Abstract
Advances in adeno-associated virus (AAV) engineering have provided exciting new tools for research and potential solutions for gene therapy. However, the lung has not received the same tailored engineering as other major targets of debilitating genetic disorders. To address this, here we engineered the surface-exposed residues AA452-458 of AAV9 capsid proteins at the three-fold axis of symmetry and employed a Cre-transgenic-based screening platform to identify AAV capsids targeted to the lung after intravenous delivery in mice. Using a custom image processing pipeline to quantify transgene expression across whole tissue images, we found that one engineered variant, AAV9.452sub.LUNG1, displays dramatically improved transgene expression in lung tissue after systemic delivery in mice. This improved transduction extends to alveolar epithelial type II cells, expanding the toolbox for gene therapy research for diseases specific to the lung.
Key Words: AAV, vector engineering, directed evolution, gene delivery, lung, lung epithelium, Adeno-associated virus, capsid library, ATII, alveolar epithelial type II
Graphical abstract

Goertsen et al. introduce an engineered AAV9 variant, AAV9.452sub.LUNG1, that has 18-fold increased transduction of mouse lung and targets alveolar epithelial type II cells. AAV9.452sub.LUNG1, derived by directed evolution of an underexplored capsid site, expands options for AAV engineering and provides a delivery vehicle for lung disease gene therapy research.
Introduction
Efforts to treat genetic disorders have been aided by the development of new techniques and technologies for gene therapy. In these efforts, adeno-associated virus (AAV) has been a preferred gene therapy vector due to its non-pathogenicity,1 stable expression in vivo,2 and amenability to genome and capsid engineering.3 The first use of AAV for gene therapy was approved in 2012 to treat lipoprotein lipase deficiency.4 Since then, clinical trials using AAVs have overwhelmingly focused on the central nervous system (CNS), eye, liver, and muscle,5,6 with other organs like lung drastically underrepresented.5 This disparity occurs despite the prevalence of lung-specific genetic disorders and their potential tractability to gene therapy.7 Clinical trials to treat cystic fibrosis, for example, demonstrated the safety of AAV administration, but had limited efficacy.7 Engineered variants with higher transduction efficiency in lung8 could help address this limitation.
Alveolar epithelial type II (ATII) cells are of particular importance in a wide array of genetic lung conditions and lung infections due to their role in host defense and repair of respiratory tissues.9 Impairment of ATII cells is shown to contribute to idiopathic pulmonary fibrosis (IPF),10,11 a debilitating chronic lung disease, for which current therapies only slow disease progression.12,13 ATII cells are also a primary target for infection by severe acute respiratory syndrome coronavirus 2,14,15 as they express the angiotensin-converting enzyme 2 receptor, and their infection and subsequent cell death is associated with cases of severe coronavirus disease 2019 illness.9,14,15 Additionally, mutations in genes producing the SP-B,16 SP-C,17,18 and ABCA319,20 proteins specific to ATII cells are associated with acute respiratory failure and interstitial lung diseases,21,22 frequently leading to death in affected patients.23 While good murine models are available for SP-B,16 SP-C,17,18 and ABCA3 deficiency,19,20 the study and treatment of other lung diseases, like IPF, are hindered by a lack of suitable animal models.13 Preclinical model development, research, and treatment options for these diseases could benefit from additional tailored gene therapy vectors efficiently targeting ATII cells.
Most gene therapy efforts using AAV rely on naturally occurring serotypes with broad, overlapping tropism,24 which can lead to decreased efficiency, safety, and applicability due to off-target transduction and immune response.25 To address this, substantial work has been dedicated to developing methods to obtain AAV variants with enhanced tissue tropism and improved efficacy.26, 27, 28, 29, 30, 31, 32 Similar to the focus of clinical trials, development of AAV variants has been devoted to the CNS,26,31,32 liver,33, 34, 35 eye,36,37 and muscle.27,38,39 The lung has received much less engineering focus,5 with, to our knowledge, one variant displaying improved efficacy in the mouse lung (AAV6.2FF),40,41 one transducing the endothelium of the pulmonary vasculature in the mouse lung (AAV2-ESGHGYF),42 one efficiently transducing human airway epithelia in culture (AAV2.5T),43 and one transducing pig airway epithelia in vivo (AAV2H22).44,45 Deriving specialized variants based on different serotypes, as described herein using AAV9, could help to expand treatment options, evade host immune response,46,47 and/or target distinct cell populations.
Variable regions in surface-exposed loops at the three-fold axis of symmetry are a locus for binding interactions with cell-surface receptors.48, 49, 50 Usually, the AAV capsid is engineered by point mutation51, 52, 53 or by peptide insertion within variable region VIII,54 between amino acids (AA) 588 and 589 of AAV capsid proteins.26,27,30,55, 56, 57, 58 This variable region is easily amenable to peptide display56,59 and it is responsible for heparan sulfate binding in AAV2.50 However, in AAV9 the surface-exposed loop containing AA455 is the furthest protruding54 and AA substitution at this site in conjunction with 588 insertion has been previously shown to yield productive variants displaying modified tropism.60 Introducing diversity into this less frequently explored region of the AAV capsid proteins could promote new receptor binding interactions, facilitating the discovery of vectors capable of elevated transduction across different targets.
Currently, validation and characterization of engineered AAV variants across tissues is a bottleneck in the AAV discovery process.61 The development of tools to consistently characterize variants across tissues will be important for the validation of unique variants derived from different screens. Here, we address this bottleneck by developing an image processing pipeline to compare transgene expression from AAV variants in whole tissue images. Using this screening tool, the CREATE method32 of positive selection, and a substitution library based on 7-mer substitution at the underexplored 452–458 site, we derive an AAV9 variant, AAV9.452sub.LUNG1, which displays markedly improved transduction of the mouse lung, and ATII cells, after systemic injection.
Results
Engineering AA 452–458 of AAV9 capsid proteins for lung targeting
Due to the prominence of the surface-exposed loop containing AA455 (Figure 1A), we theorized that the substitution of AA in this region could redirect viral tropism and facilitate cell receptor interactions in the lung. With this aim, we performed two rounds of selection with a seven AA substitution library in the AA455 loop, between AA452 and AA458 (Figure 1A) in AAV9.
Figure 1.
Capsid engineering and characterization of AAV9.452sub.LUNG1
(A) Left, AAV9 capsid surface model illustrates the location of the protruding loop structures, shown in yellow. Right, zoomed-in view shows the AA replaced by substitution between AA452-458. (B) To generate capsids specific to the mouse lung, a library of variants was created by mutating AA452–458 in the AAV9 backbone. Cre-lox sites were inserted into the viral genome to enable detection of genomes that reach Cre-recombinase containing cells. Variants were systemically administered via retro-orbital injection into Cre mouse lines. After 2 weeks of expression, relevant tissues were harvested and viral DNA isolated and sequenced. Top performing variants were subjected to a second round of selection. (C) Positively enriched variants in the lung after two rounds of selection in vivo were ranked by their average enrichment across codon and animal replicates. (D) NLS-GFP was packaged into the AAV9.452sub.LUNG1 variant under control of the ubiquitous CAG promoter and intravenously administered to mice at a dose of 5 × 1011 vg/animal. Transgene expression was assayed by fluorescence throughout lung. Comparison with the parent AAV9 (left) confirms the tropism measured by NGS. Scale bars in whole tissue images are 2 mm. Scale bars in the magnified regions are 500 μm.
To derive AAV variants specific to the lung after systemic administration, we applied the CREATE method developed by Deverman and colleagues.32 We first generated a library of AAV capsid sequences, theoretically containing all 1.28 billion 7-mer substitutions, and produced the corresponding viruses in HEK293 cells that package a replication-incompetent version of the corresponding viral genome with a polyadenylation sequence flanked by Cre/Lox sites (Figure 1B). This viral library was then intravenously injected into transgenic mouse lines (hSyn-Cre, GFAP-Cre, TH-Cre, VGAT-Cre, CHAT-Cre, and Tek-Cre) that express Cre recombinase in select tissues or cell populations. Variants capable of successfully transducing Cre+ cells have their genome flipped, enabling the recovery of that sequence. Two weeks after injection, we isolated viral genomes from adipose tissue, brain, dorsal root ganglion, heart, liver, lung, kidney, quadricep muscle, ovary, pancreas, spine, spleen, and stomach. We then sequenced the variants collected from each tissue using next-generation sequencing (NGS) and calculated their enrichment, defined as the relative abundance of the sequence found within a specific tissue divided by the relative abundance of that sequence within the injected viral library.
From a first round of selection, we selected the most enriched AA variants from each tissue and transgenic line for a second round of selection, yielding a subset of 5,840 AA variants. This collection of variants was synthesized, along with codon replicates to determine the reproducibility of each variant. The variants from each tissue and transgenic line were synthesized and produced together to avoid propagating bias due to variability in the first round of selection31 and to simplify the selection workflow. Since high levels of Tek gene expression have been previously reported in the lung relative to other tissues,62, 63, 64, 65, 66 we focused our second round of selection in Tek-Cre transgenic mice. After injecting the viral pool into Tek-Cre transgenic mice and allowing transduction and expression to occur over 2 weeks, we collected viral DNA from the lung, sequenced it by NGS, and ranked variants by their enrichment.
After two rounds of capsid selection, only 426 variants showed enrichment of both replicates in the lung of both animals. In the enrichment ranking, one variant clearly outperformed all other variants (Figure 1C): the AA motif KDNTPGR, which we call AAV9.452sub.LUNG1 and which was enriched nearly an order of magnitude more than any other variant. A retrospective analysis of the first round of selection revealed that this variant was originally included due to enrichment in the spine of hSyn-Cre animals. Additionally, the variant was present at low levels in the liver of one hSyn-Cre animal and one TH-Cre animal, but was not identified in the lung of either Tek-Cre animal after the first round. These results support performing multiple rounds of selection to decrease noise and permit variants with altered tropism to become enriched within the tissues of interest, as described previously.31,32
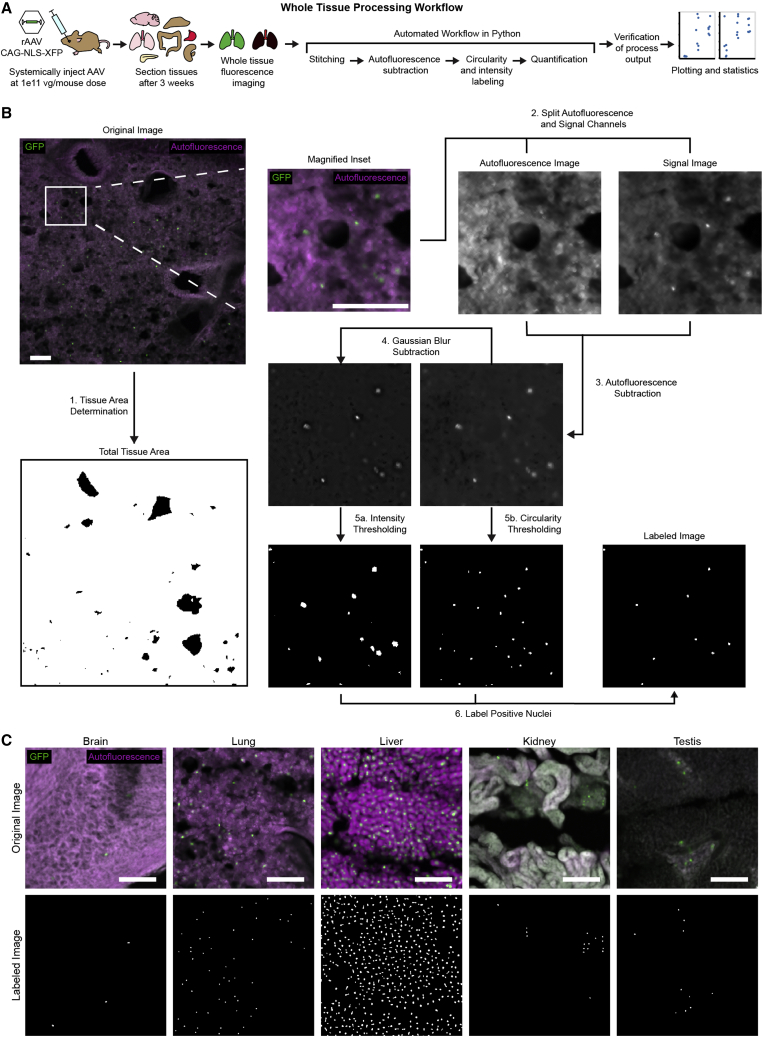
A robust image processing pipeline to quantify transduction across whole tissues
During initial efforts to characterize this variant, we realized that robust high-throughput methods were lacking for the quantification of fluorescent cells in tissue images.67 To address this, we built a simple image processing workflow using open-source Python packages to quantify nuclear-localized (NLS-)GFP signal of whole tissue images acquired by fluorescence imaging (Figures 2A and 2B). Natural autofluorescence in tissues can make it hard to discriminate real signal from noise. To combat this, we also acquired images in a channel corresponding to wavelengths (565 nm) distinct from the wavelengths (495 nm) associated with our signal to allow autofluorescence subtraction. Since we were using NLS fluorescence, we created a mask of nuclei by passing an approximately 6-μm circular template over the autofluorescence subtracted image and thresholding regions for how well they matched the template. We also smoothed the autofluorescence subtracted image by applying a Gaussian blur subtraction and used this smoothed image to identify bright regions of the image by applying a histogram-based intensity threshold. We finally compared these two maps to identify regions of the image that were both bright and circular, which we counted to determine the number of NLS-GFP+ cells. Dividing this count by the total tissue area provides a measure of transgene expression per tissue area. Segmenting the initial image using the identified cell labels allows quantification of the brightness of identified cells.
Figure 2.
Image processing pipeline for quantification of NLS-GFP expression in whole tissue sections
(A) Overview of the characterization experimental workflow. First, AAVs packaging NLS-GFP under regulation of the ubiquitous CAG promoter were systemically administered via retro-orbital injection into mice at a dose of 1 × 1011 viral genomes per animal. After 3 weeks of expression in vivo, tissues were collected, sectioned, and imaged by fluorescence microscopy, including a non-GFP channel for autofluorescence subtraction. Whole tissue images were processed using the developed pipeline. (B) An overview of the processing steps applied to obtain cell counts from tissue images. Although only a single image is displayed here, the pipeline is run over whole tissue images. First, histogram-based thresholding determines the total tissue area. Next, autofluorescence and signal images are compared to generate a subtracted image. The circularity of signal is determined by comparison to a template to generate an image mask of circular regions. Noise is decreased by subtracting Gaussian blur and the second derivative of the histogram is used to determine an intensity threshold, which is applied to generate a mask of bright regions of the image. This is combined with the circular region mask to obtain a mask of cells positive for fluorescent nuclear signal. Scale bars, 100 μm. (C) Identification of NLS fluorescent signal is shown for brain, lung, liver, kidney, and testis, with minimal parameter adjustment between tissues. Scale bars, 100 μm.
We found that this pipeline could be applied to a variety of whole tissue sections with minimal parameter adjustment, improving the consistency of quantification across tissues (Figure 2C). Applying this workflow allowed single-cell resolution biodistribution profiling across more than 690 mm3 of tissue volume, composed of 8 tissues collected from 18 mice, 6 from each of 3 AAV variants.
AAV9.452sub.LUNG1 yields increased transgene expression in mouse lung tissue after intravenous delivery
To validate the sequencing data from the Cre-recombinase selections, we produced both AAV9 and AAV9.452sub.LUNG1 packaging NLS-GFP under regulation of the strong, ubiquitous CAG promoter and intravenously injected each into mice at a dose of 5 × 1011 viral genomes per animal to test the transduction efficacy in lung tissue after systemic injection. Tissue was collected after 2 weeks of expression and compared to AAV9 (Figure 1D). The protein expression we observed in the lung was consistent with the NGS enrichments, with many more GFP positive cells with AAV9.452sub.LUNG1 than with parental AAV9.
To further characterize AAV9.452sub.LUNG1 (Figure 2A), we compared the transgene expression with AAV9 and AAV5, a natural serotype previously identified as promising for lung delivery.68 We produced each virus packaging a single stranded genome expressing CAG-NLS-GFP. These variants were intravenously injected into mice at a dose of 1 × 1011 viral genomes per animal and tissue was collected and sectioned after 3 weeks of expression. A lower dose was selected for characterization to ensure that transduction did not become saturated for quantification. Whole tissue sections from the lung, liver, brain, kidney, pancreas, spleen, intestine, and testis were imaged and compared between the three serotypes.
Qualitatively, we saw a substantial increase in AAV9.452sub.LUNG1 transgene expression in the lung compared with AAV9 and AAV5 (Figure 3A). Quantification of GFP+ nuclei per tissue area (Figure 3B) showed that AAV9.452sub.LUNG1 displays much greater expression in the lung than either AAV9 (18-fold) or AAV5 (60-fold). Both AAV9 and AAV9.452sub.LUNG1 display increased brightness relative to AAV5, with comparable brightness between AAV9 and AAV9.452sub.LUNG1 populations. We measured transgene expression in alveolar type I (ATI) and ATII cells by co-staining with α-podoplanin or α-prosurfactant protein C (proSPC), respectively (Figure 3C). We observed very little co-localization between NLS-GFP expression and ATI cell markers, while much of the transgene expression was co-localized with ATII cells (Figure 3C). Quantification of transgene expression in ATII cells revealed that AAV9.452sub.LUNG1 transduced approximately 30-fold more ATII cells than AAV9. Quantifying the fraction of GFP+ nuclei co-localized with α-proSPC indicated that the relative fraction of ATII cells expressing transgene was comparable between AAV5, AAV9, and AAV9.452sub.LUNG1. This suggests that the increased ATII transgene expression is not due to increased cell type specificity, but is instead a result of increased overall transgene expression across the whole lung.
Figure 3.
Characterization of AAV9.452sub.LUNG1 in lung tissue
(A) Qualitative comparison of transgene expression in the lung shows that there are many more cells expressing transgene after delivery with AAV9.452sub.LUNG1 than AAV5 or AAV9. Scale bar, 100 μm. (B) Quantification of NLS-GFP expression indicate that AAV9.452sub.LUNG1 displays an approximately 18-fold and an approximately 60-fold increased transgene expression compared to AAV9 (p = 0.002) and AAV5 (p = 0.002), respectively. Comparing natural serotypes AAV9 and AAV5, AAV9 yielded GFP expression in approximately three-fold more cells than AAV5 (p = 0.00004). AAV9 and AAV9.452sub.LUNG1 exhibited comparable brightness per cell (p = 0.5), while both were brighter on average than AAV5 (AAV9: p = 0.004; AAV9.452sub.LUNG1: p = 0.005). (C) Cell-type-specific antibodies show the specificity of AAV9.452sub.LUNG1. Minimal colocalization was observed with ATI cells, while there was clear colocalization with ATII cells (identified with arrowheads). Scale bars, 100 μm. (D) Quantification of co-localization between NLS-GFP and α-proSPC staining indicates that AAV9.452sub.LUNG1 transduced approximately 30-fold more ATII cells than AAV9 (p = 0.0007) and approximately 95-fold more than AAV5 (p = 0.0008). This does not appear to be due to increased specificity toward ATII cells, as the relative number of GFP + cells that co-localized with α-proSPC was unchanged compared to AAV9 (p = 0.4) and AAV5 (p = 0.5). n = 6 mice per group, mean ± standard error of the mean. Statistical significance was determined using two-sided Welch’s t tests.
We also measured the number of GFP+ nuclei per tissue area and brightness per cell in the brain, liver, kidney, pancreas, spleen, intestine, and testis (Figures 4 and S1). Quantification indicated that transgene expression of AAV9.452sub.LUNG1 was comparable with AAV9, except in the two tissues with the lowest transduction frequency, namely, the pancreas and brain. AAV9.452sub.LUNG1 transgene expression in the pancreas was significantly lower than AAV9, while in the brain it was significantly higher.
Figure 4.
Comparison of transgene expression across tissues after delivery with AAV5, AAV9, or AAV9.452sub.LUNG1
Expression of NLS-GFP was measured across the liver, testis, intestine, kidney, spleen, pancreas, and brain, with tissues ordered from highest to lowest expression efficiency. Quantification indicates that AAV9.452sub.LUNG1 was not enriched in other tissues in comparison with AAV9, except in the brain where it expressed transgene in approximately seven-fold more cells (p = 0.02). In the pancreas, AAV9.452sub.LUNG1 expressed GFP in fewer cells than AAV9 (p = 0.007). In all other tissues measured, there was no significant difference in the number of cells expressing GFP between AAV9 and AAV9.452sub.LUNG1 (liver: p = 0.5; testis: p = 0.4; intestine: p = 0.4; kidney: p = 0.9; spleen: p = 0.1). Comparing natural serotypes AAV9 and AAV5, AAV9 yielded GFP expression in more cells than AAV5 in all tissues (liver: p < 0.00001; testis: p = 0.01; kidney: p = 0.00006; spleen: p = 0.0003; pancreas: p = 0.002; brain: p = 0.00004), except intestine, where there was no significant difference (p = 0.3). n = 6 mice per group, mean ± standard error of the mean. Statistical significance was determined using two-sided Welch’s t tests. See also Figure S1.
Discussion
The lung is a critical target for gene therapy. However, very few clinical trials aimed at treating diseases in this organ using AAV have been conducted,5,6 likely due to the lack of specialized gene therapy vectors targeting this tissue. The power of directed evolution for the development of effective AAV variants with diverse tropism has been well demonstrated in other tissues.26,27,30,32 Here, we extend and adapt these methods to the lung by substitution in non-traditional sites of the AAV capsid to discover AAV9.452sub.LUNG1, a variant capable of improving transduction efficiency in the lung by approximately 18-fold and expressing transgene in approximately 30-fold more ATII cells after systemic delivery compared with AAV9.
We selected this variant in a Tek-Cre mouse line. Although high Tek gene expression levels have been measured in the mouse lung,62, 63, 64, 65, 66 recent single cell transcriptomic analysis69 did not identify Tek expression in the ATII cell population. The improved transduction of this cell type that we observed could be due to non-specific expression of Cre-recombinase in the mouse lung. Expression profiling in the adult respiratory system of the Cre transgenic line70 indicates that Cre-recombinase is abundant in the alveolus, suggesting that selection of ATII transduction was likely an unintended result of performing selections in the Tek-Cre transgenic line.
Compared with its overall improved transduction in the lung, AAV9.452sub.LUNG1 exhibited similar biodistribution to AAV9 in other tissues. The significant transgene expression in the liver mediated by AAV9 has been associated with toxicity71, 72, 73 and should be noted for future work with this vector. Natural serotypes with tropism toward the lung after system administration, such as AAV4 or AAV6,74 could be considered as alternatives during capsid evaluation.
The improved transduction yielded in the lung by AAV9.452sub.LUNG1 could enable the development of therapeutic options for lung-specific genetic disorders that currently lack effective treatments, specifically diseases where ATII cells play a central role, such as IPF or acute respiratory distress syndrome. Animal models which more closely resemble the progression of IPF in humans are needed,13 and the efficient transduction of ATII cells using this variant could enable their production. For example, the induction of pulmonary fibrosis in mice through ATII targeting and expression of Fas-associated protein with death domain75,76 could provide a useful pre-clinical model. However, we note that AAV9.452sub.LUNG1 was both derived and characterized in C57BL/6J mice, and transduction in other strains and species has not yet been measured. In addition, vector biodistribution has only been tested after systemic administration, so for developing disease models, alternative modalities of delivery such as intranasal77,78 and intratracheal79 administration could decrease off-target accumulation of viral particles in the liver or the CNS.
During the course of characterizing this AAV variant, we ran into the bottleneck of quantifying transgene expression in whole tissue images. Manual counting is subject to bias and using overall fluorescence as a proxy for the number of transduced cells can lead to data misinterpretation. These concerns led us to develop a Python-based image processing pipeline that quantifies the number, and brightness, of cells displaying NLS fluorescence per tissue area. This pipeline can be applied across tissue types with minimal adjustment, allowing unbiased and high-throughput comparison of variants. We make our code publicly available in the hope that it can aid other researchers in characterizing variants in whole tissue. Using this method, we were able to characterize the transgene expression in whole tissue images across many major organs.
Other promising, previously engineered AAV variants with lung tropism are derived from different serotypes than AAV9.452sub.LUNG1 (AAV2-ESGHGYF,42 AAV2.5T,43 and AAV2H2244 from AAV2, and AAV6.2FF40 from AAV6). The distinct antigenic footprints of these serotypes could facilitate sequential administration to avoid host immune response.47 A wide array of capsid options is particularly valuable when a treatment plan requires different interventions at different time points. The mutations in these engineered capsids are also distinct and could display further efficacy when combined, especially since one of the mutations in AAV6.2FF was previously shown to modestly improve lung targeting in AAV9.51 Future iterations of engineering with the KDNTPGR substitution could also investigate natural serotypes AAV4 and AAV6 as templates, potentially heightening their natural tropism toward lung.74 Ultimately, AAV9.452sub.LUNG1 expands the toolbox for gene delivery to the lung and displays promise in enabling lung-specific preclinical research to inform gene therapy.
Materials and methods
Plasmids
The first-round viral DNA library was generated by amplification of a section of the AAV9 capsid genome between AA 450–599 using NNK degenerate primers (Integrated DNA Technologies, Inc., IDT) to substitute AA 452–458 with all possible variations. The resulting library inserts were then introduced into the rAAV-ΔCap-in-cis-Lox plasmid via Gibson assembly as previously described.32 The resulting capsid DNA library, rAAV-Cap-in-cis-Lox, contained a theoretical diversity of approximately 1.28 billion variants at the AA level. The second-round viral DNA library was generated similarly to the first round, but instead of NNK degenerate primers at the 452–458 location, a synthesized oligo pool (Twist Bioscience) was used to generate only selected variants. This second-round DNA library contained a diversity of 5,840 variants at the AA level.
The AAV2/9 REP-AAP-ΔCap plasmid transfected into HEK293T cells for library viral production was modified from the AAV2/9 REP-AAP plasmid previously used32 by deletion of the AA between 450 and 592. This modification prevents production of a wild-type (WT) AAV9 capsid during viral library production after a plausible recombination event between this plasmid co-transfected with rAAV-ΔCap-in-cis-Lox containing the library inserts.
One rAAV genome was used in this study—pAAV-CAG-NLS-GFP (Addgene: #104061)—utilizes an ssAAV genome containing the fluorescent protein EGFP flanked by two nuclear localization sites, PKKKRKV, under control of the CAG promoter.
Viral production
Recombinant AAVs were generated according to established protocols.80 Briefly, HEK293T cells (ATCC) were triple transfected using polyethylenimine; virus was collected after 120 h from both cell lysates and media and purified over iodixanol (Optiprep, Sigma). A modified protocol was used for transfection and purification of viral libraries. First, to prevent mosaic capsid formation, only 10 ng rAAV-Cap-in-cis-Lox library DNA was transfected (per 150-mm plate) to decrease the likelihood of multiple library DNAs entering the same cell. Second, virus was collected after 60 h, instead of 120 h, to limit secondary transduction of producer cells. Finally, instead of polyethylene glycol precipitation of the viral particles from the media, as performed in the standard protocol, the medium was concentrated more than 60-fold for loading onto iodixanol.
Animals
All rodent procedures were approved by the Institutional Animal Use and Care Committee of the California Institute of Technology. Transgenic animals, expressing Cre under the control of various cell-type-specific promoters, and C57Bl/6J WT mice (000664) were purchased from the Jackson Laboratory. Transgenic mice included Tek-Cre (8863),70 Syn1-Cre (3966),81 GFAP-Cre (012,886),82 TH-Cre (008,601),83 CHAT-Cre (028,861),84 and VGAT-Cre (028,862).85 For the first round of viral library selection, we used one male and one female mouse from each transgenic line (aged 8–12 weeks), as well as a single male C57Bl/6J mouse. For the second round of selection, we used one male and female mouse from the Tek-Cre line (aged 8–12 weeks), as well as a single male C57Bl/6J mouse. For validation of individual viral variants, male C57Bl/6J mice aged 6–8 weeks were used. Mice were housed under standard conditions between 71 °F and 75 °F, 30%–70% humidity, and light cycle of 13 h on and 11 h off. Intravenous administration of rAAV vectors was performed via injection into the retro-orbital sinus.
DNA/RNA recovery and sequencing
Round 1 and round 2 viral libraries were injected into C57BL/6J and Cre-transgenic animals at a dose of 8 × 1010 vg/animal and rAAV genomes were recovered two weeks after injection, as described in the M-CREATE protocol.31 Cre-transgenic lines used for round 1 were Syn1-Cre, GFAP-Cre, TH-Cre, VGAT-Cre, CHAT-Cre, and Tek-Cre, selected to cover a wide range of cell types across tissues. For the second round of selection, only Tek-Cre animals were used. To determine the number of variants included in round 2, 0.01 times the enrichment of the top variant in each tissue was set as a threshold and variants above that threshold were included. Mice were euthanized, and most major organs were recovered, snap frozen on dry ice, and placed into long-term storage at −80 °C. Tissues collected included: brain, spinal cord, dorsal root ganglia (DRGs), liver, lungs, heart, stomach, intestines, kidneys, spleen, pancreas, testes, skeletal muscle, and adipose tissue. We homogenized 100 mg of each tissue (approximately 250 mg for brain hemispheres, <100 mg for DRGs) in Trizol (Life Technologies, 15596) using a BeadBug (Benchmark Scientific, D1036) and viral DNA was isolated according to the manufacturer’s recommended protocol. Recovered viral DNA was treated with RNase, underwent restriction digestion with SmaI (located within the ITRs) to improve later rAAV genome recovery by PCR, and purified with a Zymo DNA Clean and Concentrator kit (D4033). Viral genomes flipped by Cre-recombinase in select transgenic lines (or pre-flipped in WT animals) were selectively recovered using the following primers: 5′-CTTCCAGTTCAGCTACGAGTTTGAGAAC-3′ and 5′-CAAGTAAAACCTCTACAAATGTGGTAAAATCG-3′, after 25 cycles of 98 °C for 10 s, 60 °C for 15 s, and 72 °C for 40 s, using Q5 DNA polymerase in five 25 μL reactions with 50% of the total extracted viral DNA as a template.
After Zymo DNA purification, samples from the WT C57Bl/6J animals were serially diluted from 1:10 to 1:10,000 and each dilution further amplified around the library variable region. This amplification was done using primers: 5′-ACGCTCTTCCGATCTAATACTTGTACTATCTCTCTAGAACTATT-3′ and 5′-TGTGCTCTTCCGATCTCACACTGAATTTTAGCGTTTG-3′ and 10 cycles of 98 °C for 10 s, 61 °C for 15 s, and 72 °C for 20 s, to recover 73 bp of viral genome around and including the 21-bp variable region and add adapters for Illumina NGS. After PCR cleanup, these products were further amplified using NEBNext Dual Index Primers for Illumina sequencing (New England Biolabs, E7600), after 10 cycles of 98 °C for 10 s, 60 °C for 15 s, and 72 °C for 20 s. The amplification products were run on a 2% low melting point agarose gel (ThermoFisher Scientific, 16520050) for better separation and recovery of the 210-bp band. The dilution series was analyzed for each WT tissue and the greatest concentration dilution, which resulted in no product was chosen for further amplification of the viral DNA from the transgenic animal tissues. This process was performed to differentiate between viral genomes flipped before packaging or due to Cre in the animal. Pre-flipped viral genomes should be avoided to minimize false positives in the NGS sequencing results.
All Cre-flipped viral genomes from transgenic animal tissues were similarly amplified (using the dilutions that do not produce pre-flipped viral genomes) to add Illumina sequencing adapters and subsequently for index labeling. The amplified products now containing unique indices for each tissue from each animal were run on a low melting point agarose gel and the correct bands extracted and purified with a Zymoclean Gel DNA Recovery kit.
Packaged viral library DNA was isolated from the injected viral library by digestion of the viral capsid and purification of the contained ssDNA. These viral genomes were amplified by two PCR amplification steps, like the viral DNA extracted from tissue, to add Illumina adapters and then indices and extracted and purified after gel electrophoresis. This viral library DNA, along with the viral DNA extracted from tissue, was sent for deep sequencing using an Illumina HiSeq 2500 System (Millard and Muriel Jacobs Genetics and Genomics Laboratory, Caltech).
NGS data alignment and processing
Raw fastq files from NGS runs were processed with CREATE data analysis code (available on Github: https://github.com/GradinaruLab/mCREATE) that align the data with an AAV9 template DNA fragment containing the 21-bp diversified region between AA 452 and AA 458, for the two rounds of AAV evolution/selection. The pipeline to process these datasets involved filtering to remove low-quality reads, utilizing a quality score for each sequence, and eliminating bias from PCR-induced mutations or high GC-content. The filtered dataset was then aligned by a perfect string match algorithm and trimmed to improve the alignment quality. For the AAV engineering, read counts for each sequence were pulled out and displayed along with their enrichment score, defined as the relative abundance of the sequence found within the specific tissue over the relative abundance of that sequence within the injected viral library.
Tissue preparation and immunofluorescence
Mice were euthanized with Euthasol and transcardially perfused with ice-cold 1× PBS and then freshly prepared, ice-cold 4% paraformaldehyde (PFA) in 1× PBS. All organs were excised and post-fixed in 4% PFA at 4°C for 48 h and then sectioned at 50 μm with a vibratome. Immunofluorescence was performed on floating sections with primary and secondary antibodies in PBS containing 10% donkey serum and 0.1% Triton X-100. Primary antibodies used were Syrian hamster anti-podoplanin (1:200, Abcam, ab11936) and rabbit anti-proSPC (1:200, Sigma-Aldrich, AB3786). Primary antibody incubations were performed for 16–20 h at room temperature. Sections being stained with anti-podoplanin were washed before incubation with secondary antibody AffiniPure Goat Anti-Syrian Hamster IgG (1:200, Jackson ImmunoResearch Laboratories, Inc., 107-005-142) for 6 h at room temperature. This was followed by incubation with Alexa 647 conjugated donkey anti-goat antibody (1:200, ThermoFisher Scientific, A32849) for 6–8 h at room temperature. Sections being stained with anti-proSPC were washed and incubated with secondary Alexa 647 conjugated anti-rabbit FAB fragment antibody (1:200, Jackson ImmunoResearch Laboratories, Inc., 711-607-003) for 6–8 h at room temperature. Stained sections were then mounted with ProLong Diamond Antifade Mountant (ThermoFisher Scientific, P36970).
Imaging and quantification
The initial comparisons of transgene expression after delivery with AAV9.452sub.LUNG1 or AAV9 at a dose of 5 × 1011 viral genomes per animal (Figure 1) were imaged on a Zeiss LSM 880 confocal microscope using a Fluar 5 × 0.25 M27 objective, with matched laser powers, gains, and gamma across all samples of the same tissue. The acquired images were processed in Zen Black 2.3 SP1 (Zeiss).
The comparisons of AAV9.452sub.LUNG1 with AAV9 and AAV5 at the lower dose of 1 × 1011 viral genomes per animal were imaged on a Keyence BZ-X all-in-one fluorescence microscope at 48-bit resolution with the following objectives: PlanApo-λ 20x/0/75 (1-mm working distance) or PlanApo-λ 10x/0.45 (4-mm working distance). The filters used were BZ-X Filters OP-87763 for NLS-GFP, OP-87764 for autofluorescence, and OP-87766 for imaging tissues stained with 647 antibodies. In each tissue, exposure settings and changes to gamma or contrast were maintained across images.
All image processing was performed using our custom Python image processing pipeline, available on GitHub: https://github.com/GradinaruLab/Image_Segmentation. In brief, images are first stitched together using a custom algorithm, keeping channels distinct. An Otsu threshold with 1,024 bins is used on the autofluorescence image to determine the area within the image that is classified as tissue. The mean intensity of both the signal and the autofluorescence images are determined, and the ratio of these means is used to adjust the autofluorescence image before subtraction, to ensure that the average image intensities are matched before adjustment. On the product of subtraction, template matching with a circular template is then used to determine regions that appear nuclear using skimage.feature.match_template, outputting a binary mask of all the regions matching the circular template. Also on the product of subtraction, skimage.filters.Gaussian is applied (sigma = 5, truncation = 2) and subtracted from the image to remove large features from the background. From the background subtracted image, the second derivative of the histogram is then taken, and a predetermined value of the second derivative is used to acquire an intensity threshold for the image. This threshold is applied to generate a binary mask of bright regions before application of skimage.morphology.binary_closing to fill in partially identified intense regions. Both the circular template mask and the intensity thresholded mask are multiplied together to make a mask of colocalized regions. Finally, objects fewer than 10 pixels are removed to minimize noise. skimage.measure.label is used to determine the number of cells in this image. Custom code was used to calculate the brightness of cells within the image.
Statistics
Custom Python code was used for statistical analysis and data representation. Populations were compared using two-sided Welch’s t test, implemented using the stats module from the SciPy package. This module was implemented to compare each set of samples without assuming equal variance as: stats.ttest_ind(first population, second population, equal_var = False), where the first and second populations were the data being compared. Unless otherwise noted, all experimental groups were n = 6, determined using preliminary data and experimental power analysis. For the statistical analysis in mice and related graphs, a single data point was defined as two tissue sections per animal. For each section, the whole tissue was imaged and processed. For all statistical analyses, significance is represented as: ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001; n.s., p ≥ 0.05.
Data availability
The NGS datasets for the capsid selection reported in this article are available on the Sequence Read Archive: PRJNA821550. The vector plasmid for AAV9.452sub.LUNG1 was deposited to Addgene: #184592. The AAV9.452sub.LUNG1 sequence has been deposited on GenBank: ON959566. All other constructs and tools are available through the Beckman Institute CLOVER Center (https://clover.caltech.edu/). Python code used for this analysis is available on GitHub: https://github.com/GradinaruLab/Image_Segmentation.
Acknowledgments
We thank the entire Gradinaru laboratory for helpful discussions. We thank Dr. Deverman for helpful discussions on CREATE design and implementation as described by Deverman and colleagues.32 We thank K. Johnston and P. Almhjell for assistance with development of the image processing pipeline and C. Oikonomou for assistance editing the manuscript. We are grateful to I. Antoshechkin and the Millar and Muriel Jacobs Genetics and Genomics Core at Caltech for assistance with next generation sequencing. We thank M. Smith for helpful discussions and assistance with the bioinformatics pipeline. This work was primarily supported by Defense Advanced Research Projects Agency grant W911NF-17-2-0036 to V.G. and by National Institutes of Health (NIH) to V.G.: NIH Pioneer DP1OD025535. D.G. was supported by the National Sciences and Engineering Research Council of Canada. N.G. was supported by the Cystic Fibrosis Foundation: CFF GOEDEN19F0.
Author contributions
D.G. analyzed all data and prepared all figures with input from V.G., N.C.F., and N.G. designed and performed the variant selection experiments, D.G. characterized the variants and built the image processing pipeline. D.G. and V.G. wrote the manuscript with input from all authors. V.G. supervised all aspects of the work.
Declaration of interests
The California Institute of Technology has filed and licensed patent applications for the work described in this manuscript, with N.C.F., N.G., and V.G. listed as inventors (US Patent application no. PCT/US21/46904). V.G. is a co-founder and board member and N.C.F. and N.G. are co-founders and officers of Capsida Biotherapeutics, a fully integrated AAV engineering and gene therapy company. The remaining authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.07.010.
Supplemental information
References
- 1.Daya S., Berns K.I. Gene therapy using adeno-associated virus vectors. Clin. Microbiol. Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rivière C., Danos O., Douar A.M. Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther. 2006;13:1300–1308. doi: 10.1038/sj.gt.3302766. [DOI] [PubMed] [Google Scholar]
- 3.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R., Perry W.L., Strohl W.R. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudet D., Méthot J., Kastelein J. Gene therapy for lipoprotein lipase deficiency. Curr. Opin. Lipidol. 2012;23:310–320. doi: 10.1097/MOL.0b013e3283555a7e. [DOI] [PubMed] [Google Scholar]
- 5.Kuzmin D.A., Shutova M.V., Johnston N.R., Smith O.P., Fedorin V.V., Kukushkin Y.S., van der Loo J.C.M., Johnstone E.C. The clinical landscape for AAV gene therapies. Nat. Rev. Drug Discov. 2021;20:173–174. doi: 10.1038/d41573-021-00017-7. [DOI] [PubMed] [Google Scholar]
- 6.Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R. Gene therapy clinical trials worldwide to 2017: an update. J. Gene Med. 2018;20:e3015. doi: 10.1002/jgm.3015. [DOI] [PubMed] [Google Scholar]
- 7.Griesenbach U., Geddes D.M., Alton E.W.F.W. Gene therapy for cystic fibrosis: an example for lung gene therapy. Gene Ther. 2004;11:S43–S50. doi: 10.1038/sj.gt.3302368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liqun Wang R., McLaughlin T., Cossette T., Tang Q., Foust K., Campbell-Thompson M., Martino A., Cruz P., Loiler S., Mueller C., et al. Recombinant AAV serotype and capsid mutant comparison for pulmonary gene transfer of α-1-antitrypsin using invasive and noninvasive delivery. Mol. Ther. 2009;17:81–87. doi: 10.1038/mt.2008.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruaro B., Salton F., Braga L., Wade B., Confalonieri P., Volpe M.C., Baratella E., Maiocchi S., Confalonieri M. The history and mystery of alveolar epithelial type II cells: focus on their physiologic and pathologic role in lung. Int. J. Mol. Sci. 2021;22:2566. doi: 10.3390/ijms22052566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parimon T., Yao C., Stripp B.R., Noble P.W., Chen P. Alveolar epithelial type II cells as drivers of lung fibrosis in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 2020;21:2269. doi: 10.3390/ijms21072269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbas-Filho J.V., Ferreira M.A., Sesso A., Kairalla R.A., Carvalho C.R., Capelozzi V.L. Evidence of type II pneumocyte apoptosis in the pathogenesis of idiopathic pulmonary fibrosis (IFP)/usual interstitial pneumonia (UIP) J. Clin. Pathol. 2001;54:132–138. doi: 10.1136/jcp.54.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonella F., Stowasser S., Wollin L. Idiopathic pulmonary fibrosis: current treatment options and critical appraisal of nintedanib. Drug Des. Dev. Ther. 2015;9:6407–6419. doi: 10.2147/DDDT.S76648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tashiro J., Rubio G.A., Limper A.H., Williams K., Elliot S.J., Ninou I., Aidinis V., Tzouvelekis A., Glassberg M.K. Exploring animal models that resemble idiopathic pulmonary fibrosis. Front. Med. 2017;4:118. doi: 10.3389/fmed.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris G., Bortolasci C.C., Puri B.K., Olive L., Marx W., O’Neil A., Athan E., Carvalho A.F., Maes M., Walder K., et al. The pathophysiology of SARS-CoV-2: a suggested model and therapeutic approach. Life Sci. 2020;258:118166. doi: 10.1016/j.lfs.2020.118166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carcaterra M., Caruso C. Alveolar epithelial cell type II as main target of SARS-CoV-2 virus and COVID-19 development via NF-Kb pathway deregulation: a physio-pathological theory. Med. Hypotheses. 2021;146:110412. doi: 10.1016/j.mehy.2020.110412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark J.C., Wert S.E., Bachurski C.J., Stahlman M.T., Stripp B.R., Weaver T.E., Whitsett J.A. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. USA. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasser S.W., Burhans M.S., Korfhagen T.R., Na C.L., Sly P.D., Ross G.F., Ikegami M., Whitsett J.A. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc. Natl. Acad. Sci. USA. 2001;98:6366–6371. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasser S.W., Witt T.L., Senft A.P., Baatz J.E., Folger D., Maxfield M.D., Akinbi H.T., Newton D.A., Prows D.R., Korfhagen T.R. Surfactant protein C-deficient mice are susceptible to respiratory syncytial virus infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;297:L64–L72. doi: 10.1152/ajplung.90640.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammel M., Michel G., Hoefer C., Klaften M., Müller-Höcker J., de Angelis M.H., Holzinger A. Targeted inactivation of the murine Abca3 gene leads to respiratory failure in newborns with defective lamellar bodies. Biochem. Biophys. Res. Commun. 2007;359:947–951. doi: 10.1016/j.bbrc.2007.05.219. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald M.L., Xavier R., Haley K.J., Welti R., Goss J.L., Brown C.E., Zhuang D.Z., Bell S.A., Lu N., Mckee M., et al. ABCA3 inactivation in mice causes respiratory failure, loss of pulmonary surfactant, and depletion of lung phosphatidylglycerol. J. Lipid Res. 2007;48:621–632. doi: 10.1194/jlr.M600449-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Whitsett J.A., Weaver T.E. Hydrophobic surfactant proteins in lung function and disease. N. Engl. J. Med. 2002;347:2141–2148. doi: 10.1056/NEJMra022387. [DOI] [PubMed] [Google Scholar]
- 22.Whitsett J.A., Wert S.E., Weaver T.E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamvas A., Nogee L.M., Mallory G.B., Jr., Spray T.L., Huddleston C.B., August A., Dehner L.P., DeMello D.E., Moxley M., Nelson R., et al. Lung transplantation for treatment of infants with surfactant protein B deficiency. J. Pediatr. 1997;130:231–239. doi: 10.1016/s0022-3476(97)70348-2. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verdera H.C., Kuranda K., Mingozzi F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol. Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabebordbar M., Lagerborg K.A., Stanton A., King E.M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J.J., Messemer K.A., et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 29.Davis A.S., Federici T., Ray W.C., Boulis N.M., OʼConnor D., Clark K.R., Bartlett J.S. Rational design and engineering of a modified adeno-associated virus (AAV1)-based vector system for enhanced retrograde gene delivery. Neurosurgery. 2015;76:216–225. doi: 10.1227/NEU.0000000000000589. discussion 225. [DOI] [PubMed] [Google Scholar]
- 30.Nonnenmacher M., Wang W., Child M.A., Ren X.-Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravindra Kumar S., Miles T.F., Chen X., Brown D., Dobreva T., Huang Q., Ding X., Luo Y., Einarsson P.H., Greenbaum A., et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods. 2020;17:541–550. doi: 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian R., Xiao B., Li J., Xiao X. Directed evolution of AAV serotype 5 for increased hepatocyte transduction and retained low humoral seroreactivity. Mol. Ther. Methods Clin. Dev. 2021;20:122–132. doi: 10.1016/j.omtm.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A., et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas M., Marsic D., Li N., Zou C., Gonzalez-Aseguinolaza G., Zolotukhin I., Kumar S.R.P., Rana J., Butterfield J.S.S., Kondratov O., et al. Engineering and in vitro selection of a novel AAV3B variant with high hepatocyte tropism and reduced seroreactivity. Mol. Ther. Methods Clin. Dev. 2020;19:347–361. doi: 10.1016/j.omtm.2020.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Öztürk B.E., Johnson M.E., Kleyman M., Turunç S., He J., Jabalameli S., Xi Z., Visel M., Dufour V.L., Iwabe S., et al. scAAVengr, a transcriptome-based pipeline for quantitative ranking of engineered AAVs with single-cell resolution. Elife. 2021;10:e64175. doi: 10.7554/eLife.64175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrne L.C., Day T.P., Visel M., Strazzeri J.A., Fortuny C., Dalkara D., Merigan W.H., Schaffer D.V., Flannery J.G. In vivo-directed evolution of adeno-associated virus in the primate retina. JCI Insight. 2020;5:135112. doi: 10.1172/jci.insight.135112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarcar S., Tulalamba W., Rincon M.Y., Tipanee J., Pham H.Q., Evens H., Boon D., Samara-Kuko E., Keyaerts M., Loperfido M., et al. Next-generation muscle-directed gene therapy by in silico vector design. Nat. Commun. 2019;10:492. doi: 10.1038/s41467-018-08283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pulicherla N., Shen S., Yadav S., Debbink K., Govindasamy L., Agbandje-McKenna M., Asokan A. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Lieshout L.P., Domm J.M., Rindler T.N., Frost K.L., Sorensen D.L., Medina S.J., Booth S.A., Bridges J.P., Wootton S.K. A novel triple-mutant AAV6 capsid induces rapid and potent transgene expression in the muscle and respiratory tract of mice. Mol. Ther. Methods Clin. Dev. 2018;9:323–329. doi: 10.1016/j.omtm.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang M.H., van Lieshout L.P., Xu L., Domm J.M., Vadivel A., Renesme L., Mühlfeld C., Hurskainen M., Mižíková I., Pei Y., et al. A lung tropic AAV vector improves survival in a mouse model of surfactant B deficiency. Nat. Commun. 2020;11:3929. doi: 10.1038/s41467-020-17577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Körbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Müller O.J., et al. Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 2016;24:1050–1061. doi: 10.1038/mt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Excoffon K.J.D.A., Koerber J.T., Dickey D.D., Murtha M., Keshavjee S., Kaspar B.K., Zabner J., Schaffer D.V. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steines B., Dickey D.D., Bergen J., Excoffon K.J., Weinstein J.R., Li X., Yan Z., Abou Alaiwa M.H., Shah V.S., Bouzek D.C., et al. CFTR gene transfer with AAV improves early cystic fibrosis pig phenotypes. JCI Insight. 2016;1:e88728. doi: 10.1172/jci.insight.88728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooney A.L., Thornell I.M., Singh B.K., Shah V.S., Stoltz D.A., McCray P.B., Jr., Zabner J., Sinn P.L. A novel AAV-mediated gene delivery system corrects CFTR function in pigs. Am. J. Respir. Cell Mol. Biol. 2019;61:747–754. doi: 10.1165/rcmb.2019-0006OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bočkor L., Bortolussi G., Iaconcig A., Chiaruttini G., Tiribelli C., Giacca M., Benvenuti F., Zentilin L., Muro A.F. Repeated AAV-mediated gene transfer by serotype switching enables long-lasting therapeutic levels of hUgt1a1 enzyme in a mouse model of Crigler–Najjar Syndrome Type I. Gene Ther. 2017;24:649–660. doi: 10.1038/gt.2017.75. [DOI] [PubMed] [Google Scholar]
- 47.Calcedo R., Wilson J.M. Humoral immune response to AAV. Front. Immunol. 2013;4:1–7. doi: 10.3389/fimmu.2013.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raupp C., Naumer M., Müller O.J., Gurda B.L., Agbandje-McKenna M., Kleinschmidt J.A. The threefold protrusions of adeno-associated virus type 8 are involved in cell surface targeting as well as postattachment processing. J. Virol. 2012;86:9396–9408. doi: 10.1128/JVI.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen S., Bryant K.D., Brown S.M., Randell S.H., Asokan A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J. Biol. Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kern A., Schmidt K., Leder C., Müller O.J., Wobus C.E., Bettinger K., Von der Lieth C.W., King J.A., Kleinschmidt J.A. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 2003;77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martini S.V., Silva A.L., Ferreira D., Rabelo R., Ornellas F.M., Gomes K., Rocco P.R.M., Petrs-Silva H., Morales M.M. Tyrosine mutation in AAV9 capsid improves gene transfer to the mouse lung. Cell. Physiol. Biochem. 2016;39:544–553. doi: 10.1159/000445646. [DOI] [PubMed] [Google Scholar]
- 52.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Weigel-Van Aken K.A., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mao Y., Wang X., Yan R., Hu W., Li A., Wang S., Li H. Single point mutation in adeno-associated viral vectors -DJ capsid leads to improvement for gene delivery in vivo. BMC Biotechnol. 2016;16:1. doi: 10.1186/s12896-015-0230-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiMattia M.A., Nam H.-J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M., et al. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perabo L., Büning H., Kofler D.M., Ried M.U., Girod A., Wendtner C.M., Enssle J., Hallek M. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol. Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 56.Girod A., Ried M., Wobus C., Lahm H., Leike K., Kleinschmidt J., Deléage G., Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 1999;5:1052–1056. doi: 10.1038/12491. [DOI] [PubMed] [Google Scholar]
- 57.Müller O.J., Kaul F., Weitzman M.D., Pasqualini R., Arap W., Kleinschmidt J.A., Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 58.Yu C.-Y., Yuan Z., Cao Z., Wang B., Qiao C., Li J., Xiao X. A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther. 2009;16:953–962. doi: 10.1038/gt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michelfelder S., Varadi K., Raupp C., Hunger A., Körbelin J., Pahrmann C., Schrepfer S., Müller O.J., Kleinschmidt J.A., Trepel M. Peptide ligands incorporated into the threefold spike capsid domain to Re-direct gene transduction of AAV8 and AAV9 in vivo. PLoS One. 2011;6:e23101. doi: 10.1371/journal.pone.0023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goertsen D., Flytzanis N.C., Goeden N., Chuapoco M.R., Cummins A., Chen Y., Fan Y., Zhang Q., Sharma J., Duan Y., et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 2022;25:106–115. doi: 10.1038/s41593-021-00969-4. [DOI] [PubMed] [Google Scholar]
- 61.Brown D., Altermatt M., Dobreva T., Chen S., Wang A., Thomson M., Gradinaru V. Deep parallel characterization of AAV tropism and AAV-mediated transcriptional changes via single-cell RNA sequencing. Front. Immunol. 2021;12:730825. doi: 10.3389/fimmu.2021.730825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwama A., Hamaguchi I., Hashiyama M., Murayama Y., Yasunaga K., Suda T. Molecular cloning and characterization of mouse TIE and TEK receptor tyrosine kinase genes and their expression in hematopoietic stem cells. Biochem. Biophys. Res. Commun. 1993;195:301–309. doi: 10.1006/bbrc.1993.2045. [DOI] [PubMed] [Google Scholar]
- 63.Taichman D.B., Schachtner S.K., Li Y., Puri M.C., Bernstein A., Scott Baldwin H. A unique pattern of Tie1 expression in the developing murine lung. Exp. Lung Res. 2003;29:113–122. doi: 10.1080/01902140303767. [DOI] [PubMed] [Google Scholar]
- 64.Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huntley M.A., Lou M., Goldstein L.D., Lawrence M., Dijkgraaf G.J.P., Kaminker J.S., Gentleman R. Complex regulation of ADAR-mediated RNA-editing across tissues. BMC Genom. 2016;17:61. doi: 10.1186/s12864-015-2291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grzenda A., Shannon J., Fisher J., Arkovitz M.S. Timing and expression of the angiopoietin-1–Tie-2 pathway in murine lung development and congenital diaphragmatic hernia. Dis. Model. Mech. 2013;6:106–114. doi: 10.1242/dmm.008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alves S., Bode J., Bemelmans A.-P., von Kalle C., Cartier N., Tews B. Ultramicroscopy as a novel tool to unravel the tropism of AAV gene therapy vectors in the brain. Sci. Rep. 2016;6:28272. doi: 10.1038/srep28272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gruntman A.M., Mueller C., Flotte T.R., Gao G. Gene transfer in the lung using recombinant adeno-associated virus. Curr. Protoc. Microbiol. 2012;26 doi: 10.1002/9780471729259.mc14d02s26. Unit14D.2.1–Unit14D.2.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hurskainen M., Mižíková I., Cook D.P., Andersson N., Cyr-Depauw C., Lesage F., Helle E., Renesme L., Jankov R.P., Heikinheimo M., et al. Single cell transcriptomic analysis of murine lung development on hyperoxia-induced damage. Nat. Commun. 2021;12:1565. doi: 10.1038/s41467-021-21865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kisanuki Y.Y., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 71.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kubes P., Jenne C. Immune responses in the liver. Annu. Rev. Immunol. 2018;36:247–277. doi: 10.1146/annurev-immunol-051116-052415. [DOI] [PubMed] [Google Scholar]
- 73.Bieghs V., Trautwein C. The innate immune response during liver inflammation and metabolic disease. Trends Immunol. 2013;34:446–452. doi: 10.1016/j.it.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 74.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 75.Maeyama T., Kuwano K., Kawasaki M., Kunitake R., Hagimoto N., Matsuba T., Yoshimi M., Inoshima I., Yoshida K., Hara N. Upregulation of Fas-signalling molecules in lung epithelial cells from patients with idiopathic pulmonary fibrosis. Eur. Respir. J. 2001;17:180–189. doi: 10.1183/09031936.01.17201800. [DOI] [PubMed] [Google Scholar]
- 76.Sisson T.H., Mendez M., Choi K., Subbotina N., Courey A., Cunningham A., Dave A., Engelhardt J.F., Liu X., White E.S., et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2010;181:254–263. doi: 10.1164/rccm.200810-1615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belur L.R., Temme A., Podetz-Pedersen K.M., Riedl M., Vulchanova L., Robinson N., Hanson L.R., Kozarsky K.F., Orchard P.J., Frey W.H., 2nd, et al. Intranasal adeno-associated virus mediated gene delivery and expression of human iduronidase in the central nervous system: a noninvasive and effective approach for prevention of neurologic disease in mucopolysaccharidosis type I. Hum. Gene Ther. 2017;28:576–587. doi: 10.1089/hum.2017.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gadenstaetter A.J., Schmutzler L., Grimm D., Landegger L.D. Intranasal application of adeno-associated viruses: a systematic review. Transl. Res. 2022 doi: 10.1016/j.trsl.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 79.Santry L.A., Ingrao J.C., Yu D.L., de Jong J.G., van Lieshout L.P., Wood G.A., Wootton S.K. AAV vector distribution in the mouse respiratory tract following four different methods of administration. BMC Biotechnol. 2017;17:43. doi: 10.1186/s12896-017-0365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Challis R.C., Ravindra Kumar S., Chan K.Y., Challis C., Beadle K., Jang M.J., Kim H.M., Rajendran P.S., Tompkins J.D., Shivkumar K., et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019;14:379–414. doi: 10.1038/s41596-018-0097-3. [DOI] [PubMed] [Google Scholar]
- 81.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia A.D.R., Doan N.B., Imura T., Bush T.G., Sofroniew M.V. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 83.Savitt J.M., Jang S.S., Mu W., Dawson V.L., Dawson T.M. Bcl-x is required for proper development of the mouse: substantia nigra. J. Neurosci. 2005;25:6721–6728. doi: 10.1523/JNEUROSCI.0760-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rossi J., Balthasar N., Olson D., Scott M., Berglund E., Lee C.E., Choi M.J., Lauzon D., Lowell B.B., Elmquist J.K. Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab. 2011;13:195–204. doi: 10.1016/j.cmet.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vong L., Ye C., Yang Z., Choi B., Chua S., Lowell B.B. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS datasets for the capsid selection reported in this article are available on the Sequence Read Archive: PRJNA821550. The vector plasmid for AAV9.452sub.LUNG1 was deposited to Addgene: #184592. The AAV9.452sub.LUNG1 sequence has been deposited on GenBank: ON959566. All other constructs and tools are available through the Beckman Institute CLOVER Center (https://clover.caltech.edu/). Python code used for this analysis is available on GitHub: https://github.com/GradinaruLab/Image_Segmentation.