Abstract
Background
CC chemokine receptor 7 (CCR7) is a member of G-protein-coupled receptor family and mediates chemotactic migration of immune cells and different cancer cells induced via chemokine (C–C motif) ligand 19 (CCL19) or chemokine (C–C motif) ligand 21 (CCL21). Hence, the identification of blockade antibodies against CCR7 could lead to the development of therapeutics targeting metastatic cancer.
Methods
CCR7 was purified and stabilized in its active conformation, and antibodies specific to purified CCR7 were screened from the synthetic M13 phage library displaying humanized scFvs. The in vitro characterization of selected scFvs identified two scFvs that exhibited CCL19-competitive binding to CCR7. IgG4’s harboring selected scFv sequences were characterized for binding activity in CCR7+ cells, inhibitory activity toward CCR7-dependent cAMP attenuation, and the CCL19 or CCL21-dependent migration of CCR7+ cells.
Results
Antibodies specifically binding to purified CCR7 and CCR7+ cells were isolated and characterized. Two antibodies, IgG4(6RG11) and IgG4(72C7), showed ligand-dependent competitive binding to CCR7 with KD values of 40 nM and 50 nM, respectively. Particularly, IgG4(6RG11) showed antagonistic activity against CCR7, whereas both antibodies significantly blocked the ligand-induced migration and invasion activity of CCR7+ cancer cells.
Conclusions
Two antibody clones were successfully identified from a synthetic scFv-displaying phage library using purified recombinant CCR7 as an antigen. Antibodies specifically bound to the surface of CCR7+ cells and blocked CCR7+ cell migration. Particularly, 6RG11 showed antagonist activity against CCR7-dependent cAMP attenuation.
Keywords: antibody, CC chemokine receptor 7, GPCR, phage display, metastasis
Statement of Significance: Antibodies targeting CCR7 were identified and could serve as therapeutic reagents against cancer metastasis.
INTRODUCTION
The human CC chemokine receptor 7 (CCR7) protein belongs to the rhodopsin-type G protein-coupled receptor (GPCR) family [1]. The receptor mediates chemotactic lymphocyte processes in lymph nodes where antigen-presenting cells (APCs) present antigens to T or B-cells to facilitate adaptive immune responses [2–4]. CCR7 activation with CCL19 or CCL21 induces cAMP attenuation via the Gi-signaling pathway and induces different cell phenomena such as proliferation, apoptosis, and migration [5–9]. Specifically, CCR7 is expressed in different cancers and is involved in metastasis and invasion processes [10, 11]. It is highly expressed in nonsmall cell lung cancer, B-cell chronic lymphocytic leukemia, land lung, pancreas, breast, and hepatocellular cancers [12–18]. CCR7 activation induces PI3/Akt, MAPK/ERK, and JAK/STAT signaling in cancer cells and activates NF-κB in tumor-associated leukocytes, upregulates MMP9, and induces endothelial mesenchymal transition which contributes to metastasis [9, 19–24]. These phenomena suggest CCR7 is a potent therapeutic cancer target, especially during metastasis.
Therapeutic antibodies are advantageous in terms of high specificity, potentially low side effects, and a significantly longer half-life than chemical drugs. Therefore, an antibody that specifically blocks CCR7 signaling processes could function as a therapeutic reagent. However, limited antibodies targeting CCR7 have been developed due to difficulties preparing purified native conformation antigens. A fully human antibody R707 (IgG4), which attenuated acute graft-versus-host disease, was developed using CCR7-overexpressing cells [25]. Another CCR7-specific antibody, CAP100 (IgG1), was developed using an N-terminal CCR7 domain (amino acid sequence 36–52) peptide as antigen. CAP100 bound specifically to CCR7+ cells, inhibited ligand-dependent cAMP attenuation, exerted antibody-dependent cell-mediated cytotoxicity (ADCC), neutralized CCR7-mediated extravasation and lymph node homing [26]. Therefore, antagonistic antibodies targeting CCR7 could be developed to suppress metastasis at lymph nodes and even kill CCR7-expressing cancer cells via ADCC.
Previously, we developed a GPCR expression system in Escherichia coli membranes. Human GPCRs such as endothelin receptor A [27], lysophosphatidic acid receptor 2 [28], prostaglandin E2 receptor 4 [29], and glucagon-like peptide 1 receptor [30] were successfully expressed in active conformations. In this report, we used this system to prepare recombinant CCR7 in its active conformation. After screening a synthetic scFv-display library using purified CCR7, scFv clones specific to CCR7 were identified. IgG4s containing the complementarity determining region (CDR) sequences of selected scFvs were prepared and their binding kinetics were analyzed. We also assessed antagonistic activity to CCR7 and inhibitory activity against CCL19/CCL21-dependent migration of CCR7+ cancer cells.
MATERIALS AND METHODS
Materials
DNA sequence representing the coding region of CCR7 (National Center for Biotechnology Information (NCBI): NM_001838.4) was designed as codon-optimized sequence of E. coli and synthesized by LNCBio (Korea). The coding sequence of CCL19 (NCBI: NM_006274.3) and CCL21 (NCBI: NM_002989.4), with C-terminus Flag and H6-tags, was synthesized from LNCBio. The human CCR7 mammalian expression vector (pcDNA-CCR7) was purchased from Sino Biological (China). Restriction enzymes, DNA polymerases, and the T4 DNA ligase were obtained from New England Biolabs (UK). Amphiphilic poly-γ-glutamate (APG) was obtained from MPRAB (Korea). A synthetic scFv-displaying M13 library was kindly provided by Dr Shim at Ewha University [31]. Recombinant CCL19 and CCL21 were purchased from Peprotech (USA). Ni-NTA resin was purchased from Qiagen (Germany). The HiLoad Superdex 200 16/600 PG column and CNBr-activated sepharose 4B were purchased from GE Healthcare (USA). Horseradish peroxide (HRP)-conjugated antimouse IgG, isopropyl-β-D-thiogalactoside (IPTG), phenylmethylsulfonyl fluoride (PMSF), and sarkosyl were obtained from Sigma (USA). An ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail was purchased from GenDEPOT (USA). Opdivo, which contained the heavy chain constant sequence of IgG4, was kindly provided from Dr T. H. Kang at Department of Biopharmaceutical Chemistry, Kookmin University. All other reagents were of reagent grade. GraphPad Prism 8 software (GraphPad Inc. USA) was used for the analysis of data.
METHODS
Construction of P9-CCR7, CCL19, CCL21, and IgG4 expression vectors
The CCR7 coding region was amplified by polymerase chain reaction (PCR) using forward (5′-TATTTTCAGTCGACGATGGAATTCATGGATCTGGGTAAACCAATGAAG-3′) and reverse (5′-GTGATGGTGAGAAGCTTCGAATTCTGGAGAAAAGGTGGTAGTAGTTTC-3′) primers. The PCR product was digested with Sal I and Hind III, and then ligated at these sites of the pP9 vector [32] to generate p9-CCR7 fusion protein expression plasmid.
DNA fragments representing the coding regions of CCL19 or CCL21 with Flag and H6-tags were amplified by PCR using forward (5′-TCCAGCCTCCGGACTCTAGAGCCGCCACCATGGCCCTGCT-3′) and reverse (5′-CCGGCCTTGCCGGCCTCGAGTCATTACTTGTCGTCATCGT-3′) primers for CCL19, and forward (5′-TCCAGCCTC CGGACTCTAGAGCCGCCACCATGGCTCAGTC-3′) and reverse (5′-ATCCGG CCTTGCCGGCCTCGAGTCATTACTTGTCGTCATC-3′) primers for CCL21. PCR products were ligated at Xba I and Xho I sites into the pcDNA 3.4 vector (NEB, UK) to produce pCCL19-H6-Flag or pCCL21-H6-Flag vectors.
The sequence coding variable regions, VL and VH, were amplified via PCR. Signal sequences for each light chain or heavy chain were introduced via overlap extension PCR with light chain signal sequence (5′-GCCGCCACCATGGCCGGCTTCCCTCTCCTCCTCACCCTCCTCACTCACTGTGCAGGATCCTGGGCCCAGTCTGTGCTGACTCAGCCACCC-3′) and heavy chain signal sequence (5′-GCCGCCACCATGGAGTTTGGGCTGAGCTGGCTTTTTCTTGTGGCTATTTTAAAAGGTGTCCAGTGTGAGGTGCAGCTGTTGGAGTCTGGG-3′). The VL fragment was ligated at SgrAI and KpnI sites, and the VH fragment was ligated at NheI and AgeI sites into the pTRIOZ-hIgG4 (S228P) vector (Invivogen, USA).
P9-CCR7 purification
E. coli BL21(DE3)-RIPL cells harboring the pP9-CCR7 vector were cultured in a bioreactor (Marado-PDA, CNS, Korea) as previously described [29]. The expression of P9-CCR7 was induced by adding final 1 mM IPTG to the culture when the OD600 of culture reached 60, and the culture was further incubated for 21 h at 18°C. Cells were harvested via centrifugation and stored at −80°C. Approximately 20 g cell paste was resuspended in 100 mL buffer A (25 mM HEPES, pH 7.0) plus 1 mM PMSF and an EDTA-free protease inhibitor cocktail tablet and lysed via a microfluidizer M110P device (Microfluidics, USA). Cell debris was removed via centrifugation at 12 000 × g for 30 min, and the supernatant was further centrifuged at 100 000 × g for 1 h to pellet the membrane fraction. This fraction was solubilized in buffer B (25 mM HEPES pH 7.0 plus 1% sarkosyl) via gentle agitation for 2 h at 4°C, and precipitated materials were removed via centrifugation at 15 000 × g for 30 min. The cleared solubilized membrane fraction was loaded onto a Ni-NTA column equilibrated with buffer B, and bound P9-CCR7 was eluted in 20 mM imidazole in buffer B. The eluted fraction was mixed with 1% APG for 16 h at 4°C, after which it was purified via size exclusion chromatography (HiLoad Superdex 200 16/600 PG), previously equilibrated in buffer A plus 10% glycerol. Aliquots were stored at −80°C.
CCL19-H6-flag and CCL21-H6-flag purification
Via the ExpiFectamine™ 293 transfection kit (Thermo Fisher Scientific, USA), pCCL19-H6-Flag and pCCL21-H6-Flag vectors (50 μg) were transfected into 50 mL Expi293 cells (3 × 106 cells/mL) (American Type Culture Collection, USA) in Expi293™ expression media. Transfected cells were grown in Expi293™ expression media containing DNA complexes and enhancers for 72 h until the percentage of live cells had reduced to 60%. After cells were centrifuged at 2 000 × g for 20 min at 4°C, supernatants were loaded onto a Ni-NTA column equilibrated with Dulbecco's phosphate buffered saline (DPBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). Bound proteins were eluted in a 200 mM imidazole elution buffer. Via a desalting PD-10 column (GE Healthcare, USA), sample buffer was replaced with DPBS buffer plus 10% glycerol. Protein aliquots were stored at −80°C.
Affinity determination of purified P9-CCR7 with CCL19-H6-flag and CCL21-H6-flag
Purified CCL19-H6-Flag and CCL21-H6-Flag (1 μg) proteins were immobilized in a maxi binding 96-well plate (SPL, USA). Wells were blocked with 5% skimmed milk in DPBS, and 0–1 000 nM of purified P9-CCR7 was added. The plate was then successively treated with an anti-P9 antibody, an HRP-conjugated antimouse IgG, ultra TMB (Thermo Fisher Scientific, USA), and 1 N HCl. P9-CCR7 bound to ligands was measured via a Synergy H1 microplate reader (BioTek, USA).
Biopanning CCR7-specific scFv clones
Purified P9-CCR7 was immobilized to a bead surface using a CNBr-activated sepharose resin (GE Healthcare, USA) as per the manufacturer’s instructions. Briefly, resin powder (40 μL) was sequentially rinsed in 1 mM HCl (pH 3.0) and coupling buffer (100 mM sodium bicarbonate, pH 8.2) and incubated overnight at 4°C with purified P9-CCR7 (0.2 mg). The P9-CCR7-immobilized resin was further incubated with termination buffer (100 mM Tris-Cl, pH 8.0) for 1 h at 20°C, and then washed in DPBS via a centrifuge tube filter (Corning, USA). The first round of biopanning was initiated by incubating 20 μL P9-CCR7 immobilized beads with 200 μL of the synthetic scFv-displaying M13 phage library (~1 × 1012 CFU/mL) for 14 h at 4°C. After washing unbound phage three times in DPBS, bound phage was eluted in 200 μL elution buffer (100 mM glycine pH 2.2 plus 1% bovine serum albumin (BSA) by incubating for 10 min at 20°C, and then mixing with 40 μL neutralization buffer (1 M Tris-Cl, pH 8.0). Next, eluted phages were used to infect ER2738 cells, and subsequent rescue processes using an M13K07 helper phage were performed as previously described [31]. Successive biopanning rounds using eluted phages were repeatedly performed using these procedures, but with increased washing steps. At steps 4–7 of the biopanning process, P9-GLP1R (0.1 mg/mL) was added along with the phage solution to P9-CCR7-immobilized beads to prevent P9-specific phage enrichment.
Screening scFv clones specific to CCR7
After biopanning, eluted phages were used to infect mid-log phase ER2738 cells and infected cells were spread onto LB agar plates containing carbenicillin (50 μg/mL). After incubation, single colonies were inoculated into 750 μL SB medium plus carbenicillin (50 μg/mL) in deep 96-well plates and cultured at 37°C for 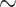 4 h until turbid. Then, 1 mM IPTG was added and cultures incubated overnight at 30°C with shaking at 180 rpm. The next day, plates were centrifuged at 3 500 × g for 20 min to remove supernatants and placed on ice to cool. To recover periplasmic fractions containing scFvs, cell pellets were resuspended in 160 μL 1× TES buffer (50 mM Tris-Cl pH 8.0, 1 mM EDTA, and 20% sucrose) and a further 240 μL 0.2× TES buffer was added. After more than 30-min incubation on ice, plates were centrifuged and the periplasmic fraction was obtained. Then, 100 μL periplasmic extracts were added to a target GPCR-immobilized 96-well maxi binding plate and incubated for 1 h at room temperature. HRP-conjugated anti-HA antibody (1:3 000) was used for the colorimetric detection of bound clones using tetramethylbenzidine (TMB) substrate.
4 h until turbid. Then, 1 mM IPTG was added and cultures incubated overnight at 30°C with shaking at 180 rpm. The next day, plates were centrifuged at 3 500 × g for 20 min to remove supernatants and placed on ice to cool. To recover periplasmic fractions containing scFvs, cell pellets were resuspended in 160 μL 1× TES buffer (50 mM Tris-Cl pH 8.0, 1 mM EDTA, and 20% sucrose) and a further 240 μL 0.2× TES buffer was added. After more than 30-min incubation on ice, plates were centrifuged and the periplasmic fraction was obtained. Then, 100 μL periplasmic extracts were added to a target GPCR-immobilized 96-well maxi binding plate and incubated for 1 h at room temperature. HRP-conjugated anti-HA antibody (1:3 000) was used for the colorimetric detection of bound clones using tetramethylbenzidine (TMB) substrate.
scFv purification
A single colony of E. coli TOP10F’ harboring a scFv clones in the pComb3X vector was grown overnight in 10 mL of SB medium plus carbenicillin (50 μg/mL). This starter culture was then transferred to 500 mL SB medium plus carbenicillin (50 μg/mL) and incubated at 37°C until the OD600 reached 0.6–0.8. After 1 mM IPTG was added, cultures were incubated overnight at 30°C. After centrifugation, the cell pellet was resuspended in 30 mL cold 1× TES buffer and 45 mL of cold 0.2× TES containing 1 mg/mL of lysozyme and 10 mM of MgCl2 was subsequently added. After incubation on ice for 30 min, the suspension was centrifuged at 10 000 × g for 20 min and the supernatant containing periplasmic fraction was obtained. The periplasmic extract was incubated with 1 mL Ni-NTA resin (Qiagen, Germany) preequilibrated in PBS buffer at 4°C for 1 h. The reacted resin was packed into a 5 mL disposable column (Thermo Scientific, USA) and washed with 10 mL wash buffer (PBS with 5 mM imidazole). ScFvs were eluted in 5 mL elution buffer (PBS with 200 mM imidazole). Eluates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Preparation of IgG4’s specific to CCR7
Variable region sequences of scFv were cloned into the pTRIOZ-hIgG4 (S228P) vector (Invivogen, USA) containing the human lambda light chain sequences, which represented IgG4 type immunoglobulin. The constructed IgG4 expression vectors were transfected as described into 50 mL Expi293 cells (3 × 106 cells/mL), cultured until viability reached < 60%, and then harvested at 2 000 × g for 20 min at 4°C. Supernatants were filtered via a syringe filter (PES, Pore 0.22 μm) (Sartorius, USA) and loaded onto CaptivA® resin (Repligen) equilibrated in DPBS. After washing in DPBS, target proteins were eluted in 100 mM glycine-HCl pH 2.7 and immediately neutralized in 1 M Tris–HCl pH 8.0. Purified proteins were diluted in DPBS to prevent precipitation and stored in 10% glycerol at −80°C.
Affinity determination of scFvs or IgG4’s to P9-CCR7
Binding affinities between CCR7 and scFvs or IgG4’s were measured via enzyme-linked immunosorbent assay (ELISA). Purified P9-CCR7 (500 ng) was immobilized to wells in a 96-well maxi binding plate and blocked using 3% BSA. The plate was treated with different scFv or IgG4 concentrations (0–1 000 nM) and incubated for 1 h at room temperature. Then, scFvs or IgG4’s bound to P9-CCR7 were detected using HRP-conjugated anti-HA antibody and HRP-conjugated anti-human Fc antibody (Thermo Fisher Scientific, USA), respectively. To identify scFv clones, which bound to the CCR7 extracellular region and competed with CCL19 for CCR7 binding, a competitive ELISA was performed. Approximately 100 nM CCL19-H6-Flag was added to a 96-well plate coated with immobilized P9-CCR7 in the presence of different purified scFv concentrations. Bound CCL19-H6-Flag was then detected using a monoclonal anti-Flag®M2 antibody (Thermo Fisher Scientific, USA) and HRP-conjugated antimouse Fc antibody (1:5 000 in DPBST) as described.
Cell culture, transfections, and flow cytometry
HEK293 cells transiently expressing CCR7 (HEK293/CCR7) were prepared by transfecting HEK293 cells with the pcDNA-CCR7 plasmid using Lipofectamine®3000 (Thermo Fisher Scientific, USA). HEK293 and HEK293/CCR7 cells were maintained in DMEM medium plus 10% fetal bovine serum (FBS) and 100 U/mL penicillin/100 μg/mL streptomycin at 37°C in 5% CO2.
For flow cytometry, cells were detached from plates using 1 mL TrypLE™ Express Enzyme (Thermo Fisher Scientific, USA). A single-cell suspension was obtained and then centrifuged at 300 × g for 3 min. After washing pellets twice in DPBS, cells were resuspended in 5 mL 3% BSA in DPBS to a final density of 1 × 106 cells/mL. They were then incubated with IgG4’s while rotating at 4°C for 1 h. After this, cells were washed twice and incubated with Alexa 488-conjugated antihuman Fc antibody (1:1 000) (Invitrogen, USA) at 4°C for 1 h. After washing three times in DPBS, fluorescence signals of cell-bound IgG were measured via a Guava™ easyCyte Flow Cytometer (Luminex, USA).
Cyclic AMP assay
HEK293/CCR7 or MDA-MB-231 cells, transfected with pcDNA-CCR7 (MDA-MB-231/CCR7), were maintained in DMEM plus 10% FBS and antibiotics. About 1 × 104 cells/well were seeded into 96-well culture plates and incubated overnight at 37°C in 5% CO2. The next day, the medium was replaced with cAMP stimulation buffer (DMEM, 0.5 mM 3-isobutyl-1-methylxanthine), and cells were incubated for a further 20 min. Different CCL19 or CCL21 (Peprotech, USA) concentrations in 2× forskolin buffer (cAMP stimulation buffer plus 40 μM forskolin), with or without IgG, were added to cells and incubated for 20 min. Ligand effects were analyzed using the cAMP-Glo™ assay kit (Promega, USA) and detected using a Synergy H1 microplate reader (BioTek).
Migration and invasion assays
MDA-MB-231 cells overexpressing CCR7 (MDA-MB-231/CCR7) were prepared by transfecting MDA-MB-231 cells with the pcDNA-CCR7 plasmid. As described, cells were grown in a medium for 20 h, the medium exchanged for a starvation medium (DMEM, 0.5% FBS, plus antibiotics), and cells were further incubated for 6 h at 37°C in 5% CO2. A Matrigel matrix was prepared by dissolving Matrigel (growth factor reduced, Corning, USA) in DMEM (200 μg/mL). This was added to permeable support (8.0 μm PET Membrane, Falcon, USA) in a 24-well tissue culture plate (Costar, USA) and incubated at 37°C for > 2 h. After solidification, DMEM in the upper chamber was removed, and 200 μL of starved cells (2 × 105) in different IgG concentrations were seeded into this chamber. The lower chamber was filled with an 800 μL starvation medium plus 30 nM CCL19 or CCL21. After incubation for 40 h, the upper chamber was wiped with cotton swabs, and migrated cells in the lower chamber were stained via a Diff Quick staining kit and enumerated via inverted microscopy. The purple-stained nuclei of migrated cells were counted using Image J software and analyzed in GraphPad Prism 8 software.
RESULTS
Preparation of P9-CCR7
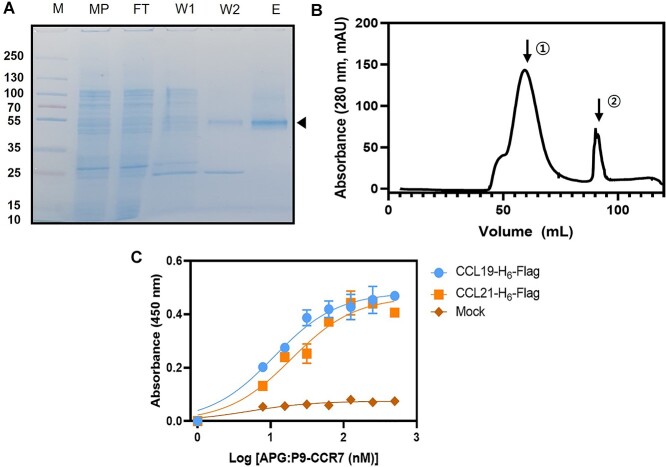
The P9 expression system in E. coli was successfully used to express human recombinant CCR7 (Supplementary Fig. 1A). The P9 sequence with a single transmembrane spanning region, which is a major envelope protein of phi6 phage [27], induced CCR7 expression in the periplasmic membrane of E. coli. Per 1 g wet E. coli cells, 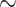 0.8 mg recombinant P9-CCR7 was recovered (Supplementary Fig. 1B). P9-CCR7 was solubilized in 1% sarkosyl and further purified via NTA chromatography (Fig. 1A). Approximately 8 mg P9-CCR7 was purified from 20 g of wet cells. Purified P9-CCR7 was stabilized in APG, an amphiphilic polymer that stabilizes different GPCRs in their native conformation [27–30]. P9-CCR7 was observed as a mono-dispersed peak (molecular weight = 350 kDa) via gel-filtration chromatography (Fig. 1B, Peak 1), accompanied by free APG whose average size was 30 kDa (Fig. 1B, Peak 2). The P9-CCR7 size suggested one to two molecules had complexed with seven to eight APG molecules.
0.8 mg recombinant P9-CCR7 was recovered (Supplementary Fig. 1B). P9-CCR7 was solubilized in 1% sarkosyl and further purified via NTA chromatography (Fig. 1A). Approximately 8 mg P9-CCR7 was purified from 20 g of wet cells. Purified P9-CCR7 was stabilized in APG, an amphiphilic polymer that stabilizes different GPCRs in their native conformation [27–30]. P9-CCR7 was observed as a mono-dispersed peak (molecular weight = 350 kDa) via gel-filtration chromatography (Fig. 1B, Peak 1), accompanied by free APG whose average size was 30 kDa (Fig. 1B, Peak 2). The P9-CCR7 size suggested one to two molecules had complexed with seven to eight APG molecules.
Figure 1.
Purification of functional P9-CCR7. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of P9-CCR7 after Ni-NTA chromatography. M = size marker, MP = soluble membrane fraction after solubilization in 1% sarkosyl, FT = flow-through from Ni-NTA chromatography, W1, W2 = wash fraction with each 0, 5 mM imidazole, respectively, E = elution fraction with each 20 mM imidazole from Ni-NTA chromatography. Purified P9-CCR7 is indicated by triangles. (B) Size exclusion chromatography of purified P9-CCR7 stabilized with APG. P9-CCR7 was eluted as a monodisperse peak (Peak 1) followed by free APG (Peak 2). (C) A binding curve of P9-CCR7 to CCL19-H6-Flag (blue dots) or CCL21-H6-Flag (orange squares). Different concentration of P9-CCR7 was added to plates containing 10 μg/mL CCL19-H6-Flag (blue dots) or CCL21-H6-Flag (orange squares) and P9-CCR7 bound to wells was measured via an anti-P9 antibody and HRP-conjugated antimouse antibody.
To confirm a properly folded conformation of purified P9-CCR7, its binding affinity with CCL19 or CCL21 was examined. Dissociation constants (KDs) for CCL19-H6-Flag or CCL21-H6-Flag to P9-CCR7 were 19 nM or 11 nM, respectively via ELISA (Fig. 1C). These values were compared favorably with EC50 values (8.8 nM or 7.8 nM, respectively) previously measured by a bioluminescence resonance energy transfer assay in CCR7+ cells [33]. This result indicated that the CCR7 region of P9-CCR7 exhibited a ligand binding activity as an active form of CCR7. Hence, the purified P9-CCR7 was subsequently used as an antigen for biopanning studies.
Biopanning phage display libraries to isolate CCR7-specific scFvs
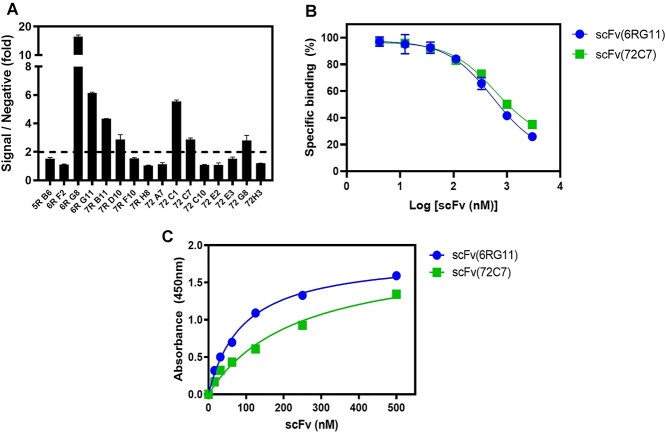
We performed biopanning of a phage library displaying synthetic scFvs [31, 34] using immobilized P9-CCR7 over seven rounds. After 384 clones were screened from eluted phages, 50 primary hits were isolated and 16 unique clones were identified from sequence analysis (Table 1). These scFvs were tested to see if they were specifically bound to the CCR7 domain in P9-CCR7 rather than the P9 sequence alone or APG using an unrelated P9-GLP1R stabilized by APG [30]. Among clones, only seven showed specific binding to CCR7 (Fig. 2A) and the rest appeared specific to the P9 region or APG. These scFvs were further purified and tested to see if they competed with CCL19 for binding to CCR7. Only two clones (6RG11 and 72C7) inhibited CCL19-H6-Flag binding to CCR7 (Fig. 2B), suggesting 6RG11 and 72C7 clones competed with CCL19 for the binding to CCR7. These scFv clones showed concentration-dependent binding to CCR7, with apparent KD values of 90 nM (6RG11) and 230 nM (72C7) (Fig. 2C). On the other hand, the remaining five clones could bind to P9-CCR7 with KD values of 80–210 nM (Supplementary Fig. 2A), but they hardly inhibit the binding of CCL19 to P9-CCR7 (Supplementary Fig. 2B). These clones may have interacted with an intracellular or extracellular region remote to the CCL19-binding site of CCR7, and they were not further investigated.
Table 1.
Biopanning statistics
| Stage | Number of clones |
|---|---|
| Screened clonesa | 384 |
| Primary hitsb | 50 |
| Unique clonesc | 16 |
| CCR7-specific clonesd | 7 |
| Ligand competitorse | 2 |
aTotal number of clones tested by ELISA after the seventh round of biopanning.
bNumber of scFv clones which bind to P9-CCR7 from initial screening 384 clones.
cNumber of scFv clones with unique sequences among primary hits.
dNumber of scFv clones which showed > 2-fold higher ratio of the absorption values from scFv bound to P9-CCR7 to P9-GLP1R.
eNumber of scFv clones inhibiting CCL19 binding to CCR7.
Figure 2.
scFv clone specificity toward CCR7. (A) Specificity of the purified scFvs toward CCR7. Sixteen scFv clones with unique sequences were purified to homogeneity and the purified scFvs (500 nM) were incubated in 96-well plates containing immobilized P9-CCR7 or P9-GLP1R, with bound scFvs detected using an HRP-conjugated anti-HA antibody. The ratio of the absorption value P9-CCR7 to P9-GLP1R values was plotted. (B) Inhibition of the interactions between CCL19 and CCR7 by two scFvs (6RG11 and 72C7). CCL19-H6-Flag (100 nM) was incubated with different scFv concentrations in a 96-well plate containing immobilized P9-CCR7, and bound CCL19-H6-Flag was measured using a monoclonal anti-Flag®M2 antibody and HRP-conjugated antimouse Fc antibody. (C) Concentration-dependent binding of scFv(6RG11) and scFv(72C7) to CCR7. Different scFv(6RG11) (blue dots) and scFv(72C7) (green squares) concentrations were added to immobilized P9-CCR7, and bound scFvs were detected using an HRP-conjugated anti-HA antibody.
Therefore, CCR7-specific scFvs were generated from the synthetic scFv-display phage library using purified P9-CCR7 as an antigen. 6RG11 and 72C7 clones were converted to IgGs and further characterized.
Preparation and characterization of CCR7-specific IgG4
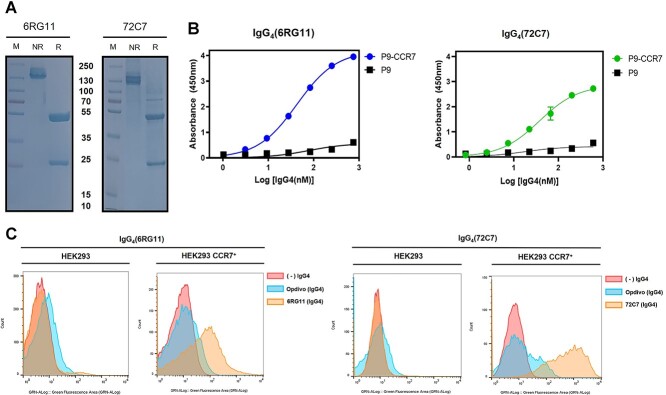
Expression vectors of human IgG4 containing the heavy and light chain variable regions of 6RG11 and 72C7 were constructed (Supplementary Fig. 3) with both IgG4-proteins expressed in Expi293 cells and purified (Fig. 3A). When the binding kinetics of purified IgG4(6RG11) and IgG4(72C7) was measured via purified P9-CCR7, both showed specific binding to CCR7 of P9-CCR7, with KD values of 40 nM and 37 nM, respectively (Fig. 3B). The affinity of purified IgG4’s increased 2.2-fold (6RG11) or 6.5-fold (72C7) when compared with the KD values of the scFv form. The enhanced affinity of the IgG4 form, when compared with the scFv form, may have resulted from the avidity effects of IgG4 which has two binding sites in a single molecule. Using flow cytometry, the specific binding of both IgG4s to CCR7 was further examined in HEK293 cells expressing CCR7. When HEK293 cells were treated with IgG4(6RG11), IgG4(72C7), or an isotype control antibody, significant binding of IgG4(6RG11) or IgG4(72C7) was not observed (Fig. 3C, left panel). Also, IgG4(6RG11) showed no significant binding with non-transfected breast cancer cells (MDA-MB-231, MCF7) in flow cytometry (Supplementary Fig. 4A), indicating HEK293, MDA-MB-231, or MCF7 cells failed to express substantial level of CCR7. However, HEK293/CCR7 cells showed strong binding signals with IgG4(6RG11) or IgG4(72C7) (Fig. 3C, right panel). This result also indicated that both IgG4s bound to the extracellular region of CCR7 expressed in the cell membrane.
Figure 3.
Specific interactions between CCR7 and IgG4(6RG11) or IgG4(72C7). (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of purified IgG4(6RG11) and IgG4(72C7). M = Size marker, NR = 6 μg of IgG4 in non-reducing conditions, R = 6 μg of IgG4 in reducing conditions plus 1 mM DTT. (B) Concentration-dependent binding of IgG4 (6RG11) and IgG4(72C7). Different purified IgG4 concentrations were added to a 96-well plate containing immobilized P9 or P9-CCR7, and bound IgG4(6RG11) (blue dots) or IgG4(72C7) (green dots) to P9-CCR7 or P9 (black squares) was measured using an HRP-conjugated antihuman Fc. (C) Cytometric analysis of IgG4(6RG11) and IgG4(72C7) using CCR7+ cells. HEK293 or CCR7-transformed HEK293 cells (HEK293/CCR7) were incubated with mock (red), 20 nM IgG4(Opdivo)(blue) as an isotype control, or 20 nM IgG4(6RG11) or IgG4(72C7) (orange), and then treated with an Alexa488-conjugated antihuman Fc antibody.
CONCLUSIONS
Blocking IgG4(6RG11) and IgG4(72C7) activity against the CCR7-signaling axis
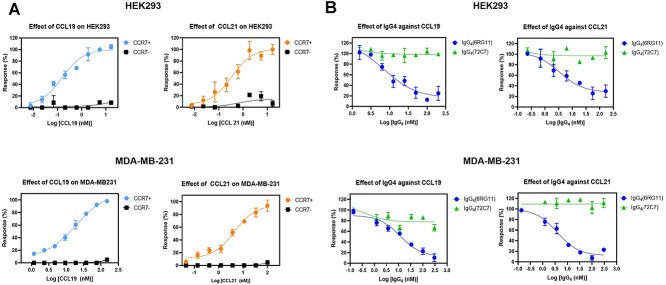
The specific binding of IgG4s to CCR7 in mammalian cells and scFv ligand-competition for CCR7 binding suggested IgG4s may have had antagonistic activity against CCR7 and prevented cell migration mediated via CCL19-CCR7 signaling. To examine if IgG4’s exhibited antagonistic activity against CCR7, cAMP levels induced via CCL19 activation were examined in CCR7+ cells. When HEK293/CCR7 or MDA-MB-231/CCR7 cells were treated with different CCL19 or CCL21 concentrations, the attenuation of forskolin-induced cAMP level was observed (Fig. 4A, color circles). However, non-transformed HEK293 or MDA-MB-231 cells failed to show cAMP attenuation by the treatment of CCL19 or CCL21 (Fig. 4A, filled squares). These results also indicated the undetectable, if any, level of CCR7 in HEK293 or MDA-MB-231 cells. When cells were treated with fixed CCL19 or CCL21 concentrations (20 nM) and different IgG4(6RG11) concentrations in the presence of forskolin, significant inhibition of cAMP attenuation induced by CCL19 or CCL21 was observed, with EC50 values of 3–12 nM (Fig 4B, blue circles). However, IgG4(72C7) failed to affect the ligand-dependent attenuation of cAMP (Fig. 4B, green triangles). Therefore, IgG4(6RG11) displayed strong antagonistic activity against CCR7 signaling, especially Gi-dependent cAMP attenuation.
Figure 4.
The antagonist activity of IgG4(6RG11) and IgG4(72C7). (A) CCL19 or CCL21-dependent cAMP attenuation in HEK293 and MDA-MB-231 cells overexpressing CCR7 in the presence of forskolin. Different concentrations of CCL19 (left panel) or CCL21 (right panel) were added to HEK293 (upper panel, black square) or HEK293/CCR7 cells (upper panel, blue or orange dots) and cAMP levels were measured. Ligand effects in MDA-MB-231 (lower panel, black squares) or MDA-MB-231/CCR7 cells (lower panel, blue or orange dots) were also measured. (B) The effects of IgG4(6RG11) and IgG4(72C7) on ligand-dependent cAMP attenuation in HEK293 and MDA-MB-231 cells overexpressing CCR7 in the presence of forskolin. Different IgG4(6RG11) (blue circles) and IgG4(72C7) (green triangles) concentrations were added to HEK293/CCR7 (upper panel) or MDA-MB-231/CCR7 cells (lower panel) in the presence of 20 nM CCL19 (left panel) or CCL21 (right panel) and cAMP levels measured.
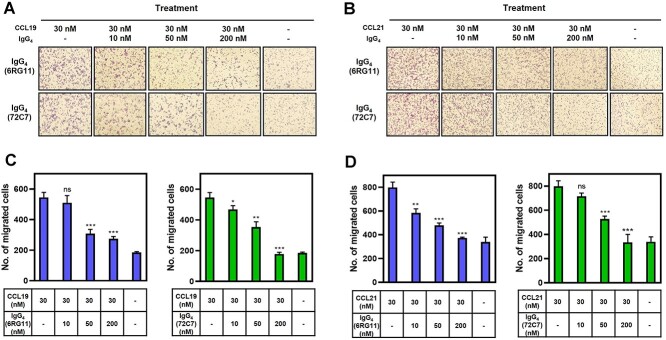
CCR7 is a chemotactic receptor and initiates cell migration [9, 10]. The effects of IgG4(6RG11) and IgG4(72C7) on CCL19- or CCL21-induced migration and invasion in MDA-MB-231/CCR7 cells were examined. Cells migrating across permeable support chambers (Matrigel-coated) were enumerated after CCL19 or CCL21 treatment in the presence of different IgG4 concentrations (Figs 5A and B, respectively). The number of migrated cells in the presence of IgG was counted and presented in Figs 5C and D. Both IgG4(6RG11) and IgG4(72C7) prevented the CCL19- or CCL21-dependent migration of CCR7+ cells. IgG4(6RG11) was slightly more potent than IgG4(72C7) at 50 nM (Fig. 5C). Notably, IgG4(72C7) displayed inhibitory activity against CCR7-dependent invasion while it failed to show antagonistic activity in terms of cAMP attenuation.
Figure 5.
Inhibition of IgG4(6RG11) and IgG4(72C7) on CCL19 or CCL21-dependent migration of MDA-MB-231/CCR7 cells. (A) Image of migrated MDA-MB-231/CCR7 cells after treatment of 30 nM CCL19 in the presence of 0, 10, 50, or 200 nM IgG4(6RG11) (upper panel) or IgG4(72C7) (lower panel). (B) The number of migrated cells in lower chambers was plotted against CCL19 and IgG4(6RG11) or IgG4(72C7) concentrations. (C) Image of migrated cells after treatment with 30 nM CCL21 in the presence of 0, 10, 50, or 200 nM IgG4(6RG11) (upper panel) or IgG4(72C7) (lower panel). (D) The number of migrated cells in lower chambers was plotted against CCL21 and IgG4(6RG11) or IgG4(72C7) concentrations (P > 0.05, no significance; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Ligand binding induces conformational changes in GPCRs, including CCR7, and generates several molecular phenomena such as G protein displacement, β-arrestin recruitment, and homo- or heterodimerization [35, 36]. In addition, CCR7, CCL19, and CCL21 ligands exert different effects on G protein activation, β-arrestin recruitment, internalization, migration, and signaling [37]. The IgG4 identified in this study has a blocking activity against ligand binding to CCR7. Although both IgG4(6RG11) and IgG4(72C7) competed with CCL19 as shown in competition assay with scFvs (Fig. 2B), only IgG4(6RG11) fully functioned as a neutral antagonistic antibody that prevented ligand binding, G protein signaling, and CCL19 or CCL21-induced cell migration and invasion. However, IgG4(72C7) failed to show any antagonistic activity toward the attenuation of forskolin-induced cAMP level, whereas it prevented CCL19 or CCL21-induced cell migration and invasion. These properties of IgG4 (72C7) suggested that it might interfere the signaling pathway of CCR7 rather than directly antagonize CCR7 [38, 39]. The detailed mechanism of these antibodies should be elucidated by further study such as epitope mapping and dissection of signaling cascade induced by these antibodies.
The identification of CCR7-specific antibodies via purified CCR7 as an antigen
CCR7 is associated with cancer and leukemia metastasis and is proposed as a therapeutic cancer target [11]. Purified full-length human CCR7 from E. coli was prepared using a P9 expression system and stabilized via APG. Several scFv clones were identified from the synthetic scFv-display phage library using purified P9-CCR7, and two clones were identified as competitors against the ligand of CCR7. The IgG forms [IgG4(6RG11) and IgG4(72C7)] of identified scFv clones displayed KD values of 30–40 nM against CCR7 and prevented the ligand-induced migration of MDA-MB-231 cells overexpressing CCR7. The expression of CCR7 mediating metastasis to lymph nodes is upregulated in breast cancer cells, but expression in breast cancer cell lines is poorly defined. Especially, CCR7 expression in MDA-MB-231 and MCF7 was detected as low copies via flow cytometry without induction [40–42]. In this study, the endogenous expression of CCR7 in MDA-MB-231 and MCF7 cells was not detected by flow cytometry and cAMP assay (Fig. 4A and Supplementary Fig. 3B). Hence, MDA-MB-231 cells overexpressing CCR7 were used for the evaluation of discovered antibodies in this study.
Previously, Cuesta-Mateos et al. reported CAP-100, an anti-CCR7 IgG1 of humanized sequence, which targeted the N-terminal domain of CCR7 (amino acids 36–52) [26]. This antibody displayed a KD of 0.8 nM against synthetic peptide antigens and an EC50 value of 10 nM against ligand-dependent cAMP attenuation [26], comparable with the EC50 value of IgG4(6RG11). Compared with the peptide antigen representing the N-terminal domain of CCR7, we used a full-length CCR7 as an antigen for identification of 6RG11 and 72C7 clones in this study. The identified clones specifically bound to CCR7 expressed in cell membrane (Figs 2 and 3) and showed antagonistic activity against CCR7 (Fig. 4). The P9-fusion system for high-level expression of full-length GPCR in E. coli and stabilization process with amphiphilic polymer could be applied for the development of therapeutic antibodies targeting GPCRs such as CCR7.
CCR7 is associated with lymphocyte or APC homing to lymph nodes [3, 4, 43–46] where adaptive immune responses typically occur. Hence, the potential toxicity of an anti-CCR7 antibody, induced by blocking CCR7 homing signals, must be carefully investigated and evaluated. After confirming antibody safety, suitable antibody applications targeting CCR7 may be used to block the migration of malignant cells overexpressing CCR7, e.g., during metastatic cancer. The antibodies identified in this study require further development such as epitope mapping, mode of action, toxicity and modification or other applications as potential therapeutics against cancer or CCR7-related disease.
FUNDING
This work was supported by the Mid-Career Research Program (2020R1A2C1101174) through the Ministry of Science and ICT, the Republic of Korea.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
DATA AND MATERIALS AVAILABILITY
All data are available upon request from the corresponding authors.
ETHICS AND CONSENT STATEMENT
Not applicable.
ANIMAL RESEARCH STATEMENT
No animal was used in this study.
Supplementary Material
Contributor Information
Moon-Sung Jang, Department of Biopharmaceutical Chemistry, Kookmin University, Seoul 02707, Republic of Korea; Antibody Research Institute, Kookmin University, Seoul 02707, Republic of Korea.
Nurain Syahirah Binti Ismail, Department of Biopharmaceutical Chemistry, Kookmin University, Seoul 02707, Republic of Korea; Antibody Research Institute, Kookmin University, Seoul 02707, Republic of Korea.
Yeon Gyu Yu, Department of Biopharmaceutical Chemistry, Kookmin University, Seoul 02707, Republic of Korea; Antibody Research Institute, Kookmin University, Seoul 02707, Republic of Korea.
References
- 1. Raju, R, Gadakh, S, Gopal, Pet al. Differential ligand-signaling network of CCL19/CCL21-CCR7 system. Database (Oxford) 2015; 2015: bav106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bryce, SA, Wilson, RAM, Tiplady, EMet al. ACKR4 on stromal cells scavenges CCL19 to enable CCR7-dependent trafficking of APCs from inflamed skin to lymph nodes. J Immunol 2016; 196: 3341–53. [DOI] [PubMed] [Google Scholar]
- 3. Choi, H, Song, H, Jung, YW. The roles of CCR7 for the homing of memory CD8+ T cells into their survival niches. Immune Netw 2020; 20: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Förster, R, Davalos-Misslitz, AC, Rot, A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol 2008; 8: 362–71. [DOI] [PubMed] [Google Scholar]
- 5. Corbisier, J, Galès, C, Huszagh, Aet al. Biased signaling at chemokine receptors. J Biol Chem 2015; 290: 9542–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heng, BC, Aubel, D, Fussenegger, M. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol Adv 2013; 31: 1676–94. [DOI] [PubMed] [Google Scholar]
- 7. Lacalle, RA, et al. Chemokine receptor signaling and the hallmarks of cancer. Int Rev Cell Mol Biol 2017; 331: 181–244. [DOI] [PubMed] [Google Scholar]
- 8. Okada, T, Cyster, JG. CC chemokine receptor 7 contributes to Gi-dependent T cell motility in the lymph node. J Immunol 2007; 178: 2973–8. [DOI] [PubMed] [Google Scholar]
- 9. Rizeq, B, Malki, MI. The role of CCL21/CCR7 chemokine axis in breast cancer progression. Cancers (Basel) 2020; 12: 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mishan, MA, Ahmadiankia, N, Bahrami, AR. CXCR4 and CCR7: two eligible targets in targeted cancer therapy. Cell Biol Int 2016; 40: 955–67. [DOI] [PubMed] [Google Scholar]
- 11. Salem, A, Alotaibi, M, Mroueh, Ret al. CCR7 as a therapeutic target in cancer. Biochim Biophys Acta Rev Cancer 2021; 1875: 188499. [DOI] [PubMed] [Google Scholar]
- 12. Maekawa, S, Iwasaki, A, Shirakusa, Tet al. Association between the expression of chemokine receptors CCR7 and CXCR3, and lymph node metastatic potential in lung adenocarcinoma. Oncol Rep 2008; 19: 1461–8. [PubMed] [Google Scholar]
- 13. Mashino, K, Sadanaga, N, Yamaguchi, Het al. Expression of chemokine receptor CCR7 is associated with lymph node metastasis of gastric carcinoma. Cancer Res 2002; 62: 2937–41. [PubMed] [Google Scholar]
- 14. Nakata, B, Fukunaga, S, Noda, Eet al. Chemokine receptor CCR7 expression correlates with lymph node metastasis in pancreatic cancer. Oncology 2008; 74: 69–75. [DOI] [PubMed] [Google Scholar]
- 15. Schimanski, CC, Bahre, R, Gockel, Iet al. Chemokine receptor CCR7 enhances intrahepatic and lymphatic dissemination of human hepatocellular cancer. Oncol Rep 2006; 16: 109–13. [PubMed] [Google Scholar]
- 16. Takanami, I. Overexpression of CCR7 mRNA in nonsmall cell lung cancer: correlation with lymph node metastasis. Int J Cancer 2003; 105: 186–9. [DOI] [PubMed] [Google Scholar]
- 17. Wiley, HE, Gonzalez, EB, Maki, Wet al. Expression of CC chemokine receptor-7 and regional lymph node metastasis of B16 murine melanoma. J Natl Cancer Inst 2001; 93: 1638–43. [DOI] [PubMed] [Google Scholar]
- 18. Ghobrial, IM, Bone, ND, Stenson, MJet al. Expression of the chemokine receptors CXCR4 and CCR7 and disease progression in B-cell chronic lymphocytic leukemia/small lymphocytic lymphoma. Mayo Clin Proc 2004; 79: 318–25. [DOI] [PubMed] [Google Scholar]
- 19. El-Ghonaimy, EA, et al. Positive lymph-node breast cancer patients–activation of NF-κB in tumor-associated leukocytes stimulates cytokine secretion that promotes metastasis via C-C chemokine receptor CCR7. FEBS J 2015; 282: 271–82. [DOI] [PubMed] [Google Scholar]
- 20. Liu, FY, et al. CCR7 regulates cell migration and invasion through MAPKs in metastatic squamous cell carcinoma of head and neck. Int J Oncol 2014; 45: 2502–10. [DOI] [PubMed] [Google Scholar]
- 21. Shannon, LA, Calloway, PA, Welch, TPet al. CCR7/CCL21 migration on fibronectin is mediated by phospholipase Cgamma1 and ERK1/2 in primary T lymphocytes. J Biol Chem 2010; 285: 38781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang, J, Zhang, X, Thomas, SMet al. Chemokine receptor 7 activates phosphoinositide-3 kinase-mediated invasive and prosurvival pathways in head and neck cancer cells independent of EGFR. Oncogene 2005; 24: 5897–904. [DOI] [PubMed] [Google Scholar]
- 23. Xu, B, Zhou, M, Qiu, Wet al. CCR7 mediates human breast cancer cell invasion, migration by inducing epithelial-mesenchymal transition and suppressing apoptosis through AKT pathway. Cancer Med 2017; 6: 1062–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang, W, Tu, G, Lv, Cet al. Matrix metalloproteinase-9 is up-regulated by CCL19/CCR7 interaction via PI3K/Akt pathway and is involved in CCL19-driven BMSCs migration. Biochem Biophys Res Commun 2014; 451: 222–8. [DOI] [PubMed] [Google Scholar]
- 25. Fowler, KA, Vasilieva, V, Ivanova, Eet al. R707, a fully human antibody directed against CC-chemokine receptor 7, attenuates xenogeneic acute graft-versus-host disease. Am J Transplant 2019; 19: 1941–54. [DOI] [PubMed] [Google Scholar]
- 26. Cuesta-Mateos, C, Juárez-Sánchez, R, Mateu-Albero, Tet al. Targeting cancer homing into the lymph node with a novel anti-CCR7 therapeutic antibody: the paradigm of CLL. MAbs 2021; 13: 1917484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee, K, Jung, Y, Lee, JYet al. Purification and characterization of recombinant human endothelin receptor type a. Protein Expr Purif 2012; 84: 14–8. [DOI] [PubMed] [Google Scholar]
- 28. Han, SG, Baek, SI, Son, TJet al. Preparation of functional human lysophosphatidic acid receptor 2 using a P9(*) expression system and an amphipathic polymer and investigation of its in vitro binding preference to G(α) proteins. Biochem Biophys Res Commun 2017; 487: 103–8. [DOI] [PubMed] [Google Scholar]
- 29. Kim, NH, et al. Functional expression of human prostaglandin E2 receptor 4 (EP4) in E. coli and characterization of the binding property of EP4 with G(α) proteins. Biochem Biophys Rep 2021; 25: 100871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kang, S, Kim, NH, Yu, YG. Identification of novel positive allosteric modulators of GLP1R that stimulate its interaction with ligands and G(α) subunits. Biochem Biophys Res Commun 2021; 583: 162–8. [DOI] [PubMed] [Google Scholar]
- 31. Yang, HY, Kang, KJ, Chung, JEet al. Construction of a large synthetic human scFv library with six diversified CDRs and high functional diversity. Mol Cells 2009; 27: 225–35. [DOI] [PubMed] [Google Scholar]
- 32. Han, SG, Baek, SI, Lee, WKet al. Overexpression and functional stabilization of recombinant human lysophosphatidic acid receptor 1 using an amphiphatic polymer. Bull Korean Chem Soc 2017; 38: 63–9. [Google Scholar]
- 33. Lim, HD, Lane, JR, Canals, Met al. Systematic assessment of chemokine signaling at chemokine receptors CCR4, CCR7 and CCR10. Int J Mol Sci 2021; 22: 4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ju, MS, Ahn, HM, Han, SGet al. A human antibody against human endothelin receptor type a that exhibits antitumor potency. Exp Mol Med 2021; 53: 1437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luttrell, LM. Reviews in molecular biology and biotechnology: transmembrane signaling by G protein-coupled receptors. Mol Biotechnol 2008; 39: 239–64. [DOI] [PubMed] [Google Scholar]
- 36. Goupil, E, Laporte, SA, Hébert, TE. Functional selectivity in GPCR signaling: understanding the full spectrum of receptor conformations. Mini Rev Med Chem 2012; 12: 817–30. [DOI] [PubMed] [Google Scholar]
- 37. Hjortø, GM, et al. Differential CCR7 targeting in dendritic cells by three naturally occurring CC-chemokines. Front Immunol 2016; 7: 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hauser, MA, Legler, DF. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J Leukoc Biol 2016; 99: 869–82. [DOI] [PubMed] [Google Scholar]
- 39. Webb, DR, Handel, TM, Kretz-Rommel, Aet al. Opportunities for functional selectivity in GPCR antibodies. Biochem Pharmacol 2013; 85: 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Molino, D, Del Barrio, Iet al. Contribution of heparan sulphate binding in CCL21-mediated migration of breast cancer cells. Cancer 2021; 13: 3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pan, M-R, Hou, M-F, Chang, H-Cet al. Cyclooxygenase-2 up-regulates CCR7 via EP2/EP4 receptor signaling pathways to enhance lymphatic invasion of breast cancer cells. J Biol Chem 2008; 283: 11155–63. [DOI] [PubMed] [Google Scholar]
- 42. Fang, L-W, Kao, Y-H, Chuang, Y-Tet al. Ets-1 enhances tumor migration through regulation of CCR7 expression. BMB Rep 2019; 52: 548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Campbell, JJ, Murphy, KE, Kunkel, EJet al. CCR7 expression and memory T cell diversity in humans. J Immunol 2001; 166: 877–84. [DOI] [PubMed] [Google Scholar]
- 44. Förster, R, Schubel, A, Breitfeld, Det al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 1999; 99: 23–33. [DOI] [PubMed] [Google Scholar]
- 45. López-Giral, S, Quintana, NE, Cabrerizo, Met al. Chemokine receptors that mediate B cell homing to secondary lymphoid tissues are highly expressed in B cell chronic lymphocytic leukemia and non-Hodgkin lymphomas with widespread nodular dissemination. J Leukoc Biol 2004; 76: 462–71. [DOI] [PubMed] [Google Scholar]
- 46. Sánchez-Sánchez, N, Riol-Blanco, L, Rodríguez-Fernández, JL. The multiple personalities of the chemokine receptor CCR7 in dendritic cells. J Immunol 2006; 176: 5153–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.