSE, daptomycin-non-susceptible enterococci; CLABSI, central line-associated bloodstream infection; POD, post-operative day; PICC, peripherally inserted central catheter; LLQ, left lower quadrant; ST, sequence type; MDR, multidrug resistant; p1–5, plasmids 1–5
Keywords: Case report, Vancomycin-resistant Enterococcus faecium, Daptomycin-resistance, Linezolid-resistance, Whole genome sequencing, Synergy antimicrobial resistance testing
Introduction
Nosocomial infections caused by multidrug resistant (MDR) Enterococcus faecium (Efm) are a significant challenge to patients and clinicians. As an important part of the gastrointestinal (GI) microbiota, enterococci may be exposed to serial courses of antimicrobials and persist, in part, because of the remarkable plasticity of their genome [1]. Antimicrobial resistance (AMR) in enterococci arises through both genomic mutation and acquisition of mobile elements [2]. Most enterococci demonstrate low-level intrinsic resistance to beta-lactam compounds, moderate resistance to aminoglycosides and high level resistance to most cephalosporins and clindamycin [3], [4]. Natural ampicillin resistance in Efm is attributed to the chromosomally-encoded penicillin-binding protein (PBP5) [5] which can encode a low-affinity allele (pbp5-R) responsible for ampicillin resistance in the dominant hospital-associated Efm clade [6]. Vancomycin resistance, driven by expression of multiple gene clusters (van operons), results in the replacement of terminal D-alanine residues of peptidoglycan precursors with either D-lactate or D-serine [4].
Treatment of vancomycin-resistant Efm (VRE) requires the use of antimicrobial agents including linezolid (LZD) and daptomycin (DAP) [7]. Though overall resistance to either remains rare (< 1 % linezolid and < 2 % daptomycin) [8], [9], resistance emerged shortly after each antimicrobial was clinically introduced [7], [9]. Specifically, resistance to linezolid is linked to mutations in 23S ribosomal RNA genes or acquisition of cfr or optrA, which encode a ribosomal methyltransferase and an ATP-binding cassette transporter, respectively [9]. Daptomycin resistance is linked to mutations in two major groups of genes [4], [8]. The first (liaFSR and yycFGHIJ) encodes regulatory pathways that coordinate stress responses in the bacterial cell envelope. The second encodes enzymes that metabolize phospholipids, including glycerophosphoryl diester phosphodiesterase (gdpD) and cardiolipin synthetase (cls) [10]. The molecular understanding of both linezolid and daptomycin resistance is an active area of investigation [9], [10].
Though rare (< 1 %), the prevalence of daptomycin-, linezolid-, and vancomycin-resistant Efm (DLVRE) is increasing [8], [9]. Immunosuppression, neutropenia, receipt of an invasive medical procedure, and antimicrobial exposure increase a patient’s risk for development of DLVRE [11]. There are limited data on efficacious treatment strategies for DLVRE. For daptomycin-non-susceptible enterococci (DNSE), combination therapy with daptomycin and a beta-lactam antibacterial is believed to act synergistically wherein the beta-lactam alters the charge on the bacterial cell membrane which improves daptomycin binding and bactericidal activity [12], [13], [14]. In vitro studies have demonstrated synergy between daptomycin and ampicillin, ceftriaxone, cefepime, ertapenem and ceftaroline (CPT) [15]. Of the many combinations tested in vitro, ceftaroline significantly lowered the minimal inhibitory concentration (MIC) of daptomycin the most and showed the greatest enhancement in daptomycin binding [15]. Additionally, daptomycin and ceftaroline have also been employed successfully to treat E. faecalis endocarditis [13]. Ultimately, data are limited on the clinical impact of dual antimicrobial therapy on outcome of DNSE and DLVRE infections.
Here, we report successful treatment of breakthrough DLVRE centrtal line-associated bloodstream infection (CLABSI) in the setting of a protracted polymicrobial intraabdominal (IA) abscess. Additionally, we applied whole genome sequencing (WGS) to characterize the genetic changes underlying linezolid and daptomycin resistance in this case.
Case presentation
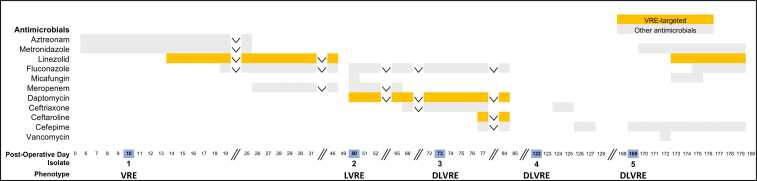
A 64-year-old female with a history of small bowel obstruction and multiple laparoscopic abdominal surgeries presented to our institution for elective incisional hernia repair. The patient underwent lysis of adhesions and small bowel resection with mesh closure. Post-operatively, the patient’s course was complicated by the development of multiple loculated IA abscesses (1.5 × 6.6 cm, 2.0 × 3.3 cm, and 2.9 × 11.9 cm) and an enteric leak at the anastomotic site. A drain was placed into the largest pocket and aspiration cultures (60 mL of feculent material) obtained on post-operative day (POD) 10 were consistent with a polymicrobial abscess including Escherichia coli, Enterococcus faecalis, coagulase-negative staphylococcus, Streptococcus constellatus, Candida albicans and vancomycin-resistant Efm (Table 1, Isolate 1, VRE). Given an allergy to penicillin, the patient was initially treated with aztreonam and metronidazole. The patient was ultimately transitioned to culture-directed therapy (meropenem, fluconazole, and linezolid) for four weeks (Fig. 1). At the end of her antimicrobial course, clinical improvement was achieved, though several small IA and subcutaneous fluid collections (1.9 × 1.8 cm, 6.0 × 0.9 cm, 3.0 × 1.9 cm, 3.7 × 1.0 cm, and 2.0 × 0.7 cm) remained. Given the patient's prolonged exposure to antimicrobial therapy and in the context of her clinical and radiographic improvement, a decision was made to stop all broad-spectrum antimicrobials.
Table 1.
Minimal inhibitory concentrations (MICs µg/mL) for the patient’s isolates of Efm as reported by the clinical microbiology laboratory.
| Antibacterial | Isolate 1 (VRE) |
Isolate 2 (LVRE) |
Isolate 3a (DLVRE) |
Isolate 4a (DLVRE) |
Isolate 5a (DLVRE) |
|---|---|---|---|---|---|
| Source | Peritoneal Fluid | Abdominal Fluid | Blood & PICC | LLQ Abdominal Wall | Abdominal Abscess |
| Ampicillin | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb |
| Daptomycin | – | 2 Sc | 12 Rc | 16 Rc | 4 Sc |
| Linezolid | 2 Sb | 128 Rc | ≥ 256 Rc | ≥ 8 Rb | 2 Sb |
| Ciprofloxacin | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb |
| Levofloxacin | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb |
| Erythromycin | ≥ 8 Rb | ≥ 8 Rb | ≥ 8 Rb | 4 Ib | ≥ 8 Rb |
| Gentamicin (synergy) | SYN-Sb | SYN-Sb | SYN-Sb | SYN-Sb | SYN-Sb |
| Streptomycin (synergy) | SYN-Sb | SYN-Sb | SYN-Sb | SYN-Sb | SYN-Sb |
| Nitrofurantoin | 64 Ib | 64 Ib | 64 Ib | 32 Sb | 64 Ib |
| Tetracycline | ≥ 16 Rb | ≥ 16 Rb | ≥ 16 Rb | ≥ 16 Rb | ≥ 16 Rb |
| Tigecycline | ≤ 0.12 Sb | ≤ 0.12 Sb | ≤ 0.12 Sb | ≤ 0.12 Sb | ≤ 0.12 Sb |
| 0.094 NIc | 0.125 NIc | ||||
| Vancomycin | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb | ≥ 32 Rb |
| Ceftaroline | 32 NIc |
R, resistant; S, susceptible; I, intermediate, NI, no interpretation; –, not reported.
Sequenced isolate.
MICs determined by Vitek-2 AST GP75.
MICs determined by MIC strip testing assays.
Fig. 1.
Timeline of antimicrobial treatment andE. faeciumisolate recovery. Post-operative day and Efm isolate recovery profiles (V, vancomycin; L, linezolid, D, daptomycin; bottom) are shown together with antibacterial and antifungal treatment regimens. Vancomycin-resistant enterococci (VRE) targeted antimicrobials are highlighted in orange and other antimicrobials are presented in light grey. Isolate post-operative day recovery dates are highlighted in blue. Inverted carrots signify that the antimicrobial agent in question was continued throughout the time period in question. Two vertical forward slashes denote time breaks.
Four days after stopping antimicrobials (POD 50), the patient developed abdominal pain and leukocytosis of 26,600 cells/µL. An abdominal CT scan revealed multiple new, loculated rim-enhancing fluid collections (8.7 × 4.0 × 11.2 cm and 5.7 × 5.8 × 11 cm) and persistent small bowel enteric leak. She underwent an excision laparotomy with abdominal washout and IA cultures ultimately demonstrated a new linezolid resistant-VRE (Table 1, Isolate 2, LVRE). Meropenem, fluconazole and dose-optimized daptomycin (9.6 mg/kg/day) were initiated and continued for two weeks until ultimately transitioning to ceftriaxone, fluconazole and daptomycin. On POD 73, following an episode of emesis directly onto an existing peripherally inserted central catheter (PICC), the patient developed a fever. Blood cultures from the periphery and PICC line both revealed new daptomycin resistant-LVRE (Table 1, Isolate 3, DLVRE). Ceftriaxone was discontinued and ceftaroline was added when the daptomycin non-susceptibility results became available. The PICC line was removed and blood cultures cleared the following day. After one week of combination antimicrobial therapy, the patient had a decrease in the size of the IA abscess (2.4 × 2.3 cm), resolution of leukocytosis, reduction of fevers and was discharged home off antimicrobials on POD 86 with an abdominal drain in place.
Five weeks after discharge (POD 122), the patient was readmitted with left lower quadrant (LLQ) abdominal pain and increased drainage from the abdominal drain site. CT scan revealed an additional abdominal wall fluid collection (8.1 × 1.0 × 0.9 cm) which was again drained. Cultures were polymicrobial and grew Klebsiella oxytoca, Enterobacter cloacae, Streptococcus anginosus, E. coli, E. faecalis and DLVRE (Table 1, Isolate 4). The patient was diagnosed with an enterocutaneous fistula and briefly treated with ceftriaxone followed by cefepime. To spare the patient additional prolonged courses of broad-spectrum antimicrobials in the setting of clinical stability, the patient was discharged home off all antimicrobials on POD 128.
On POD 169 the patient was readmitted following a fall. While hospitalized, the patient developed a new leukocytosis to 16,400 cells/µL, LLQ abdominal tenderness and erythema. An abdominal CT scan demonstrated a new 5.8 cm abscess in the left anterior abdominal wall. The abscess was drained and cultures grew Pseudomonas aeruginosa, K. oxytoca, E. cloacae, C. albicans and DLVRE (Table 1, Isolate 5). The patient received a 10-day course of antimicrobials including cefepime, linezolid, metronidazole and fluconazole. Of note, Isolate 5 was initially reported as susceptible to linezolid (MIC of 2 µg/mL on Vitek2 platform). However, upon post-hoc laboratory resistance testing, Isolate 5 was determined to be resistant to linezolid with an MIC of 12 µg/mL (Table 2, Isolate 5). Despite this discrepancy which was unknown at the time of treatment, the patient’s clinical status improved. After one week of inpatient observation off antimicrobials, the patient was discharged home on POD 187. A detailed course of antimicrobial therapy is outlined in Fig. 1.
Table 2.
Post-hoc laboratory MICs (µg/mL) for the patient’s isolates of Efm.
| Antibacterial | Isolate 3 (DLVRE) |
Isolate 4 (DLVRE) |
Isolate 5 (DLVRE) |
|---|---|---|---|
| Source | Blood & PICC | LLQ Abdominal Wall | Abdominal Abscess |
| Daptomycin | 32 Ra‡ | 16 Ra | 32 Ra‡ |
| Linezolid | 48 Rb‡ | > 256 Rb‡ | 12 Rb‡ |
R, resistant. aMICs determined by microbroth dilution according to CLSI protocols. bMICs determined by MIC strip testing assays (Liofilchem®). ‡Results discordant between clinical microbiology laboratory and post-hoc laboratory MIC testing.
Post-hoc resistance testing reveals no synergy between DPT and CPT
Given that the patient’s CLABSI was resistant to daptomycin (DLVRE, Isolate 3) and developed while on daptomycin and ceftriaxone combination therapy, we made the clinical decision to treat with daptomycin and ceftaroline combination therapy based several studies suggesting improved synergistic effect between these agents for the treatment of daptomycin non-susceptible enterococci [12], [13], [14], [15]. However, we did not have synergy AMR data available at the time of clinical decision making. Therefore, in a post-hoc analysis, we sought to confirm that synergy between ceftaroline and daptomycin existed for this isolate. First, we repeated both linezolid and daptomycin MIC testing. Using microbroth dilution, we determined the daptomycin MICs of Isolates 3, 4 and 5 to be 32, 16 and 32 µg/mL respectively. The MICs of Isolates 3 and 5 were higher than that reported by the clinical microbiology laboratory and the linezolid MICs differed from those reported by the clinical microbiology laboratory using different methodology (Table 2). For Isolate 3, we performed microbroth dilution checkerboard experiments with daptomycin and ceftaroline. We observed no synergy between ceftaroline and daptomycin as the MIC of Isolate 3 remained 32 µg/mL despite addition of ceftaroline.
Genomic evaluation of antimicrobial resistance in Isolate 3
We used genomics to understand the mechanisms behind evolving drug-resistance in our DLVRE isolates. Unfortunately, Isolates 1 and 2 had been previously discarded. WGS of Isolate 3 confirmed the identity of a sequence type (ST) 584 Efm. ST584 is a member of the pandemic clonal complex 17 that includes a collection of hospital-acquired Efm STs [16]. The complete genome of Isolate 3 included a single chromosome of 2.8 Mbp and 5 plasmids ranging from 228.3 kbp to 1.9 kbp. Plasmid sequences were similar to previously described Efm plasmids, although present in different configurations. For example, the majority of the sequence present in plasmids 2 and 4 has been described in substantially larger plasmids reaffirming that Efm plasmids are highly modular [2] (Supplementary Table S1).
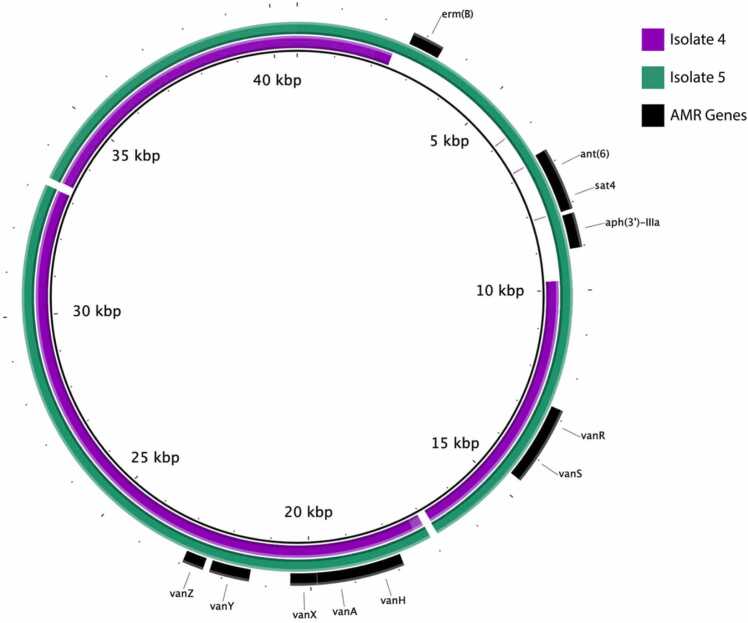
Examination of the complete genome of Isolate 3 for known antimicrobial resistance determinants reasonably explained the observed susceptibility pattern (Table 3). Isolate 3 contained the low-affinity pbp5-R allele known to be associated with ampicillin resistance in Efm [4], [6]. Plasmid 3 (p3) contained a vanA operon (Fig. 2), explaining the observed vancomycin resistance. Daptomycin resistance was attributed to the presence of chromosomal mutations in liaS (Thr120Ala), liaR (Trp73Cys) and cls (Asp13Ile). Other resistance mechanisms identified are described in Table 3.
Table 3.
Genomic mechanisms of antimicrobial resistance identified in Isolates 3, 4, and 5.
| Antibacterial | Resistance determinants | Location in Isolate 3 | Isolate 3 | Isolate 4 | Isolate 5 |
|---|---|---|---|---|---|
| Ampicillin | pbp5 (Val24Ala, Ser27Gly, Arg34Gln, Gly66Glu, Ala68Thr, Glu85Asp, Glu100Gln, Lys144Gln, Thr172Ala, Leu177Ile, Asp204Gly, Ala216Ser, Thr324Ala, Met485Ala, Asn496Lys, Ala499Thr, Glu525Asp, Glu629Val, Pro667S) | Chromosome | + | + | + |
| Linezolid | 23S rRNA gene G2576T | Chromosome | ~ 3/6 copies | ~ 3/6 copies | ~ 2/6 copies |
| Daptomycin | liaS (Thr120Ala) | Chromosome | + | + | + |
| liaR (Trp73Cys) | Chromosome | + | + | + | |
| cls (Asp13Ile) | Chromosome | + | + | + | |
| Vancomycin | vanHAX | Plasmid 3 | + | + | + |
| vanSR | Plasmid 3 | + | + | + | |
| vanZY | Plasmid 3 | + | + | + | |
| Fluoroquinolones | gyrA (Ser83Tyr) | Chromosome | + | + | + |
| parC (Ser80Arg) | Chromosome | + | + | + | |
| Aminoglycosides | aac(6′)-I | Chromosome | + | + | + |
| aph(3′)-IIIa | Plasmid 3 | + | – | + | |
| ant(6)-Ia | Plasmid 3 | + | – | + | |
| Trimethoprim | dfrF | Chromosome | + | + | + |
| dfrG | Chromosome | + | + | + | |
| Tetracyclines | tetL | Chromosome | + | + | + |
| tetM | Chromosome | partial and complete copy | partial copy | partial copy | |
| Streptothricin | sat4 | Plasmid 3 | + | – | + |
| Macrolides | msrC | Chromosome | + | + | + |
| ermB | Plasmid 3 | + | – | + |
Fig. 2.
Alignment of plasmid 3 from Isolates 4 and 5 to the vancomycin resistance plasmid 3 of Isolate 3. Regions present in Isolate 4 and 5 draft genomes are indicated in purple and green respectively. Identified resistance genes are indicated in black.
The mechanism of linezolid resistance for Isolate 3 was not immediately apparent from interrogation of its complete genome sequence. A read-based analysis estimated that Isolate 3 contained three copies of the variant G2576T allele in the six copy 23S rRNA gene (48 % of aligned reads contained the G2576T variant allele, Table 3). The absence of these variants in the complete genome may be an artifact of the assembly method interacting with a multi-copy gene. The level of linezolid resistance has been shown to correlate with the number of variant alleles, particularly G2576T, in the 23S rRNA gene [9], [17]. Other well characterized linezolid resistance determinants were not identified (Table 3).
Evolution of Enterococcus faecium during protracted infection
Given the patient’s repeated isolation of DLVRE, we chose to perform short-read sequencing on two additional Efm Isolates, Isolates 4 and 5. Isolates 4 and 5 were clonal with Isolate 3, possessing a total of 2 and 16 single nucleotide variants (SNVs) with Isolate 3 respectively (Supplementary Tables S2, S3). All SNVs present in Isolate 5 were previously observed in Isolate 4. Altogether, these findings strongly suggest persistent infection or colonization by a specific DLVRE clone.
Isolates 4 and 5 contained the same ampicillin, vancomycin, and daptomycin resistance elements as Isolate 3. Similar to Isolate 3, Isolate 4 contained an estimated three copies (56 %) of the G2576T 23S rRNA gene allele. Isolate 5, on the other hand, contained an estimated two copies (33 %) of the G2576T allele (Table 2). As G2576T allele copy number correlates with level of linezolid resistance [17], this potentially explains the lower linezolid MIC of Isolate 5, but there may be other contributing factors or additional mutations given the substantially different linezolid MICs between isolates 3 and 4 which have similar percentage of reads containing the G2576T variant alleles (Table 1).
Additionally, we noted the absence of a substantial portion of plasmid 3 in Isolate 4 (Fig. 2, Supplementary Table S2), resulting in the loss of multiple resistance genes (Table 2, Fig. 2). This 7.4 kbp deletion was flanked by IS1216 family transposase genes, suggesting that it was located on a mobile element. Interestingly, this sequence was present in a temporally later isolate, Isolate 5 (Fig. 2). This implies that multiple subpopulations of this ST584 DLVRE clone developed during protracted infection, as an intact p3 would have to have been present in the DLVRE population when Isolate 4 was collected to be found subsequently in Isolate 5.
Discussion and conclusions
Limited data exist on appropriate antibacterial choice in DLVRE bacteremia in the setting of protracted IA abscess [7]. In this challenging case, it is notable that our patient developed breakthrough daptomycin resistance and bacteremia while receiving high-dose daptomycin (9.6 mg/kg) in conjunction with beta-lactam therapy (meropenem followed by ceftriaxone). Given our limited options for therapy and based on prior clinical and experimental data [12], [13], [14], [15], we elected to transition to ceftaroline in combination with daptomycin for synergy once the patient developed DLVRE bacteremia. While this patient improved rapidly, cleared her blood cultures and had a decrease in the size of her IA abscess, post-hoc laboratory analysis, unlike previous reports, did not demonstrate synergy between daptomycin and ceftaroline for this patient’s DLVRE isolate [12], [13], [14], [15]. Therefore, we believe that the patient most likely cleared her cultures once the source (an indwelling PICC line) was removed. More broadly available rapid MIC synergy testing would have been clinically useful in crafting this patient’s antimicrobial regimen and minimizing ineffective antimicrobial exposures. Overall, the patient received a total of 57 days of broad-spectrum antimicrobials for protracted IA abscesses during her initial hospitalization. Therefore, this case also underscores the challenges that clinicians face when deploying culture-directed therapy in patients with protracted and uncontrolled infectious reservoirs. In settings where complete source control is not achievable, there is no clear answer on antimicrobial choice or duration. Following initial improvement, the patient remained persistently colonized with DLVRE as evidenced by its repeated recovery in the months following treatment. Larger studies are needed for further evaluation of daptomycin and beta-lactam antimicrobial combination therapy for DLVRE infections.
By deploying WGS in this limited series of DLVRE isolates, we identified possible genetic explanations for vancomycin, daptomycin and linezolid resistance. The molecular mechanisms underpinning the evolution of daptomycin and linezolid resistance in this case are limited by the lack of access to Isolates 1 and 2, but, by sequencing Isolates 4 and 5, we confirmed that the patient was persistently colonized over a protracted period with clonal subpopulations of DLVRE. This work, in conjunction with others, demonstrates the value of antimicrobial synergy testing and WGS during prolonged infection with VREfm to provide improved molecular understanding of the genomic changes responsible for acquired AMR, preferably at the time of antimicrobial decision-making [18]. Our post-hoc laboratory MIC testing revealed discrepancies in daptomycin and linezolid MICs for Isolates 3, 4 and 5 reinforcing the clinical challenges of selecting active antimicrobial regimens in cases of DLVRE infection. Additionally, we noted plasmid heterogeneity between Isolates 3 and 5 and Isolate 4 related to loss of a large plasmid segment on p3 containing several AMR genes. This implies that there were subpopulations of Efm that developed during infection. The concept of a single species bacterial “cloud of diversity” in prolonged infection is not new, but the impact of this diversity on successful antimicrobial treatment and infection outcome is clinically underappreciated [1], [19]. We have shown here that subpopulations possess different armamentariums of AMR genes and may vary in clinically relevant degrees of resistance. These observations are often missed by traditional phenotypic testing of an isolated colony, potentially masking small subpopulations that harbor more difficult-to-treat antimicrobial resistant phenotypes and contributing to suboptimal antimicrobial therapy decisions.
With the increasing rates of MDR Efm infections, it is critical that we understand how Efm adapts to prolonged antimicrobial pressure and recognize that traditional clinical resistance testing may not mirror population level AMR phenotypes. Further research is needed to elucidate the best course of treatment for patients with VRE requiring long-term antimicrobial therapy especially for those who develop daptomycin resistance despite dose-optimized daptomycin therapy [20]. In scenarios such as these, there is an urgent need for more rapid implementation of synergy MIC testing paired with genomic analysis to support real-time clinical decision making.
CRediT authorship contribution statement
Nathan Pincus: Conceptualization, Software, Formal analysis, Data curation, Writing – original draft, Writing – review & editing, Visualization. Tejas Joshi: Conceptualization, Writing – original draft, Writing – review & editing, Visualization. Samuel Gatesy: Methodology, Investigation, Resources, Writing – original draft, Writing – review & editing. Omar Al-Heeti: Conceptualization, Writing – original draft, Writing – review & editing. W. Justin Moore: Conceptualization, Writing – original draft, Writing – review & editing, Supervision. Kelly Bachta: Conceptualization, Validation, Investigation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition. All authors reviewed the results and approved the final version of the manuscript.
Ethics approval
This research study (STU00214816) was submitted for review to the Northwestern University Institutional Review Board and was deemed a study that does not include factors necessitating patient consent. Further IRB review and approval was, therefore, not required.
Consent
This case report was discussed openly with the patient in question and the patient provided verbal and written informed consent for its publication. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Funding
This work was supported by grants from the National Institutes of Health (NIH)/National Institute of General Medical Sciences (NIGMS) (T32 GM008152 awarded to N.B.P), an American Cancer Society (ACS) Clinician Scientist Development Grant (#134251-CSDG-20-053-01-MPC, awarded to K.E.R.B.), and a Northwestern University Emerging and Re-emerging Pathogen Program (EREPP) grant (awarded to K.E.R.B.). This work was also supported by the Northwestern Memorial Clinical Microbiology Laboratory and the NUSeq Core Facility which is generously supported by the NCI CCSG P30 CA060553 award to the Robert H. Lurie Comprehensive Cancer Center. This work was supported in part through the computational resources and staff contributions provided by the Genomics Compute Cluster which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, Feinberg's Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University's high-performance computing facility, with the purpose to advance research in genomics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
N.P., T.J., S.W.M.G., O.A., W.J.M, and K.E.R.B declare that they have no conflicts of interest.
Acknowledgements
We would like to thank members of the Hauser and Ozer laboratories for their valuable comments during numerous discussions of this manuscript and the clinical microbiology laboratory at Northwestern Memorial Hospital, notably Dr. Chao Qi and Michael Malczynski, for their support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.idcr.2022.e01593.
Appendix A. Supplementary material
Supplementary material
Availability of data and materials
Sequencing and genome assemblies have been deposited to NCBI under BioProject accession no. PRJNA787599, with Isolate 3 denoted NMVRE-001 (SAMN23828484), Isolate 4 denoted NMVRE-002 (SAMN23828912), and Isolate 5 denoted NMVRE-003 (SAMN23828937). Reads are available under SRA accessions SRR17230443 to SRR17230446. Assemblies are available under GenBank accessions GCA_021228615.1, GCA_021364775.1, and GCA_021364755.1.
References
- 1.Dubin K.A., Mathur D., McKenney P.T., et al. Diversification and evolution of vancomycin-resistant Enterococcus faecium during intestinal domination. Infect Immun. 2019;87(7):e00102–e00119. doi: 10.1128/IAI.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arredondo-Alonso S., Top J., McNally A., et al. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. mBio. 2020;11(1):e03284–19. doi: 10.1128/mBio.03284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Solache M., Rice L.B. The Enterococcus: a model of adaptability to its environment. Clin Microbiol Rev. 2019;32(2) doi: 10.1128/CMR.00058-18. [e00058-18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias C.A., Murray B.E. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana R., Aldegheri M., Ligozzi M., et al. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1994;38(9):1980–1983. doi: 10.1128/aac.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietta E., Montealegre M.C., Roh J.H., Cocconcelli P.S., Murray B.E. Enterococcus faecium PBP5-S/R, the missing link between PBP5-S and PBP5-R. Antimicrob Agents Chemother. 2014;58(11):6978–6981. doi: 10.1128/AAC.03648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias C.A., Contreras G.A., Murray B.E. Management of multidrug-resistant enterococcal infections. Clin Microbiol Infect. 2010;16(6):555–562. doi: 10.1111/j.1469-0691.2010.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelesidis T., Humphries R., Uslan D.Z., Pegues D.A. Daptomycin nonsusceptible enterococci: an emerging challenge for clinicians. Clin Infect Dis. 2011;52(2):228–234. doi: 10.1093/cid/ciq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi R., Qin T., Fan W., Ma P., Gu B. The emerging problem of linezolid-resistant enterococci. J Glob Antimicrob Resist. 2018;13:11–19. doi: 10.1016/j.jgar.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Arias C.A., Panesso D., McGrath D.M., et al. Genetic basis for in vivo daptomycin resistance in enterococci. N Engl J Med. 2011;365(10):892–900. doi: 10.1056/NEJMoa1011138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene M.H., Harris B.D., Nesbitt W.J., et al. Risk factors and outcomes associated with acquisition of daptomycin and linezolid-nonsusceptible vancomycin-resistant enterococcus. Open Forum Infect Dis. 2018;5(10):ofy185. doi: 10.1093/ofid/ofy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakoulas G., Rose W., Nonejuie P., et al. Ceftaroline restores daptomycin activity against daptomycin-nonsusceptible vancomycin-resistant Enterococcus faecium. Antimicrob Agents Chemother. 2014;58(3):1494–1500. doi: 10.1128/AAC.02274-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakoulas G., Nonejuie P., Nizet V., et al. Treatment of high-level gentamicin-resistant Enterococcus faecalis endocarditis with daptomycin plus ceftaroline. Antimicrob Agents Chemother. 2013;57(8):4042–4045. doi: 10.1128/AAC.02481-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang Y.C., Chen P.Y., Lin C.Y., et al. A retrospective clinical comparison of daptomycin vs daptomycin and a beta-lactam antibiotic for treating vancomycin-resistant Enterococcus faecium bloodstream infections. Sci Rep. 2018;8(1):1632. doi: 10.1038/s41598-018-19986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith J.R., Barber K.E., Raut A., et al. Beta-Lactam combinations with daptomycin provide synergy against vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J Antimicrob Chemother. 2015;70(6):1738–1743. doi: 10.1093/jac/dkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee T., Pang S., Abraham S., Coombs G.W. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. J Glob Antimicrob Resist. 2019;16:36–47. doi: 10.1016/j.jgar.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Marshall S.H., Donskey C.J., Hutton-Thomas R., Salata R.A., Rice L.B. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2002;46(10):3334–3336. doi: 10.1128/AAC.46.10.3334-3336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran T.T., Panesso D., Gao H., et al. Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob Agents Chemother. 2013;57(1):261–268. doi: 10.1128/AAC.01454-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilambi G.S., Nordstrom H.R., Evans D.R., et al. Evolution of vancomycin-resistant Enterococcus faecium during colonization and infection in immunocompromised pediatric patients. Proc Natl Acad Sci USA. 2020;117(21):11703–11714. doi: 10.1073/pnas.1917130117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contreras G.A., Munita J.M., Simar S., et al. Contemporary clinical and molecular epidemiology of vancomycin-resistant enterococcal bacteremia: a prospective multicenter cohort study (VENOUS I) Open Forum Infect Dis. 2022;9(3):ofab616. doi: 10.1093/ofid/ofab616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Sequencing and genome assemblies have been deposited to NCBI under BioProject accession no. PRJNA787599, with Isolate 3 denoted NMVRE-001 (SAMN23828484), Isolate 4 denoted NMVRE-002 (SAMN23828912), and Isolate 5 denoted NMVRE-003 (SAMN23828937). Reads are available under SRA accessions SRR17230443 to SRR17230446. Assemblies are available under GenBank accessions GCA_021228615.1, GCA_021364775.1, and GCA_021364755.1.