Abstract
Notch/Delta signaling regulates numerous cell-cell interactions that occur during development, homeostasis, and in disease states. In many cases, the Notch/Delta pathway mediates lateral inhibition between cells to specify alternative fates. Here, we provide new tools for use in C. elegans to investigate feedback between the Notch receptor LIN-12 and the ligand LAG-2 (Delta) in vivo . We report new, endogenously tagged strains to visualize LAG-2 protein and lag-2 transcription, which we combined with endogenously tagged LIN-12 to visualize Notch and Delta dynamics over the course of a stochastic Notch-mediated cell fate decision. To validate these tools in a functional context, we demonstrated that our endogenous lag-2 transcriptional reporter was expressed in ectopic anchor and primary vulval precursor cells after auxin-mediated depletion of LIN-12. This toolkit provides new reagents for the C. elegans research community to further investigate Notch/Delta signaling mechanisms and functions for this pathway in vivo .
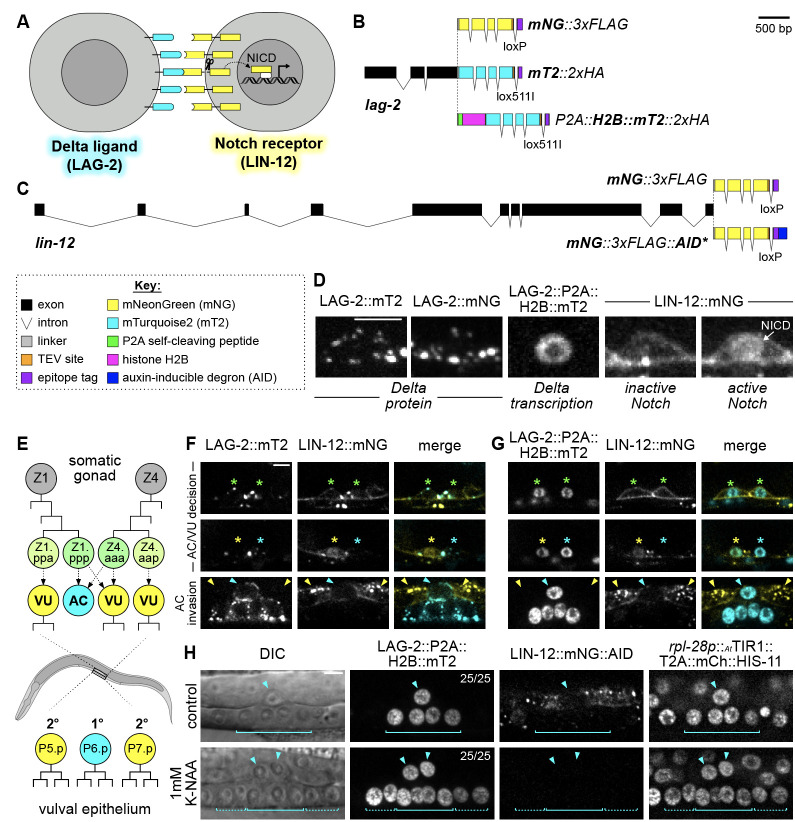
Figure 1. A toolkit to simultaneously visualize endogenous Delta and Notch dynamics in vivo .
( A ) Simplified schematic of Notch-Delta signaling. Binding of the Delta-like ligand LAG-2 (cyan) to the Notch receptor LIN-12 (yellow) results in cleavage of the Notch intracellular domain (NICD) by ɣ-secretase (represented by scissors) and NICD translocation to the nucleus. ( B-C ) Schematics of the endogenously tagged lag-2 ( B ) and lin-12 ( C ) alleles used in this study. The lag-2 transcriptional reporter was generated by including a self-cleaving viral peptide (P2A) and histone H2B (HIS-58) between LAG-2 and mT2 (Ahier & Jarriault, 2014). ( D ) Representative micrographs showing localization of the tagged LAG-2 and LIN-12 alleles in anchor cell/ventral uterine cell (AC/VU) precursors. mT2- and mNG-tagged LAG-2 form puncta on the cell membrane while the transcriptional reporter (LAG-2::P2A::H2B::mT2) is nuclear-localized. LIN-12::mNG is expressed on the cell membrane and exhibits nuclear localization of the Notch intracellular domain (NICD) upon activation. ( E ) Cartoon of AC/VU cell fate specification (top) as well as primary (1°) and secondary (2°) fated vulval precursor cells (VPCs) (bottom) in a wild-type animal. ( F-G ) Micrographs depicting LAG-2::mT2 ( F ) or LAG-2::P2A::H2B::mT2 ( G ) in concert with LIN-12::mNG reveal Delta and Notch dynamics during the AC/VU decision pre- and post-specification as well as during AC invasion. Asterisks: AC/VU precursors; arrowheads, specified AC and VU cells with colors matching panel E . ( H ) Endogenous lag-2 transcriptional reporter expression marks AC and 1° VPC fates in animals harboring LIN-12::mNG::AID and a ubiquitously expressed TIR1::T2A::mCh::HIS-11 transgene. Depleting LIN-12 by treatment with auxin (1 mM K-NAA) leads to specification of a second AC and ectopic 1° VPCs that express LAG-2::P2A::H2B::mT2. Solid bracket, P6.p-derived primary-fated VPCs; dotted bracket, ectopic primary-fated VPCs. Scale bars: 5 µm.
Description
The Notch signal transduction pathway represents a widely conserved mechanism of cell-cell communication utilized throughout the Metazoa (Gazave et al ., 2009). One of the most notable functions for Notch signaling is lateral inhibition, where negative intercellular feedback specifies distinct fates in ligand- and receptor-expressing cells that contact each other (reviewed in Sjöqvist & Andersson, 2019). In a simplified view of Notch signal transduction, Notch ligands and receptors are both transmembrane proteins that activate signaling at cell contacts. The activated Notch receptor then undergoes ɣ-secretase mediated cleavage to release the Notch intracellular domain (NICD), which translocates into the nucleus and acts as a transcriptional co-activator ( Figure 1A ) (Struhl et al ., 1993). For decades, C. elegans has been used as a model investigate functions and mechanisms of Notch signaling. C. elegans possesses four canonical Notch ligands in the Delta Serrate LAG-2 family named lag-2, apx-1, arg-1, and dsl-1 (Henderson et al ., 1994, Greenwald & Kovall, 2013). C. elegans also possesses two Notch receptors, glp - 1 and lin - 12 , with both unique and redundant functions (Lambie & Kimble, 1991; Greenwald & Kovall, 2013).
To investigate Notch/Delta signaling dynamics in vivo , we sought to generate a toolkit to visualize feedback between LIN-12 and LAG-2, which mediates several well-characterized cell fate decisions during larval development (Greenwald et al ., 1983; Wilkinson et al ., 1994; Levitan & Greenwald, 1998). Because endogenous LIN-12 (Pani et. al., 2022) and LAG-2 (Gordon et. al., 2019) have previously both been tagged with the bright green/yellow fluorescent protein mNeonGreen (mNG) (Shaner et. al., 2013), it is not possible to distinguish them in the same animal using existing tools. To make it possible to visualize endogenously tagged LAG-2 and LIN-12 simultaneously, we tagged LAG-2 with the cyan fluorescent protein mTurquoise2 (mT2) (Goedhart et. al., 2012) ( Figure 1B,C ). LAG-2::mT2, and LAG-2::mT2; LIN-12::mNG animals were morphologically indistinguishable from wild-type and did not exhibit the cell fate defects characteristic of lag-2 or lin-12 mutants (n ≥ 50 animals examined). Tagged LAG-2::mT2 and LAG-2::mNG protein localized primarily to punctae on the cell surface, which made it challenging to identify the cellular source of expression in contexts where multiple neighboring cells may express LAG-2 ( Figure 1D ). To visualize lag-2 expression with easily discernible cellular resolution, we also generated an endogenous transcriptional reporter by inserting a self-cleaving peptide (P2A) and histone H2B (HIS-58) between LAG-2 and mT2 to generate a bicistronic gene that produces both unlabeled LAG-2 and a nuclear H2B::mT2 marker from the same transcript ( Figure 1B ). Unlike traditional transgenic reporters made using somewhat arbitrary promoter fragments, endogenous transcriptional reporters are more indicative of native expression patterns as they reflect the chromatin landscape and full complement of cis-regulatory elements acting on the endogenous gene (Rojas-Fernandez et al ., 2015). All of the alleles used here also contain epitope tags (3xFLAG or 2xHA) that can be used for biochemical assays ( Figure 1B,C ).
To visualize Notch/Delta signaling dynamics during development, we paired LIN-12::mNG with each of the lag-2 alleles. These combinations allowed us to visualize LIN-12 activity in concert with LAG-2 protein or lag-2 transcription during key cell fate specification events in vivo . LIN-12 and LAG-2 play important roles in development of the hermaphroditic reproductive system through a lateral inhibition event that specifies distinct fates (Greenwald et al ., 1983). In the somatic gonad, specification of the anchor cell (AC) and ventral uterine cells (VU) relies on a stochastic Notch signaling event between two initially equipotent cells, Z1.ppp and Z4.aaa ( Figure 1E ). Consistent with expectations, we found that LAG-2 and LIN-12 are first expressed in both AC/VU precursors without visually discernible differences between cells ( Figure 1F,G ). During the course of AC and VU cell fate specification, we observed that LIN-12 becomes localized to the nucleus in the presumptive VU where it is activated and is lost from the presumptive AC, while LAG-2 expression becomes restricted to the AC ( Figure 1F,G ) (Wilkinson et al ., 1994). Although asymmetry in LAG-2::P2A::H2B::mT2 signal is detectable during AC/VU specification, there is a delayed reduction in H2B::mT2 fluorescence that is likely due to perdurance of the histone H2B (Toyama et al ., 2013) ( Figure 1G ). However, this discrepancy resolves over time, and at later developmental stages we observed strong LAG-2::P2A::H2B::mTurquoise2 signal in the AC with no detectable signal in VU cells ( Figure 1G ). This difference in perdurance between proteins suggests a productive approach may be to utilize LAG-2::mT2 to visualize real-time changes in LAG-2 protein levels in parallel with the transcriptional reporter to visualize the identities of LAG-2 expressing cells.
In order to validate our toolkit for reading out molecular phenotypes after functional manipulations, we perturbed Notch signaling by degrading endogenous LIN-12::mNG::AID using the TIR1-AID system (Zhang et al ., 2015; Martinez et al ., 2020). Degrading LIN-12::mNG::AID (Pani et. al., 2022) using a ubiquitously expressed TIR1 transgene (Hills-Muckey et al ., 2022) caused uterine and vulval cell patterning defects consistent with known functions for Notch/Delta-mediated lateral inhibition in AC/VU decision and vulval precursor cell (VPC) fates ( Figure 1E, H ) (Greenwald et al. , 1983). In control animals, we observed expression of the lag-2 transcriptional reporter in the single AC and the primary-fated VPC descendants of P6.p. Following treatment with auxin (1 mM K-NAA in solid media) to degrade LIN-12, we observed the phenotype of two ACs and ectopic primary VPCs expressing the lag-2 transcriptional reporter at 100% penetrance (n = 25) ( Figure 1H ). These phenotypes recapitulate those previously reported, where both AC/VU precursors adopt the default AC state, and P5.p and P7.p adopt the primary VPC fate in the absence of Notch signaling (Greenwald et al ., 1983).
In summary, here we expand the toolkit for studying Notch/Delta signaling in C. elegans by generating additional tools to visualize endogenous LAG-2 protein and lag-2 transcription in vivo . The strains reported here make it possible for the first time to observe endogenous Notch (LIN-12) and Delta (LAG-2) dynamics simultaneously in living animals, which we expect will facilitate increasingly precise efforts to dissect Notch/Delta signaling mechanisms and functions. It is our hope that these reagents will be useful to the C. elegans community and others.
Methods
Strain maintenance:
Animals were reared under standard conditions and cultured at either 20°C (for imaging at the L2 stage) or 25°C (for imaging at the L3 stage) (Brenner, 1974). Animals were synchronized through alkaline hypochlorite treatment of gravid adults to isolate eggs (Porta-de-la-Riva et al. , 2012). The genotypes of all strains used in this study can be found in the Strain Table.
CRISPR/Cas9 injections:
The endogenous lag-2 locus was edited via CRISPR/Cas9 mediated genome engineering using the self-excising cassette (SEC) method to facilitate screening and the guide RNA targeting sequence 5’ GAAGAATCTAGACATAGTGAC 3’ (Dickinson et al. , 2013; Dickinson et al. , 2015; Gordon et. al., 2019). The repair templates for lag-2 were generated by first cloning homology arms, synthesized by PCR using genomic DNA as a template, into pDD315 (mTurquoise2). The LAG-2::mTurquoise2 plasmid was then modified by digestion with the restriction enzyme Bsu36I and assembled with an amplified P2A::H2B sequence through Gibson cloning.
Auxin-inducible degradation:
Nematode growth media plates were prepared with the synthetic auxin, K-NAA, for a final concentration of 1 mM, as previously described (Martinez & Matus, 2020). Synchronized L1 stage animals were plated on auxin plates seeded with OP50 E. coli .
Live-cell imaging :
Micrographs were collected on a Hamamatsu ORCA EM-CCD camera mounted on an upright Zeiss AxioImager A2 with a Borealis-modified CSU10 Yokagawa spinning disk scan head (Nobska Imaging) using 440 nm, 514 nm, and 561 nm Vortran lasers in a VersaLase merge and a Plan-Apochromat 100×/1.4 (NA) Oil DIC objective. MetaMorph software (Molecular Devices) was used for microscopy automation. Animals were mounted into a drop of M9 on a 5% Noble agar pad containing approximately 10 mM sodium azide anesthetic and topped with a coverslip.
Image processing:
Images were processed using Fiji/ImageJ (v.2.0.0) (Schindelin et al. , 2012). Panels D and H feature maximum intensity projections, whereas panels F and G show single plane micrographs. Schematics of gene loci were generated using sequences from WormBase (Harris et al. , 2020) and the Exon-Intron Graphic Maker (http://wormweb.org/exonintron). Figures were assembled in Adobe Illustrator (v.26.0.2).
Reagents
Strains:
|
Strain |
Genotype |
Source |
|
DQM1049 |
lin-12 ( ljf31 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG]) III; lag-2 ( bmd204 [ lag-2 ::mTurquoise2^lox511I^2xHA]) V. |
This study |
|
DQM1051 |
lin-12 ( ljf31 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG]) III; lag-2 ( bmd202 [ lag-2 ::P2A::H2B::mTurquoise2^lox511I^2xHA]) V. |
This study |
|
DQM1066 |
lin-12 ( ljf33 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG::AID*]) III; lag-2 ( bmd204 [ lag-2 ::mTurquoise2^lox511I^2xHA]) V; cshIs128 [ rpl-28p :: At TIR1::T2A::mCherry::HIS-11)] II. |
This study |
|
DQM1068 |
lin-12 ( ljf33 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG::AID*]) III; lag-2 ( bmd204 [ lag-2 ::mTurquoise2^lox511I^2xHA]) V; cshIs140 [ rpl-28p :: At TIR1(F79G)::T2A::mCherry::HIS-11] II. |
This study |
|
DQM1070 |
lin-12 ( ljf33 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG::AID*]) III; lag-2 ( bmd202 [ lag-2 ::P2A::H2B::mTurquoise2^lox511I^2xHA]) V; cshIs128 [ rpl-28p :: At TIR1::T2A::mCherry::HIS-11)] II. |
Pani et al., 2022 |
|
DQM1072 |
lin-12 ( ljf33 [ lin-12 ::mNeonGreen[C1]^loxP^3xFLAG::AID*]) III; lag-2 ( bmd202 [ lag-2 ::P2A::H2B::mTurquoise2^lox511I^2xHA]) V; cshIs140 [ rpl-28p :: At TIR1(F79G)::T2A::mCherry::HIS-11] II. |
Pani et al., 2022 |
|
LP469 |
lag-2 ( cp193 [ lag-2 ::mNeonGreen[C1]^loxP^3xFLAG]) V. |
Gordon et al., 2019 |
Plasmids:
|
Plasmid |
Description |
Source |
|
pAP031 |
lag-2 sgRNA plasmid |
Gordon et al., 2019 |
|
pTNM062 |
lag-2 mTurquoise2^SEC^2xHA repair plasmid |
This study |
|
pTNM114 |
lag-2 P2A::H2B::mTurquoise2^SEC^2xHA repair plasmid |
This study |
Acknowledgments
Acknowledgments
We thank Michael A.Q. Martinez for providing valuable feedback on the manuscript. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
NIH R01GM121597 (D.Q.M.); NIH R35GM142880 (A.M.P.); Damon Runyon-Rachleff Innovator award DRR-47-17 supported in part by the Damon Runyon Cancer Research Foundation (D.Q.M.); NIH predoctoral fellowship F31HD100091 (T.N.M.-K.); Stony Brook University Presidential Critical Research Funds (T.N.M.-K.)
References
- Ahier A, Jarriault S. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics. 2013 Dec 20;196(3):605–613. doi: 10.1534/genetics.113.160846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attner MA, Keil W, Benavidez JM, Greenwald I. HLH-2/E2A Expression Links Stochastic and Deterministic Elements of a Cell Fate Decision during C. elegans Gonadogenesis. Curr Biol. 2019 Aug 8;29(18):3094–3100.e4. doi: 10.1016/j.cub.2019.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974 May 1;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013 Sep 1;10(10):1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015 Jun 3;200(4):1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, Lapébie P, Richards GS, Brunet F, Ereskovsky AV, Degnan BM, Borchiellini C, Vervoort M, Renard E. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009 Oct 13;9:249–249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Payne SG, Linden-High LM, Pani AM, Goldstein B, Hubbard EJA, Sherwood DR. Ectopic Germ Cells Can Induce Niche-like Enwrapment by Neighboring Body Wall Muscle. Curr Biol. 2019 Feb 21;29(5):823–833.e5. doi: 10.1016/j.cub.2019.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW Jr, Royant A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun. 2012 Mar 20;3:751–751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983 Sep 1;34(2):435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Greenwald I, Kovall R. Notch signaling: genetics and structure. WormBook. 2013 Jan 17;:1–28. doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, Lee RYN, Muller HM, Nakamura C, Nuin P, Paulini M, Raciti D, Rodgers FH, Russell M, Schindelman G, Auken KV, Wang Q, Williams G, Wright AJ, Yook K, Howe KL, Schedl T, Stein L, Sternberg PW. WormBase: a modern Model Organism Information Resource. Nucleic Acids Res. 2020 Jan 8;48(D1):D762–D767. doi: 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994 Oct 1;120(10):2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hills-Muckey K, Martinez MAQ, Stec N, Hebbar S, Saldanha J, Medwig-Kinney TN, Moore FEQ, Ivanova M, Morao A, Ward JD, Moss EG, Ercan S, Zinovyeva AY, Matus DQ, Hammell CM. An engineered, orthogonal auxin analog/AtTIR1(F79G) pairing improves both specificity and efficacy of the auxin degradation system in Caenorhabditis elegans. Genetics. 2022 Feb 4;220(2) doi: 10.1093/genetics/iyab174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie EJ, Kimble J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development. 1991 May 1;112(1):231–240. doi: 10.1242/dev.112.1.231. [DOI] [PubMed] [Google Scholar]
- Levitan D, Greenwald I. LIN-12 protein expression and localization during vulval development in C. elegans. Development. 1998 Aug 1;125(16):3101–3109. doi: 10.1242/dev.125.16.3101. [DOI] [PubMed] [Google Scholar]
- Martinez MAQ, Kinney BA, Medwig-Kinney TN, Ashley G, Ragle JM, Johnson L, Aguilera J, Hammell CM, Ward JD, Matus DQ. Rapid Degradation of Caenorhabditis elegans Proteins at Single-Cell Resolution with a Synthetic Auxin. . G3 (Bethesda) 2020 Jan 7;10(1):267–280. doi: 10.1534/g3.119.400781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MAQ, Matus DQ. Auxin-mediated Protein Degradation in Caenorhabditis elegans . . Bio Protoc. 2020 Apr 20;10(8) doi: 10.21769/BioProtoc.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani, AM; Gibney, TV; Medwig-Kinney, TN; Matus, DQ; Goldstein, B (2022). A new toolkit to visualize and perturb endogenous LIN-12/Notch signaling in C. elegans. microPublication Biology. 10.17912/micropub.biology.000603 [DOI] [PMC free article] [PubMed]
- Porta-de-la-Riva M, Fontrodona L, Villanueva A, Cerón J. Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp. 2012 Jun 10;(64):e4019–e4019. doi: 10.3791/4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Fernandez A, Herhaus L, Macartney T, Lachaud C, Hay RT, Sapkota GP. Rapid generation of endogenously driven transcriptional reporters in cells through CRISPR/Cas9. Sci Rep. 2015 Apr 29;5:9811–9811. doi: 10.1038/srep09811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jun 28;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods. 2013 Mar 24;10(5):407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöqvist M, Andersson ER. Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev Biol. 2017 Sep 29;447(1):58–70. doi: 10.1016/j.ydbio.2017.09.032. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993 Jul 30;74(2):331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013 Aug 29;154(5):971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson HA, Fitzgerald K, Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994 Dec 30;79(7):1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015 Nov 9;142(24):4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]