Abstract
SigB, a newly discovered alternative sigma factor of Staphylococcus aureus, has been shown to play an important role in stress responses and the regulation of virulence factors. The rsbW (orf159) gene is immediately upstream of sigB. Its gene product is homologous to Bacillus subtilis RsbW which under appropriate conditions binds to B. subtilis SigB and functions as an anti-sigma factor or negative posttranslational regulator. To define the function of S. aureus RsbW, both the S. aureus SigB and RsbW proteins were expressed in Escherichia coli and purified. Cross-linking experiments with these purified proteins revealed that RsbW was capable of specific binding to SigB. In an in vitro transcription runoff assay, RsbW prevented SigB-directed transcription from the sar P3 promoter, a known SigB-dependent promoter, and the inhibitory activity of RsbW was found to be concentration dependent. We also identified SigB promoter consensus sequences upstream of the genes encoding alkaline shock protein 23 and coagulase and have demonstrated SigB and RsbW dependence for the promoters in vitro. These results show that RsbW is a protein sequestering anti-sigma factor of S. aureus SigB and suggest that SigB activity in S. aureus is regulated posttranslationally.
Staphylococcus aureus is a major human pathogen that causes a variety of diseases ranging from minor skin ailments to life-threatening deep infections, such as endocarditis, meningitis, arthritis, and toxic shock syndrome (26, 36, 39). The frequencies of both community- and hospital-acquired staphylococcal infections have increased steadily with little change in overall mortality (16). Treatment of these infections has become more difficult because of the emergence of multidrug-resistant strains (16, 37).
The virulence of S. aureus is dependent upon its ability to respond to a wide range of host conditions during the infection process. Transcriptional regulators such as sigma factors are likely to play an important role in the bacterial adaptive responses needed for pathogenesis (29). Indeed, alternative sigma factors have been correlated with virulence in several pathogenic species (13, 17, 18). Recently, an S. aureus alternative sigma factor gene known as sigB was identified within a four-gene operon (22, 40). With strong primary amino acid similarity to SigB of Bacillus subtilis, S. aureus SigB has been evaluated as a stress response and stationary-phase sigma factor and has been shown to be induced during stationary phase and upon heat shock (22). Additionally, interruption of the S. aureus sigB gene causes increased sensitivity to hydrogen peroxide during the stationary growth phase (23). By in vitro transcription S. aureus SigB has further been shown to participate in the transcription of the sar locus (12), which is itself a key regulator of virulence gene expression (7, 8, 27). Furthermore, the expression of lipase and thermonuclease, which play important roles in abscess formation, have been associated with SigB control (23). Thus, SigB appears to participate directly and indirectly in the expression of S. aureus virulence genes.
While most sigma factors are themselves transcriptionally regulated, posttranslational control by other proteins known as anti-sigma factors also plays an important role in controlling their activity in some instances (4, 5, 31). Anti-sigma factor proteins bind and sequester a specific sigma factor, thus blocking transcription initiation (5, 19). The B. subtilis RsbW protein has been shown to function as an anti-sigma factor of the stress response regulator SigB (4, 14, 19). The B. subtilis rsbW gene is located immediately upstream of the sigB gene, and the two are cotranscribed. B. subtilis RsbW and SigB demonstrate specific binding by column chromatography and coimmunoprecipitation with monoclonal antibodies to either protein (4). B. subtilis RsbW also efficiently blocked SigB-dependent transcription in vitro. Recent DNA sequence analyses of the S. aureus sigB operon revealed four complete open reading frames (rsbU, rsbV, rsbW, and sigB) with significant predicted amino acid homology and gene arrangement to rsbU, rsbV, rsbW, and sigB in B. subtilis (22, 40). These similarities suggest that RsbW of S. aureus is an anti-sigma factor of S. aureus SigB. In this report we demonstrate that S. aureus RsbW binds to S. aureus SigB and inhibits SigB-dependent transcription. We also identify two possible new members of the S. aureus SigB regulon by demonstrating that the genes for S. aureus alkaline shock protein 23 and coagulase show SigB and RsbW dependence in vitro.
MATERIALS AND METHODS
Strains and plasmids.
The TA cloning vector pCRII was purchased from Invitrogen Corp. (Carlsbad, Calif.). Escherichia coli BL21(DE3) and the vector pET32b, used for protein overexpression, were obtained from Novagen (Madison, Wis.). Isolation and purification of plasmids were performed by using the Qiagen system (Qiagen, Inc., Chatsworth, Calif.). S. aureus strains ATCC 29213 and 8325-4 were used as sources of chromosomal DNA for the PCR amplifications.
Construction of plasmids for overexpressing sigB and rsbW (orf159).
A DNA fragment encoding 776 bp of the sigB gene was amplified by PCR with primers SAF005 (5′-ATCCATGGCGAAAGAGTCGAAATC-3′) and SAR003 (5′-CGGATCCTATTGATGTGCTGCTTCTTG-3′); this PCR product was cloned into pCRII, and the resulting plasmid was designated pEM101. The sigB-overexpressing plasmid, pEM102, was the product of cloning the NcoI-BamHI-digested fragment from pEM101 with the same enzymes into pET32b.
pEM201 was constructed by cloning a 486-bp fragment containing the rsbW (orf159) gene into pCRII. Oligonucleotides ASAF001 (5′-CCATGGATGCAATCTAAAGAAGATTTT-3′) and ASAR002 (5′-GGATCCTTAACTGATTTCGACTCTTTCGGC-3′) were used to amplify this PCR product. An NcoI-BamHI-digested fragment from pEM201 was inserted into pET32b digested with the same enzymes to create the rsbW expression vector, pEM202.
Purification of SigB and RsbW fusion proteins.
pET32b-based, His6-thioredoxin (Trx) fusion proteins were expressed and purified according to the recommendations of the manufacturer (Clontech Laboratories, Inc., Palo Alto, Calif.) with some modifications. E. coli BL21(DE3) transformed with pEM102, pEM202, and pET32b (generating strains EMBL1, EMBL2, and EMBL3, respectively) was grown in 250 ml of Luria-Bertani medium containing ampicillin (100 μg/ml) at 37°C until the culture reached an optical density at 600 nm (OD600) of between 0.6 to 0.8. Induction with isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM was conducted for 3 h, and then the cells were harvested, suspended in 10 ml of lysis buffer (50 mM NaH2PO4, 10 mM Tris-HCl, 100 mM NaCl; pH 8), and lysed by sonication. After centrifugation at 12,000 rpm for 30 min, the resulting supernatant was loaded onto a 2-ml column of metal affinity resin (Clontech) equilibrated with sonication buffer. After exposure to excess wash buffer (50 mM NaH2PO4, 100 mM NaCl; pH 7), the Trx-tagged protein was eluted with elution buffer (50 mM NaH2PO4, 20 mM PIPES [piperazine-N,N1-bis(2-ethanesulfonic acid)], 100 mM NaCl; pH 6) and dialyzed against a solution of 10 mM Tris-HCl (pH 8), 50 mM KCl, 10 mM MgCl2, 0.4 mM, dithiothreitol, and 20% glycerol. Protein concentrations were determined with the Coomassie reagent (Pierce, Rockford, Ill.). Purified protein was divided into aliquots and stored at −70°C.
[35S]methionine labeling of SigB and RsbW.
EMBL1, EMBL2, and EMBL3 cells were grown at 37°C in 20 ml of M9 minimal medium supplemented with 0.2% glucose, vitamin B1 (1 μg/ml), and ampicillin (75 μg/ml) to an OD600 of 0.5; IPTG was then added to 1 mM. After 30 min, rifampin was added to a final concentration of 200 μg/ml for an additional hour. Then, 1-ml aliquots of culture were labeled with 20 μCi of [35S]methionine for 5 min and chased with unlabeled excess methionine for an additional 5 min. The labeled cells were then collected by centrifugation, washed, and frozen at −70°C. Cell pellets from 1 ml of [35S]methionine-labeled cells were resuspended in 0.5 ml of lysis buffer and lysed by repeated freeze-thaw steps. Particulate matter was removed by centrifugation.
Preparation of crude cell lysates from EMBL3 cells.
Cultures (25-ml each) of EMBL3 cells (harboring empty vector) were grown to an OD600 of 0.6, induced with IPTG for 3 h, harvested, and frozen as pellets at −70°C. Before use, the pellets were suspended in lysis buffer and sonicated. After centrifugation at 14,000 rpm for 15 min, the resulting supernatant was collected.
Chemical cross-linking reaction.
Chemical cross-linking was carried out in 50-μl reaction mixtures containing 1 mM ethylene glycol-bis(succinimidylsuccinate) (EGS) (Pierce), 5 to 10 μg of extract containing 35S-labeled proteins from recombinant E. coli, and unlabeled proteins at the following concentrations: RsbW, 0.25 to 0.5 μg; SigB, 0.75 to 1.5 μg; crude EMBL3 extract, 150 μg; or lysis buffer. The cross-linking reactions were allowed to proceed for 3 h on ice and were terminated by the addition of sodium dodecyl sulfate (SDS) gel loading dye and l-lysine to 20 mM (final concentration). Samples were boiled and separated by electrophoresis in SDS–10% polyacrylamide slab gels. Gels were stained with Coomassie brilliant blue R-250, impregnated with a scintillation fluor, dried, and analyzed by autoradiography.
In vitro transcription assay.
For RNA polymerase holoenzyme reconstitution, purified SigB and E. coli core RNA polymerase (Epicentre Technologies, Madison, Wis.) were coincubated at 37°C for 30 min. For inhibition experiments, SigB was preincubated with purified RsbW, albumin, or dilution buffer prior to incubation with the core RNA polymerase. Single runoff in vitro transcription reactions were conducted by the sequential addition of template DNA, nucleotides including [α-32P]CTP, and a mixture of heparin and unlabeled CTP. Final concentrations in 40 μl of reaction mixture were as follows: 10 mM Tris-HCl (pH 8), 50 mM KCl, 10 mM MgCl2, 0.4 mM dithiothreitol, 0.25 mM ATP, 0.25 mM GTP, 0.25 mM TTP, 10 μCi of [α-32P]CTP, 0.25 mM CTP, 500 μg of heparin per ml, and 1.0 U of RNase inhibitor. Finally, 40 μl of formamide loading dye was added to the sample. After being boiled, the samples were loaded and electrophoresed in a 6% denaturing polyacrylamide gel. Gels were analyzed immediately by autoradiography. Template DNA fragments containing S. aureus promoters were prepared as follows: sar P3 (349 bp) was amplified with primers sarF01 (5′-GTATAGACACTTTAACGTGCT-3′) and sarR02 (5′-ACAGTGATTGTATTTCTGGGT-3′); sar P1 (339 bp) was amplified with primers sarF03 (5′-AAAGCGTTGATTTGGGTAGTA-3′) and sarR04 (5′-AGCACGTTAAAGTGTCTATAC-3′); sar P2 (321 bp) was amplified with primers sarF05 (5′-TCGAAACATTTAATTGCGCTA-3′) and sarR06 (5′-ACCTCCCTATTTGATGCATCT-3′); asp23 (320 bp) was amplified with aspF1 (5′-GACTCTACACAACAAGTGATT-3′) and aspR2 (5′-AGTTTGATTGTCGTATGCTTG-3′); and coa (300 bp) was amplified with coaF1 (5′-CAAAAAGATAGTTAATGCTTTGTT-3′) and coaR2 (5′-AGTCTTCCAAATAATATAGAGCTG-3′). Each DNA fragment was gel purified prior to use in runoff assays.
RESULTS
Purification of SigB and RsbW proteins.
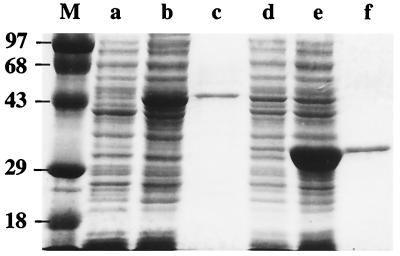
A pET32b-based expression vector called pEM102 was constructed in which the T7 promoter was fused to the S. aureus sigB gene. Induction of E. coli BL21(DE3) harboring pEM102 with IPTG led to high-level expression of soluble SigB fusion protein, as can be seen in Fig. 1 (lane b). Affinity column chromatography gave Trx-tagged SigB protein which migrated at an estimated molecular mass of 44 kDa on SDS-polyacrylamide gel electrophoresis (PAGE) and was 90% pure (Fig. 1, lane c). The deduced molecular mass of untagged SigB is 29.4 kDa and that of the Trx-SigB fusion protein is 41.1 kDa (the mass of the Trx moiety is 11.7 kDa).
FIG. 1.
Overexpression and purification of recombinant SigB and RsbW proteins. SDS–10% PAGE gel analysis (25) of fractions from the purification of SigB and RsbW. Lane M contains the molecular mass markers (masses in kilodaltons are shown on the left). Cell extracts from E. coli BL21(DE3) harboring pEM102 (Trx-SigB overexpression) grown without IPTG induction (lane a), in the presence of IPTG (lane b), and the affinity-purified Trx-SigB fusion protein (lane c) are shown. Cell extracts from E. coli BL21(DE3) harboring pEM202 (Trx-RsbW overexpression) grown without IPTG induction (lane d), in the presence of IPTG (lane e), and with the affinity-purified Trx-RsbW fusion protein (lane f) are shown.
RsbW protein was obtained by the same method, yielding large amounts of soluble fusion protein in an IPTG-dependent manner (Fig. 1, lane e versus lane d). The purified Trx-tagged RsbW protein migrated at an estimated molecular mass of 33 kDa on SDS-PAGE and was 90% pure (see Fig. 1, lane f). The deduced molecular mass of untagged RsbW is 17.9 kDa, and the deduced total mass of the fusion protein is 29.6 kDa.
Formation of SigB-RsbW complexes by chemical cross-linking.
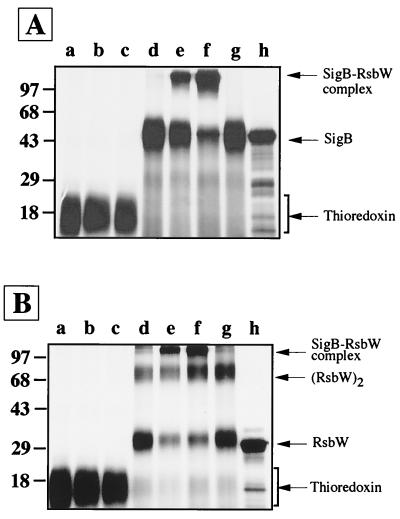
After preparing Trx-tagged SigB and RsbW proteins in radioactive and nonradioactive forms by E. coli overexpression, we tested for direct protein-protein interactions between SigB and RsbW by using EGS. EGS is a bifunctional chemical cross-linking reagent which reacts to form covalent bonds with lysine residues spaced no more than 11 Å apart. As may be seen in Fig. 2A, 35S-labeled, tagged SigB migrates at 44 kDa on an SDS-polyacrylamide gel (Fig. 2A, lane h) and does not form high-molecular-mass complexes in the presence of EGS (Fig. 2A, lane d). Incubation of radioactive, tagged SigB extracts with different amounts of unlabeled, purified RsbW in the presence of EGS generated high-molecular-mass SigB-containing complexes of over 97 kDa on an SDS–10% polyacrylamide gel (lane e and f). The amount of radioactive SigB appearing in a high-molecular-weight complex correlated directly with the amount of RsbW added to the mixture, suggesting that it is a SigB-RsbW covalent aggregate.
FIG. 2.
Specific interaction between SigB and RsbW. (A) 35S-labeled, unfused Trx protein incubated in the presence of EGS with lysis buffer (lane a) or with 0.25 μg (lane b) or 0.5 μg (lane c) of unlabeled RsbW are shown. Also shown are 35S-labeled, Trx-tagged SigB incubated in the presence of EGS alone (lane d) or with 0.25 μg (lane e) or 0.5 μg (lane f) of unlabeled RsbW in the presence of EGS. Lanes g and h show 35S-labeled, Trx-tagged SigB incubated in the presence of EGS with cell extract from E. coli EMBL3 (lane g) and 35S-labeled, Trx-tagged SigB incubated in the absence of EGS with 0.5 μg of RsbW (lane h). (B) 35S-labeled, unfused thioredoxin (Trx) protein incubated in the presence of EGS with lysis buffer (lane a) or with 0.75 μg (lane b) or 1.5 μg (lane c) of unlabeled SigB. Remaining lanes show 35S-labeled, Trx-tagged RsbW incubated in the presence of EGS alone (lane d) or with 0.75 μg (lane e) or 1.5 μg (lane f) of unlabeled SigB in the presence of EGS; 35S-labeled Trx-tagged RsbW incubated in the presence of EGS with cell extract from E. coli EMBL3 (lane g); and 35S-labeled, Trx-tagged RsbW incubated in the absence of EGS with 1.5 μg of unlabeled SigB (lane h). Molecular masses in kilodaltons are shown at the left.
To evaluate the specificity of complex formation, we tested the ability of protein extracts lacking RsbW to complex with radiolabeled, tagged SigB. Whole-cell extracts from EMBL3 (vector alone strain of E. coli expressing only the Trx tag) incubated in the presence of EGS with radioactive, tagged SigB failed to produce high-molecular-weight complexes (Fig. 2A, lane g). To control for the Trx tag on SigB, we also tested the ability of the radiolabeled Trx polypeptide alone to cross-link with RsbW. When 0.75 or 1.5 μg of purified tagged RsbW was incubated with radioactive Trx-containing extracts in the presence of EGS, high-molecular-weight adducts did not form (Fig. 2A, compare lane a [no EGS] with lanes b and c [with EGS]). This experiment excludes the possibility that interactions between RsbW and the Trx tag are sufficient for cross-linking to occur.
Additionally, we performed the converse experiment in which radiolabeled, tagged RsbW-containing E. coli extracts were allowed to interact with nonradioactive, purified SigB in the presence of cross-linker. E. coli extracts containing 35S-labeled, tagged RsbW produced a new 68-kDa band after EGS treatment (Fig. 2B, lane d), while in the absence of EGS only a 33-kDa species was seen (Fig. 2B, lane h). This suggests that tagged RsbW exists as a dimer, although we cannot exclude the possibility that it binds to another protein of similar size derived from E. coli. RsbW also appeared to be able to form higher self-aggregates, as may be seen by the faint high-molecular-weight bands in Fig. 2B, lanes d and g. When purified, tagged SigB was added to the extract containing radioactive, tagged RsbW in the presence of EGS, high-molecular-mass complexes running above the 97-kDa marker on an SDS–10% polyacrylamide gel were observed (Fig. 2B, lanes e and f). The intensity of the RsbW-containing complexes was dependent upon the amount of tagged SigB added, strongly suggesting that it is a SigB-RsbW covalent aggregate. As before, the specificity for complex formation was tested by incubating radioactive Trx with unlabeled SigB. No high-molecular-weight complexes were formed in these control experiments (Fig. 2B, lanes a to c). These results indicate that S. aureus SigB and RsbW undergo a relatively specific protein-protein interaction in vitro and that conjugates between the two proteins may be trapped by using the chemical cross-linker EGS.
Inhibition of SigB-directed transcription of sar by RsbW.
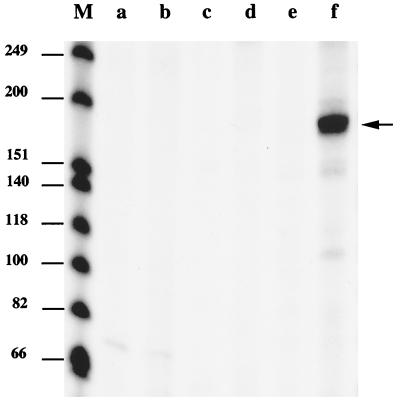
PCR products (300 to 350 bp) corresponding to S. aureus promoters were used as templates for in vitro transcription runoff assays. We first tested the three sar operon promoters, including sar P3 (producing the sarC transcript), which has been shown to be SigB dependent. The expected sizes of the transcripts from sar P1, sar P2, or sar P3 are 140, 167, or 194 bases, respectively. As shown in Fig. 3, the sar P1 and sar P2 promoters failed to direct transcription by core polymerase alone or holoenzyme EςB (core polymerase reconstituted with Trx-tagged SigB) (Fig. 3, lanes a to d). On the other hand, EςB generated a transcript at 190 bases (Fig. 3, lane f), whereas core enzyme alone failed to transcribe from the sar P3 promoter (Fig. 3, lane e). In our single-round transcription assay, we occasionally observed low-abundance bands smaller than the anticipated runoff product when large amounts of SigB (≥0.15 μg) were added to the core enzyme (e.g., Fig. 3, lane f). These bands may result from stutter products or from nonspecific binding due to the saturated state of the promoter region when EςB is present at a high concentration. We also tested whether the Trx tag present on the SigB protein may have interfered with in vitro transcription. Untagged SigB which had had the recombinant Trx tag removed by treatment with enterokinase was found to be of equal potency with Trx-tagged sigma factor in the in vitro transcription assay (data not shown). Hence, the presence of the Trx tag on SigB appears to have little effect on the activity of SigB.
FIG. 3.
In vitro transcription analysis of the sar operon. Lane M, 32P-labeled DNA size markers. The sizes of the individual DNA fragments in bases are indicated on the left. DNA templates (0.02 μg) for the transcription reaction containing the sar P1, P2, or P3 promoters were added to the reaction mixtures. Lanes a and b were from mixtures containing P1; lanes c and d were from mixtures containing P2; and lanes e and f were from mixtures containing P3. The transcription reaction mixtures contained 0.4 U of E. coli core RNA polymerase preincubated with 0.58 μg of Trx-tagged SigB protein (lanes b, d, and f) or with dialysis buffer (lanes a, c, and e). The products of transcription were subjected to electrophoresis on a 6% denaturing polyacrylamide gel and visualized by autoradiography. The arrow indicates the position of the 190-base sar P3 transcript.
To examine whether RsbW would inhibit SigB-directed transcription, we preincubated tagged SigB with various amounts of purified, tagged RsbW before it was incubated with core RNA polymerase. As may be seen in Fig. 4, transcription generated by EςB was inhibited by the addition of RsbW (Fig. 4, lanes c to e). When RsbW was added to SigB in an equimolar amount, transcription was completely prevented (Fig. 4, lane e). However, preincubation of SigB with an excess of albumin did not affect the transcription (Fig. 4, lane f). Similarly, preformed EςB was resistant to the inhibitory effect of RsbW (Fig. 4, lane g).
FIG. 4.
Inhibition of SigB-directed transcription from sar P3 by RsbW. The transcription reaction mixtures contained 0.4 U of E. coli core RNA polymerase and 0.02 μg of DNA template containing the sar P3 promoter. Lanes: a, core RNA polymerase incubated with dialysis buffer; b, core RNA polymerase incubated with 0.58 μg of Trx-tagged SigB; c to e, Trx-tagged SigB incubated with increasing amounts (0.03, 0.12, and 0.36 μg in lanes c, d, and e, respectively) of the Trx-tagged RsbW fusion protein prior to the addition of core RNA polymerase; f, Trx-tagged SigB incubated with an excess amount (18 μg) of bovine serum albumin instead of RsbW prior to the addition of core RNA polymerase; g, Trx-tagged SigB incubated with core RNA polymerase prior to the addition of 0.36 μg of RsbW. The products of transcription were subjected to electrophoresis on a 6% denaturing polyacrylamide gel and visualized by autoradiography. The arrow indicates the position of the anticipated sar transcript. Lane M, 32P-labeled DNA size markers as in Fig. 3.
Transcription of genes encoding alkaline shock protein 23 and coagulase is SigB dependent and inhibited by RsbW.
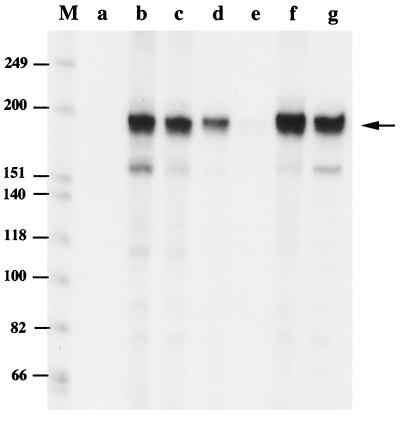
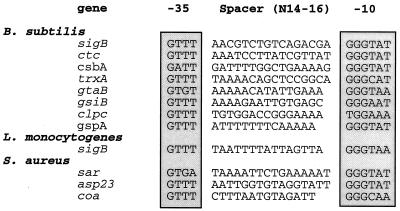
We screened all S. aureus genes available in the GenBank as of November 1998 for the presence of sequences resembling the B. subtilis SigB promoter consensus (Fig. 5). The search identified putative SigB promoter sequences upstream of the asp23 gene which encodes alkaline shock protein 23, and coa gene which encodes coagulase (21, 33) (Fig. 5). To test the SigB dependence of these promoters, we PCR amplified these promoters from S. aureus genomic DNA; SigB-directed in vitro transcription from the asp23 and coa promoter templates was calculated to produce 144 and 200 base transcripts, respectively. As shown in Fig. 6, core RNA polymerase reconstituted with SigB produced a 150-base transcript from the asp23 putative promoter (Fig. 6, lane b), while core enzyme alone failed to generate a transcript (Fig. 6, lane a). In the same manner, EςB produced a 210-nucleotide transcript from the coa gene (Fig. 6, lane f). When an equimolar amount of RsbW was mixed with SigB prior to the addition of core polymerase, SigB-dependent asp23 and coa transcription was inhibited almost completely (Fig. 6, lanes c and g). Excess quantities of albumin as a control protein did not influence the transcription of asp23 and coa (Fig. 6, lanes d and h).
FIG. 5.
SigB promoter sequences from B. subtilis, L. monocytogenes, and S. aureus. The putative promoter sequences of the asp23 gene and the coa gene are aligned with those of known SigB-dependent promoters from B. subtilis, L. monocytogenes, and S. aureus. In B. subtilis and L. monocytogenes the SigB-dependent promoters control the sigB operon in part.
FIG. 6.
SigB-directed transcription of asp23 and coa and inhibition by RsbW. The transcription reaction mixtures contained 0.4 U of E. coli core RNA polymerase and 0.02 μg of DNA templates containing the putative promoter region of asp23 (lanes a to d) and coa (lanes e to h). Lanes: a and e, no addition; b and f, 0.58 μg of Trx-tagged SigB that had been preincubated with dialysis buffer; c and g, 0.58 μg of Trx-tagged SigB that had been preincubated with 0.36 μg of Trx-RsbW fusion protein; d and h, 0.58 μg of Trx-tagged SigB that had been preincubated with an excess amount (18 μg) of bovine serum albumin. The products of transcription were subjected to electrophoresis on a 6% denaturing polyacrylamide gel and visualized by autoradiography. The arrows indicate the positions of the anticipated transcripts. Lane M, 32P-labeled DNA size markers as in Fig. 3.
DISCUSSION
Anti-sigma factors are posttranslational transcription regulators which, under appropriate cellular conditions, bind to their cognate sigma factor and block sigma factor association with core RNA polymerase. As a result, anti-sigma factors inhibit transcription from a given regulon by inhibiting the action of a specific sigma factor (5). In this study we determined that the gene product of the rsbW (orf159) gene is an anti-sigma factor of S. aureus SigB. Our data show that RsbW and SigB are capable of direct protein-protein interaction, as documented by cross-linking experiments, and that RsbW is a specific inhibitor of SigB-directed transcription in vitro.
Our cross-linking experiments show the RsbW and SigB interaction to be relatively specific and indicate that RsbW exists as a dimer in solution, while SigB is monomeric. This agrees well with the results of SpoIIAB, an anti-sigma factor of B. subtilis SigF (9, 15) and UsfX, an anti-sigma factor of Mycobacterium tuberculosis SigF (unpublished data), respectively. In spite of its ability to dimerize, RsbW inhibits SigB with 1:1 stoichiometry, since our in vitro transcription experiments indicate that equimolar concentrations of RsbW completely blocked SigB-directed transcription.
Based on sequence comparison, the sar P3 promoter fits the consensus for SigB-dependent promoters of B. subtilis (2). We found that, biochemically, the sar P3 promoter is recognized by SigB, confirming the results reported by Deora et al. (12). Additionally, we showed that RsbW prevented SigB-directed transcription from the sar P3 promoter in a concentration-dependent manner.
The similar organization of the sigB operons in S. aureus and B. subtilis suggests analogous roles for the RsbV regulatory proteins in addition to RsbW (22, 40). In B. subtilis, RsbV is an anti-anti-sigma factor which competes with SigB for binding to anti-sigma factor RsbW—a mechanism dubbed partner switching (1, 14). An increased requirement for SigB in B. subtilis is governed by an increase in polycistronic transcription of the rsbV-rsbW-sigB-rsbX operon, and posttranslational mechanisms subsequently determine whether SigB is active. During normal exponential phase, RsbW inactivates RsbV by phosphorylation, promoting the formation of RsbW-SigB complexes. In response to stress or starvation, on the other hand, RsbV is dephosphorylated and captures the RsbW to form RsbV-RsbW complexes leading to the release of active SigB (14, 41). In addition to the operon transcription and partner-switching mechanisms for modulating sigma factor activity observed in B. subtilis, it has recently been proposed that the S. aureus sigB gene may also be expressed as a monocistronic message, independent of its upstream regulators during stationary phase (22). Thus, while some elements of SigB regulatory control appear to be conserved across species, important differences may be present. Recently, additional sigB-like operons have been discovered in Listeria monocytogenes (3, 38) and M. tuberculosis (10, 11, 30). The L. monocytogenes and M. tuberculosis operons show similarities in amino acid sequence and gene organization to the S. aureus and B. subtilis sigB operons, although the M. tuberculosis SigB-like operon lacks RsbV and RsbX homologues (11). In view of the apparent differences among these gram-positive bacteria, it will be important to evaluate the function of each SigB-like system independently.
Recent studies have characterized S. aureus SigB as a major regulator of the stress response against environmental changes such as heat shock, oxidative stress, and acid stress (6, 22, 23); however, the SigB-dependent genes responsible for these physiologic adaptations have not been identified. The asp23 gene encoding alkaline shock protein 23 was suggested as a possible target gene of SigB because the expression of Asp23 was affected by deletion of the entire rsbV-rsbW-sigB-rsbX operon in S. aureus (23). Expression of Asp23 is strongly induced upon pH upshift (24). Our in vitro transcription data support the notion that asp23 is a member of the S. aureus SigB regulon, although the physiological role of this stress response protein remains uncertain.
We have also found that the coa gene encoding coagulase is recognized by S. aureus SigB in vitro. Coagulase has been one of the most reliable determinants for the differentiation of S. aureus from other, less-virulent staphylococci. Since several reports have indicated that coagulase-deficient mutants of S. aureus are attenuated in experimental infections in mice, coagulase is considered a virulence determinant in the pathogenesis of S. aureus infections (20, 28, 34, 35). In culture, coagulase is preferentially expressed in early to late exponential phase. While coagulase deficiency did not appear to influence the course of valvular infection in the rat endocarditis model (32), coagulase was important for the establishment of lung infection in a model promoting pulmonary abscess formation (34). These different observations imply that coagulase expression may participate in later stages of infection and that SigB-dependent genes may be important for the abscess formation and survival in a purulent, microaerophilic environment. The role of S. aureus sigB in pathogenicity has been examined in one study with the mouse subcutaneous abscess model. While the analysis revealed no difference in virulence between a sigB mutant and the corresponding parent strain (6), it has been noted that the wild-type strain used (S. aureus 8325-4) contains an 11-bp deletion in the regulatory gene rsbU (22, 23) and may itself be attenuated.
As the S. aureus SigB regulon contains genes which have been associated with virulence, studies to clarify the role of SigB in the infection process and to identify more of the genes under its control may offer valuable insight into staphylococcal adaptive mechanisms. Since RsbW is a natural inhibitor of a virulence-associated transcription factor, it is possible that pharmacologic analogues of RsbW could have novel antibacterial properties against this important medical pathogen.
ADDENDUM
We recently tested whether RsbW had an inhibitory effect on in vitro transcription from the sar locus by a holoenzyme other than EςB. We found that commercially available E. coli ς70 (Epicentre Technologies, Madison, Wis.) associated with E. coli core RNA polymerase by the same methods described above was able to direct in vitro transcription from the S. aureus sar P2 promoter. Preincubation with purified RsbW or with albumin did not affect the level of Eς70-directed in vitro transcription from this promoter. These results provide additional support for the conclusion that RsbW is a specific anti-sigma factor for S. aureus SigB.
ACKNOWLEDGMENTS
We thank Richard Novick for providing S. aureus strains. We are grateful to Miki Miyazaki for excellent technical assistance and to Jennifer Doetsch for assistance in manuscript preparation.
This work was supported in part by NIH grants AI36973 and AI37856.
REFERENCES
- 1.Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. doi: 10.1016/0092-8674(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 2.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker L A, Cetin M, Hutkins R W, Benson A K. Identification of the gene encoding the alternative sigma factor ςB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180:4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown K L, Hughes K T. The role of anti-sigma factors in gene regulation. Mol Microbiol. 1995;16:397–404. doi: 10.1111/j.1365-2958.1995.tb02405.x. [DOI] [PubMed] [Google Scholar]
- 6.Chan P F, Foster S J, Ingham E, Clements M O. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Ying P. Regulation of α- and β-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decatur A L, Losick R. Three sites of contact between the Bacillus subtilis transcription factor ςF and its antisigma factor SpoIIAB. Genes Dev. 1996;10:2348–2358. doi: 10.1101/gad.10.18.2348. [DOI] [PubMed] [Google Scholar]
- 10.DeMaio J, Zhang Y, Ko C, Young D B, Bishai W R. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMaio J, Zhang Y, Ko C, Bishai W R. The Mycobacterium tuberculosis sigF gene is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tubercle Lung Dis. 1997;78:3–12. doi: 10.1016/s0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 12.Deora R, Tseng T, Misra T K. Alternative transcription factor ςSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan L, Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc Natl Acad Sci USA. 1993;15:2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emori T G, Gayner R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative ς factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillen K L, Hughes K T. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes K T, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 20.Jonsson P, Lindberg M, Haraldsson I, Wadström T. Virulence of Staphylococcus aureus in a mouse mastitis model: studies of alpha hemolysin, coagulase, and protein A as possible virulence determinants with protoplast fusion and gene cloning. Infect Immun. 1985;49:765–769. doi: 10.1128/iai.49.3.765-769.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaida S, Miyata T, Yoshizawa Y, Kawabata S, Morita T, Igarashi H, Iwanaga S. Nucleotide sequence of the staphylocoagulase gene: its unique COOH-terminal 8 tandem repeats. J Biochem. 1987;102:1177–1186. doi: 10.1093/oxfordjournals.jbchem.a122156. [DOI] [PubMed] [Google Scholar]
- 22.Kullik I, Giachino P. The alternative sigma factor sigma B in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 23.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin-resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 27.Manna A C, Bayer M G, Cheung A L. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. 1998;180:3828–3836. doi: 10.1128/jb.180.15.3828-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masuda S. An efficient method for the isolation of a mutant with an extremely low producibility of coagulase from a Staphylococcus aureus strain. Microbiol Immunol. 1983;27:801–805. doi: 10.1111/j.1348-0421.1983.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 29.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michele T M, Ko C, Bishai W R. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob Agents Chemother. 1999;43:218–225. doi: 10.1128/aac.43.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min K T, Hilditch C M, Diederich B, Errington J, Yudkin M D. ςF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-ς factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 32.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, Francois P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phonimdaeng P, O’Reilly M, Nowlan P, Bramley A J, Foster T J. The coagulase of Staphylococcus aureus 8325-4. Sequence analysis and virulence of site-specific coagulase-deficient mutants. Mol Microbiol. 1990;4:393–404. doi: 10.1111/j.1365-2958.1990.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 34.Sawai T, Tomono K, Yanagihara K, Yamamoto Y, Kaku M, Hirakata Y, Koga H, Tashiro T, Kohno S. Role of coagulase in a murine model of hematogenous pulmonary infection induced by intravenous injection of Staphylococcus aureus enmeshed in agar beads. Infect Immun. 1997;65:466–471. doi: 10.1128/iai.65.2.466-471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seki K, Ogasawara M, Sakurada J, Murai M, Masuda S. Altered virulence of a pleiotropic Staphylococcus aureus mutant with low producibility of coagulase and other factors in mice. Microbiol Immunol. 1989;33:981–990. doi: 10.1111/j.1348-0421.1989.tb03156.x. [DOI] [PubMed] [Google Scholar]
- 36.Sheagren J N. Staphylococcus aureus: the persistent pathogen. N Engl J Med. 1984;310:1368–1373. doi: 10.1056/NEJM198405243102107. [DOI] [PubMed] [Google Scholar]
- 37.Speller D C E, Johnson A P, James D, Marples R R, Charlett A, George R C. Resistance to methicillin and other antibiotics in isolates of Staphylococcus aureus from blood and cerebrospinal fluid, England and Wales, 1989–95. Lancet. 1997;350:323–325. doi: 10.1016/s0140-6736(97)12148-1. [DOI] [PubMed] [Google Scholar]
- 38.Wiedmann M, Arvik T J, Hurley R J, Boor K J. General stress transcription factor ςB and its role in acid tolerance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson B J. Biology. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 1–38. [Google Scholar]
- 40.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]