Abstract
African swine fever (ASF) is a dangerous infectious disease of domestic pigs and wild boar caused by African swine fever virus (ASFV). In Vietnam, the ASF epidemic is gradually turning into an endemic status with several recovered pigs post infection, but there were not many studies evaluating the role of these pigs in the epidemiological context in Vietnam. The aim of this study was to evaluate the viral antigen distribution and lesions in recovered pigs post ASFV infection. Ten pigs recovered from ASF at 6 weeks of age were monitored and assessed for anti-ASFV antibodies and viremia until slaughter. The five major organs (lung, liver, spleen, kidney, and lymph nodes) of these pigs were evaluated for microscopic lesions and viral antigen distribution. Anti-ASFV antibody was consistently observed to be high (S/P% ≥ 80) until slaughter, while viremia levels were very high (7 log10 copies/mL) at 6 weeks of age and gradually decreased to undetectable levels at 12 weeks of age (6th week post-infection). At slaughter, the ASFV-associated lesions in the organs of these pigs were mild and nonspecific. Seven out of ten pigs recovering from ASF still carried the virus in surveyed organ tissues, although not in the serum. These findings suggest that ASF-recovered pigs may be potential carriers of the virus, contributing to the increased complexity in the current endemic status in Vietnam.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11250-022-03229-0.
Keywords: African swine fever, Antigen presence, Lesions, Vietnam
Introduction
African swine fever (ASF) is a serious infectious disease that causes great economic losses to the swine industry due to its high mortality rate. The causative agent is African swine fever virus (ASFV) which is a large double-stranded virus of the genus Asfivirus, family Asfarviridae (Sánchez-Vizcaíno et al. 2012). Although the rate of transmission is slow, when infected with completely susceptible swine herds, ASF often breaks out in an acute form with a high mortality rate of 90–100% (Sánchez-Cordón et al. 2018). The transmission route of the disease is complex, associated with human activities and the pork food supply chain (Bellini et al. 2016). In which, humans are considered to be the main transmitters of the disease in domestic pigs (Chenais et al. 2019). ASF was first reported on a farm in Kenya in 1921 (Montgomery 1921). After that, the disease has spread to many countries in Africa and Europe over the past several decades (Mighell and Ward 2021; Sánchez-Vizcaíno et al. 2012). In August 2018, ASF was officially reported in China and later in Asian countries. The rapid spread of ASF across China and other Asian countries led to the huge loss of pig populations and reflected in the food supply chain (Woonwong et al. 2020).
At present, ASF is reported to be endemic in many regions of the world, including most of sub-Saharan Africa, the island of Sardinia, and parts of the Caucasus region and Eastern Europe (Brown and Bevins 2018; Costard et al. 2013; Kivumbi et al. 2021). In Vietnam, as various ASFV variants have been found, ASF outbreaks are occurring in small and scattered patterns (GSO 2021; Nguyen et al. 2022; Tran et al. 2021). This seems to show that ASF is gradually moving from pandemic to endemic status in Vietnam. Although ASF is a very serious disease due to its high mortality, there was a large portion of pigs that could overcome and continue to thrive after infection (Ståhl et al. 2019). This phenomenon has also been recorded in couple of outbreaks in Vietnam. There is a current trend that ASFV-infected farms often hold and raise recovered pigs in an attempt to regain lost costs and shorten the time to re-herd. The retention of these pigs should be assessed for their ability to carry ASFV, which is the potential source of ASFV for transmission to uninfected pigs and may contribute to endemic status in Vietnam. Studies on the role of ASFV carriers of wild boars have been performed. In particular, some wild boar species in Africa can be infected with ASFV but not exhibit any clinical signs (Jori and Bastos 2009; Sánchez-Vizcaíno et al. 2012). These pigs had low tissue viral load and low or undetectable viremia which were not sufficient to transmit ASFV through direct or indirect tick-borne contact (Jori and Bastos, 2009). Besides, the study of Blome et al. (2012) showed no indication of chronic disease or ASFV carrier states among adult wild boards (Blome et al. 2012). Most of the experiments have demonstrated a limited potential of genotype II ASFV isolates to cause persistent infection which generates ASFV carriers (Nurmoja et al. 2017; Schulz et al. 2021). Furthermore, the role of carrier domestic pigs as an ASFV source has been reported in Kenya and Uganda (Abworo et al. 2017; Sánchez-Cordón et al. 2018). Domestic carriers may be related to causing ASF outbreaks but their possibility of ASF transmission to naïve pigs was not proven (Sánchez-Cordón et al. 2018).
Therefore, this study was conducted in ten recovered pigs from an outbreak of commercial farm in Vietnam. The pigs were monitored for viremia and antibodies in the serum until slaughter and evaluated for the presence of ASFV DNA and ASFV antigen–positive cells in major organ tissues. The objective of this study was to determine the possibility of ASFV carrier in recovered pigs and then clarify their role in ASF epidemiology in Vietnam.
Materials and method
Farm history and study design
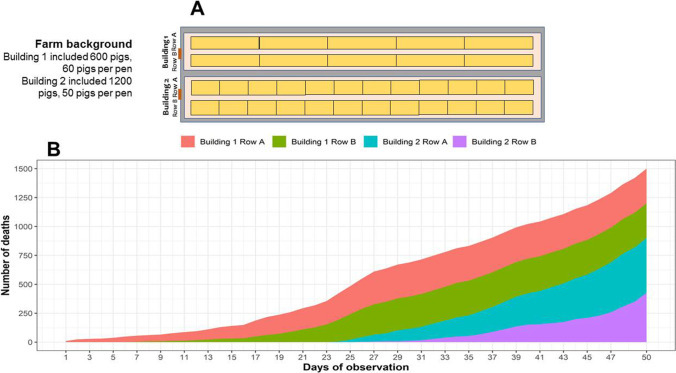
The study was designed in a commercial pig farm keeping 1800 weaned pigs (6 weeks old), Yorkshire x Landrace x Duroc breed. The farm had an opened door system (Fig. 1A) and two buildings with a total of 34 pens. In particular, building 1 had 10 pens (60 pigs per pen) while building 2 had 24 pens (50 pigs per pen). On August 1st, 2019, seven pigs from a middle pen of building 1 showed clinical manifestation suspected ASF such as refusal to eat, fever, cyanosis, and lethargy. We also observed that skin redness and hemorrhages appear starting from the ears, under the neck, and the abdomen and then spread to the whole body. These pigs were then sampled and tested for ASF, classical swine fever (CSF), and porcine reproductive and respiratory syndrome (PRRS). Two days later, the infection and outbreak of ASF were confirmed positive by testing of quantitative real-time polymerase chain reaction (qPCR) and polymerase chain reaction (PCR). On August 3rd, 2019, the survey farm recorded the first ten weaned pigs died with enlarged spleen lesions and severe hemorrhage in several internal organs such as the lung, liver, kidney, and lymph nodes. Besides, there were some clinical signs appearing scattered as black stools, hematuria, and conjunctival hemorrhage. Within 50 days of the follow-up (August 3rd–September 21st, 2019), 93.75% of all pigs on the farm died of disease (1687 heads). The average pig mortality rate of the farm within 1 day is 1.87%, approximately 34 pigs per day. The cumulative mortality rate of 50% in the population fell on day 30th of outbreak (Fig. 1B). The data was also noted that pens containing sick pigs have a tendency to spread ASFV to adjacent pens with healthy pigs. During the outbreak, we identified that some cases of pigs died suddenly without specific clinical signs. Although the majority of ASFV-infected pigs would die, there were a number of pigs showing signs of recovery, in which ten pigs were kept for rearing and laboratory testing until slaughter. The serum of these pigs was collected every 2 weeks to assess the titer of anti-ASFV antibody (ELISA) and viremia (qPCR). At the time of slaughter (24 weeks of age), these ten pigs were healthy and weighed at 103–122 kg. Then, their five organ tissues were collected and fixed in buffered formalin 10% to evaluate for the presence of ASFV and microscopic lesions. The remaining pigs in outbreak were culled on September 22nd, 2019, according to the regulations of Vietnam Department of Animal Health.
Fig. 1.
A Map of pens in the survey farm. B Cumulative number of deaths in each row of two buildings during the ASF outbreak
Detection of ASFV genome by qPCR
Serum or 1 g of tissue samples was extracted by using the Wizard® Genomic DNA Purification Kit (Promega) according to the manufacturer’s instructions. The extracted products were stored at − 20 °C and used as raw material for q qPCR reaction. qPCR reaction of our study used a pair of primers, probe, and thermal cycle guided from OIE (2019).
The total volume of a qPCR reaction was 20 µL consisting of 10 µL SensiFAST™ Probe No-ROX (Bioline, the UK), 0.5 µL primer and probe (5 pmol), 5 µL sample DNA, and 3.5 µL nuclease free water. The reaction mixture was then processed by the use of the Mygo Pro Real-time PCR instrument. The thermal process includes 1 cycle of 95 °C, 5 min; 40 cycles of 95 °C, 15 s; and 60 °C, 40 s. A positive sample result would yield a sigmoid gain curve, showing the number of cycles with a fluorescence signal level of Ct < 40. A negative sample maintains the fluorescence signal at background fluorescence, and the instrument does not report any Ct values (Ct ≥ 40).
Anti-ASFV antibody detection by enzyme-linked immunosorbent assay
Serum samples were stored at − 20 °C. The study used the ID Screen® African Swine Fever Indirect kit (ID.Vet, France) in order to evaluate the levels of antibody against ASFV. The implementation process was conducted strictly according to the manufacturer’s instructions.
Assessment of microscopic lesion by hematoxylin and eosin stain
Tissue samples of the organs of ten recovered pigs (liver, spleen, kidney, lung, and lymph nodes) were processed to paraffin block and carried out H&E staining according to the routine procedure of the Department of Infectious Diseases and Veterinary Public Health – Nong Lam University, Ho Chi Minh City.
ASFV antigen detection by immunohistochemistry
IHC slides were dehydrated by immersion in xylene and decreasing alcohol, respectively. The slides were treated at 37 °C for 20 min with proteinase K (GenDEPOT, USA) and then incubated overnight at 4 °C with primary antibody Rb anti-ASFV antiserum (Alpha Diagnostic, USA). After being washed in PBS for 10 min, the slides were incubated with Goat anti-rabbit AP (Dako, Denmark) secondary antibody at 37 °C for 1 h. The ImPACT Vector Red kit (Vector laboratories, USA) was used to show the reaction color. Positive cells (the presence of p30 protein) showed a red color in the cytoplasm and were observed under an optical microscope. Positive cells were counted and averaged over 3 microfields at 400 × magnification. The degree of positivity was scored on a scale of 0 (no positive cells), 1 + (1–10 positive cells), 2 + (10–30 positive cells), 3 + (30–100 positive cells), and 4 + (> 100 positive cells) (Oh et al. 2021).
Phylogenetic analysis
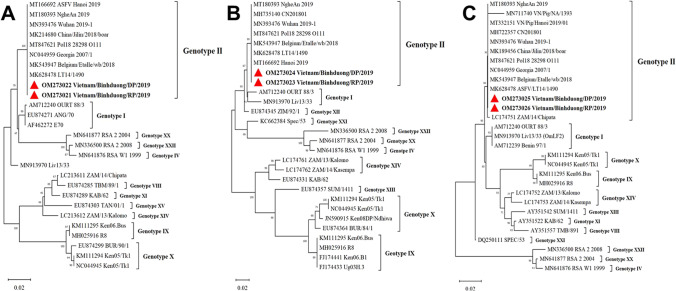
The partial genes encoding the p30, p54, and p72 proteins of two ASFV strains from one in ten recovered pig and a sick pig dying from ASFV were amplified according to previous studies (Bastos et al. 2003; Gallardo et al. 2009; Rowlands et al. 2008). The genome copies were sent to sequence at Macrogen laboratory (Korea). Sequences of these two strains were compared with reference strains of ASFV published on GenBank (NCBI). The phylogenetic tree was constructed by the neighbor-joining method (maximum composite likelihood model) on Mega X software with a bootstrap of 1000. The nucleotide sequences of two strains in the study were deposited in GenBank under the accession numbers OM273021-26 (Table S1).
Statistic analysis
We constructed a standard curve by using ten-fold serial dilutions of the quantitated ASFV DNA template. The number of genomic copies calculated from qPCR results was transformed to log10 of ASFV copies/mL serum. All statistical analyses in this study were performed by the use of R 4.1.2 software (http://www.r-project.org/).
Result
The viremia and antibody in recovered pigs
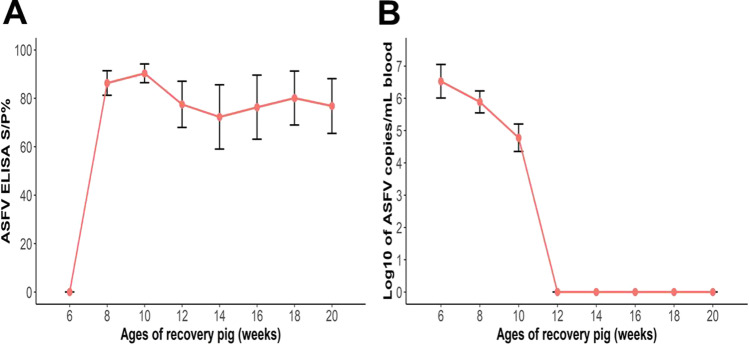
All recovered pigs showed no antibody at the time of infection (6th week of age) (Fig. 2A). These pigs became clinically healthy 2 weeks later (8th week of age), and anti-ASFV antibody levels were detected at high level (S/P% > 80). In the following weeks of age, the levels of antibody of these pigs remained high until slaughter time (approximately S/P% = 80). The viremia reported at the time of infection was very high (7 log10 copies/mL serum). However, the amount of virus in the serum gradually decreased in the 8th and 10th weeks with approximately 6 and 5 log10/mL serum, respectively. From 12 weeks of age until slaughter time, the virus was not detected in the serum of these pigs (Fig. 2B).
Fig. 2.
Levels of anti-ASFV antibodies (A) and viremia in serum (B) of recovered pigs after infection
Microscopic lesions of recovered pigs
The microscopic lesions in 5 organs (lung, liver, spleen, kidney, and lymph node) in recovered pigs were mild and nonspecific. Fibrous proliferation, mononuclear cell infiltration, hemorrhage, and edema were commonly found with mild severity in these pigs.
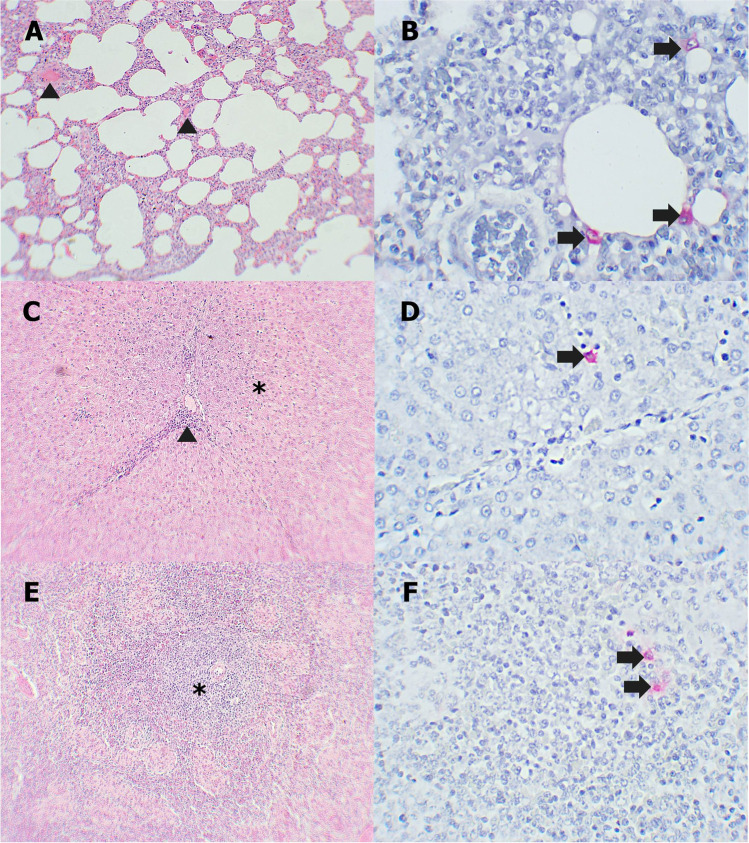
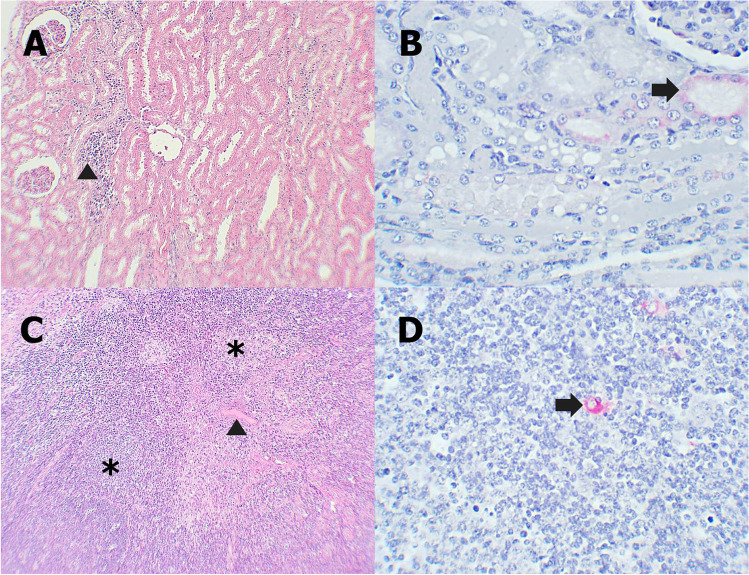
Fibrotic tissue proliferation, mild edema, and hemorrhage were observed commonly in the lung of recovered pigs (Fig. 3A). Some patchy areas of pulmonary consolidation and mild mononuclear cell infiltration in alveolar walls appeared in some pigs. Besides, there were some areas of hepatocyte necrosis, infiltration of mononuclear cells in hepatic parenchyma and periportal veins (Fig. 3C). The spleens showed mild to moderate local proliferation of fibrous tissue and mild lymphoid depletion in red pulp (Fig. 3E). Kidneys showed multifocal tubular necrosis and mononuclear cell infiltration at the renal cortex (Fig. 4A). The lymph nodes of pigs exhibited mild edema and hemorrhage in the medulla, fibroblast proliferation, and thickening of capsule, and mild lymphoid depletion was seen (Fig. 4C).
Fig. 3.
A, C, E Microscopic lesions of recovered pigs in the lung, liver, and spleen. B, D, F Immunohistochemical staining of recovered pigs in the lung, liver, and spleen. A Mild edema in alveolar walls (arrowhead). B Positive staining of interstitial mononuclear cells (arrow). C Hepatocyte necrosis (asterisk) and mononuclear cells infiltration around the portal veins (arrowhead). D Kuffer cells have cytoplasmic immunolabeling (arrow). E Mild lymphoid depletion in white pulp (asterisk) and red pulp. F Positive staining of macrophages in red pulp (arrow)
Fig. 4.
A, C Microscopic lesions of recovered pigs in the kidney and lymph node. B, D Immunohistochemical staining of recovered pigs in kidney and lymph node. A Diffuse mononuclear cell infiltration in the renal cortex (arrowhead). B Tubular epitheliums have cytoplasmic immunolabeling (arrow). C Mild edema in medulla (arrowhead) and mild lymphoid depletion (asterisk). D Positive staining of macrophages in medulla (arrow)
The presence of ASFV DNA and ASFV antigen–positive cells in the tissues
Results of qPCR showed that 7/10 recovered pigs had the presence of ASFV DNA in organ tissues (Table 1). Particularly, four pigs (no. 3, 4, 7, and 10) showed viral genomes in all five organs, while three pigs (no. 2, 6, and 8) recorded only in some organs. However, three pigs (no. 1, 5, and 9) did not detect the viral DNA in the surveyed tissues. Besides, the amount of viral DNA in these positive tissue samples was mostly low (Ct > 29).
Table 1.
qPCR and IHC results on five organ tissues of ten recovered pigs
| Organ | Lung | Liver | Spleen | Kidney | Lymph node | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Assay Recovered pig |
qPCR (Ct) | IHC | qPCR (Ct) | IHC | qPCR (Ct) | IHC | qPCR (Ct) | IHC | qPCR (Ct) | IHC |
| No. 1 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| No. 2 | 32.13 | nd | nd | nd | 36.51 | nd | 35.55 | nd | nd | nd |
| No. 3 | 29.11 | 1 + | 30.67 | 1 + | 32.16 | 1 + | 33.54 | 1 + | 31.98 | 1 + |
| No. 4 | 31.16 | 1 + | 34.42 | nd | 32.06 | nd | 32.25 | nd | 32.04 | nd |
| No. 5 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| No. 6 | 31.54 | nd | nd | nd | 34.25 | nd | 34.36 | nd | nd | nd |
| No. 7 | 30.16 | 1 + | 33.37 | nd | 33.15 | nd | 33.63 | nd | 33.50 | nd |
| No. 8 | 31.35 | 1 + | 32.05 | 1 + | nd | nd | 33.25 | nd | 33.26 | nd |
| No. 9 | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| No. 10 | 30.16 | 1 + | 30.67 | 1 + | 31.85 | 1 + | 32.75 | nd | 32.72 | nd |
| Proportion | 7/10 | 5/10 | 5/10 | 3/10 | 6/10 | 2/10 | 7/10 | 1/10 | 5/10 | 1/10 |
nd not detected; 1 + : 1–10 positive cells/1 mm
IHC results showed that only a few of tissues recorded the presence of ASFV antigen (Table 1). Only 12/50 samples of all recovered pigs recorded the presence of cells. The amount of ASFV antigen–positive cells in such tissues was recorded at a low level (< 10 cells/1 mm3).
Phylogenetic analysis
Phylogenetic analysis and comparison of nucleotide sequences to reference strains showed that two isolates in the study belonged to genotype II (Fig. 5). These strains shared 100% nucleotide homology in sequences encoding p30, p54, and p72. Besides, they had a very high nucleotide identity of p30, p54, and p72 sequences with strains firstly reported from Vietnam and neighboring countries including China.
Fig. 5.
Phylogenetic analysis based on genetic sequences of genes encoding p30, p54, and p72. A p30 phylogenetic tree, B p54 phylogenetic tree, and C p72 phylogenetic tree. The maximum likelihood approach was used for construction of phylogenetic trees in MEGA X software (https://www.megasoftware.net/). Numbers along branches indicate bootstrap values > 50% (1000 replicates). The two red triangles are ASFV isolates from our study. Scale bars indicate nucleotide substitutions per site
Discussion
Montgomery, who was the first to describe ASF, identified it as an acute hemorrhagic disease with a mortality rate of up to 100% (Montgomery 1921). However, mortality ranges from 3 to 100% depending on the virulence of the virus strains (Blome et al. 2013). Highly virulent strains of ASFV often cause acute and subacute forms of the disease that can kill 100% of pigs within 1–8 days of infection (Mebus 1988; Sánchez-Vizcaíno et al. 2012). Our study showed that the ASFV strain belongs to Genotype II and had a close relationship with ASFV strains from Georgia and China. In fact, ASFV strains circulating in Vietnam have been confirmed as highly virulent ASFV strains and may have originated in China (Nguyen et al. 2021; Tran et al. 2020; Van Phan Le et al. 2019). At the beginning of ASF occurrence in Vietnam, it caused severe impacts with a very high mortality rate (Oh et al. 2021). During outbreak of the study, the highest number of pigs dying in 1 day was only 5% (81 heads); most infected pigs died. The observation of virus transmission in the surveyed farm is consistent with previous studies that ASFV spread is usually slow with the main route of transmission being through direct contact (Guinat et al. 2014). Genetic analysis was performed to confirm the presence of the virus which has no mutations in its genes to adapt to its hosts. In spite of long persistence in the tissues of recovered pigs, we did not observe any difference in the ASFV gene encoding p30, p54, and p72. This supports an idea proposed previously by Dixon et al. (2020) that ASFV is a very stable double-stranded DNA virus with very low mutagenicity.
Although the formation of an antibody response following ASFV infection has been documented, the virus-neutralizing capacity of these antibodies is poorly understood. In acute cases, pigs infected with highly virulent ASFV strains often die before specific antibodies are produced (OIE 2019). However, anti-ASFV antibodies remain an important immune component that protects pigs after ASFV infection (Sereda et al. 2015). Many studies have identified antibodies against p30, p54, and p72 proteins that play important roles in immunity against ASFV. Anti-p72 and anti-p54 antibodies suppressed the attachment of ASFV to host cells, while anti-p30 antibodies prevented the penetration of virions into the cells (Sereda et al. 2015). Antibodies can appear 7–10 days after infection and can persist for months, even years (OIE, 2019). Petrov et al. (2018) also found that in pigs infected with a medium virulent strain Netherlands’86, anti-p73 antibodies could be detected in sera from all experimental pigs 4–9 days and maintained until the end of the experiment (day 63). Walczak et al. (2021) identified that specific antibodies against ASFV strain Pol18_28298_O111 (Poland) were detected 9–18 days after infection and 2 to 5 days after the detection of viremia.
Our study also recorded very high viremia at the first day of observation (6th week of age) and the viremia gradually decreased to undetectable threshold of qPCR at 12 weeks of age. Time to record viremia in infected pigs in our study was approximate 42 days, shorter than the time recorded in the previous study of 91 days (Petrov et al. 2018). The difference in viremia duration may be due to the virulence of ASFV and naturally infected pigs may not determine the exact time. ASFV strain in the study was high virulent while Netherlands’86 strain in Petrov’s experiment was determined to be a moderately virulent strain. In addition, the duration of viremia and serology has been noted to differ between virus strains in the experiment of de Carvalho Ferreira et al. (de Carvalho Ferreira et al. 2012). Experimental pigs infected with Brazilian strain showed very severe disease but viremia and excretion patterns could only be followed for 9 days. Meanwhile, in the low virulent viruses as Malta and Netherlands group, viremia and excretion patterns persisted up to 70 days (de Carvalho Ferreira et al. 2012). Presumably, high and prolonged antibody levels after infection helped shorten the duration of viremia so that from 12 weeks of age until slaughter time, no virus was detected in the serum.
Unlike the ASF-specific lesions in acute form, recovered pigs exhibited a less severe degree. Our findings also support the descriptions of Pornthummawat et al. (2021). The microscopic lesions of pigs in this study are quite similar with lesions recorded in the chronic form of ASF in previous studies (Ganowiak 2012; Salguero 2020). These findings showed that these recovered pigs may develop to chronic infection or these lesions are sequelae of previous acute infection.
Our study also showed that recovered pigs had small amounts of viral genome and scattered antigen in organ tissues. In the study of Pornthummawat et al. (2021), blood and tissue samples of all 4 convalescent pigs did not detect viral genome, but found a small number of ASFV-infected cells. Their study found that small amounts of antigen were detected in the lung (less than 10 infected cells), tonsil, and stomach (less than 30 infected cells). Although ASFV was not detected in the serum of recovered pigs 1 week after recovery, it is still potentially present in low amounts in some organ tissues of some pigs. An idea previously proposed by Abworo et al. (2017) is that there are carrier pigs which lack clinical signs of ASF or detectable ASFV in blood but have ASFV sequestered in tissues. The complete similarity in the genotypes of ASFV in recovered and dead pigs suggests that ASFV may remain infectious and cause new outbreaks. Costard et al. (2009) described that domestic pigs recovering ASF seem to extend the duration of carrying ASFV to 6 months or more and may transmit ASFV to susceptible pigs through direct contact or ingestion of infected meat products. According to Eblé et al. (2019), clinically healthy convalescent pigs following infection with the Netherlands’86 strain may be the source of a new acute infection in fully susceptible pigs. This report is quite contrary to that of Petrov et al. suggesting that pigs that survived infection with the Netherlands’86 strain completely cleared the virus from blood and tissues after 91 days of infection without any transmission to pigs exposed to them (Petrov et al. 2018). Ståhl et al. (2019) showed that there are two types of pigs that survive ASFV infection: pigs developing a persistent infection and pigs that complete recover and live healthy. The role of such carriers is not evident in the epidemiology (Ståhl et al. 2019). Our study also recorded three pigs with no viral genome and antigen detected in all tissues. This finding is also in line with the study reported by Nurmoja et al. (2017) that not all surviving pigs become ASFV carriers.
Conclusion
Viremia in ten recovered pigs gradually decreased and lasted for 6 weeks post infection. Meanwhile, anti-ASFV antibodies remained high until slaughter. ASFV-associated lesions were observed to be mild but common in multiple organs in these pigs. At the time of slaughter, although no virus was detected in the serum, low levels of viral genome and antigen were detected in the tissues of seven recovered pigs. Therefore, we concluded that pigs recovered from ASFV may still carry the virus at low levels in their organs and be a potential contributor to endemic status in Vietnam.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful for the technical support that we received from the Department of Infectious Diseases and Veterinary Public Health – Nong Lam University.
Author contribution
D.T.D., O.T., and D.C.L. designed the study. D.C.L. and H.T.N. performed experiments. D.C.L., H.T.N., and O.T. analyzed the data. D.T.D., O.T., and D.C.L. wrote the manuscript.
Data availability
The p30, p54, and p72 sequences of two ASFV strains identified in this study have been deposited in GenBank under the accession numbers OM273021–OM273026.
Declarations
Ethics approval
The study was conducted in compliance with the institutional rules for the care and use of laboratory animals and using a protocol approved by the Ministry of Agriculture and Rural Development (MARD) Vietnam (TCVN 8402:2010).
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Danh Cong Lai and Taehwan Oh have equal contribution in this study.
References
- Abworo EO, Onzere C, Amimo JO, Riitho V, Mwangi W, Davies J, Blome S, Bishop RP. Detection of African swine fever virus in the tissues of asymptomatic pigs in smallholder farming systems along the Kenya-Uganda border: Implications for transmission in endemic areas and ASF surveillance in East Africa. Journal of General Virology. 2017;98:1806–1814. doi: 10.1099/jgv.0.000848. [DOI] [PubMed] [Google Scholar]
- Bastos AD, Penrith M-L, Cruciere C, Edrich J, Hutchings G, Roger F, Couacy-Hymann E, Thomson GR. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Archives of Virology. 2003;148:693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- Bellini S, Rutili D, Guberti V. Preventive measures aimed at minimizing the risk of African swine fever virus spread in pig farming systems. Acta Veterinaria Scandinavica. 2016;58:1–10. doi: 10.1186/s13028-016-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blome S, Gabriel C, Dietze K, Breithaupt, A., and Beer, M. (2012). High virulence of African swine fever virus caucasus isolate in European wild boars of all ages. Emerging Infectious Diseases18. [DOI] [PMC free article] [PubMed]
- Blome S, Gabriel C, Beer M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Research. 2013;173:122–130. doi: 10.1016/j.virusres.2012.10.026. [DOI] [PubMed] [Google Scholar]
- Brown VR, Bevins SN. A review of African swine fever and the potential for introduction into the United States and the possibility of subsequent establishment in feral swine and native ticks. Frontiers in Veterinary Science. 2018;5:11. doi: 10.3389/fvets.2018.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenais E, Depner K, Guberti V, Dietze K, Viltrop A, Ståhl K. Epidemiological considerations on African swine fever in Europe 2014–2018. Porcine Health Management. 2019;5:1–10. doi: 10.1186/s40813-018-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costard S, Wieland B, De Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK. African swine fever: how can global spread be prevented? Philosophical Transactions of the Royal Society b: Biological Sciences. 2009;364:2683–2696. doi: 10.1098/rstb.2009.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costard S, Mur L, Lubroth J, Sanchez-Vizcaino J, Pfeiffer DU. Epidemiology of African swine fever virus. Virus Research. 2013;173:191–197. doi: 10.1016/j.virusres.2012.10.030. [DOI] [PubMed] [Google Scholar]
- de Carvalho Ferreira H, Weesendorp E, Elbers A, Bouma A, Quak S, Stegeman J, Loeffen W. African swine fever virus excretion patterns in persistently infected animals: a quantitative approach. Veterinary Microbiology. 2012;160:327–340. doi: 10.1016/j.vetmic.2012.06.025. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Stahl K, Jori F, Vial L, Pfeiffer DU. African swine fever epidemiology and control. Annual Review of Animal Biosciences. 2020;8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- Eblé P, Hagenaars T, Weesendorp E, Quak S, Moonen-Leusen H, Loeffen W. Transmission of African Swine Fever Virus via carrier (survivor) pigs does occur. Veterinary Microbiology. 2019;237:108345. doi: 10.1016/j.vetmic.2019.06.018. [DOI] [PubMed] [Google Scholar]
- Gallardo C, Mwaengo DM, Macharia JM, Arias M, Taracha EA, Soler A, Okoth E, Martín E, Kasiti J, Bishop RP. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes. 2009;38:85–95. doi: 10.1007/s11262-008-0293-2. [DOI] [PubMed] [Google Scholar]
- Ganowiak, J. (2012). Patho-anatomical studies on African swine fever in Uganda. Sveriges lantbruksuniversitet57.
- GSO (2021). Livestock of pig has recovered. General Statistics Office - Ministry of Planning and Investment Vietnam.
- Guinat C, Reis AL, Netherton CL, Goatley L, Pfeiffer DU, Dixon L. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Veterinary Research. 2014;45:1–9. doi: 10.1186/s13567-014-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jori F, Bastos AD. Role of wild suids in the epidemiology of African swine fever. EcoHealth. 2009;6:296–310. doi: 10.1007/s10393-009-0248-7. [DOI] [PubMed] [Google Scholar]
- Kivumbi C, Yona C, Hakizimana J, Misinzo G. An assessment of the epidemiology and socioeconomic impact of the 2019 African swine fever outbreak in Ngara district, western Tanzania. Veterinary and Animal Science. 2021;14:100198. doi: 10.1016/j.vas.2021.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebus CA. African swine fever. Advances in Virus Research. 1988;35:251–269. doi: 10.1016/S0065-3527(08)60714-9. [DOI] [PubMed] [Google Scholar]
- Mighell E, Ward MP. African Swine Fever spread across Asia, 2018–2019. Transboundary and Emerging Diseases. 2021;68:2722–2732. doi: 10.1111/tbed.14039. [DOI] [PubMed] [Google Scholar]
- Montgomery RE. On a form of swine fever occurring in British East Africa (Kenya Colony) Journal of Comparative Pathology and Therapeutics. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- Nguyen HQ, Nguyen DM, Nguyen NM, Nguyen DN, Luu HQ, Do DT. Genetic analysis of African swine fever virus based on major genes encoding p72, p54 and p30. The Journal of Agriculture and Development. 2021;20:18–25. doi: 10.52997/jad.3.03.2021. [DOI] [Google Scholar]
- Nguyen, V. T., Cho, K.-h., Mai, N. T. A., Park, J.-Y., Trinh, T. B. N., Jang, M.-K., Nguyen, T. T. H., Vu, X. D., Nguyen, T. L., and Nguyen, V. D. (2022). Multiple variants of African swine fever virus circulating in Vietnam. Archives of Virology, 1–4. [DOI] [PubMed]
- Nurmoja I, Petrov A, Breidenstein C, Zani L, Forth J, Beer M, Kristian M, Viltrop A, Blome S. Biological characterization of African swine fever virus genotype II strains from north-eastern Estonia in European wild boar. Transboundary and Emerging Diseases. 2017;64:2034–2041. doi: 10.1111/tbed.12614. [DOI] [PubMed] [Google Scholar]
- Oh, T., Nguyen, T. M., Ngo, T. T. N., Thinh, D., Nguyen, T. T. P., Do, L. D., and Do, D. T. (2021). Long‐term follow‐up of convalescent pigs and their offspring after an outbreak of acute African swine fever in Vietnam. Transboundary and Emerging Diseases. [DOI] [PubMed]
- OIE OIE Terrestrial Manual. OIE Chapter. 2019;3(8):1. [Google Scholar]
- Petrov A, Forth J, Zani L, Beer M, Blome S. No evidence for long-term carrier status of pigs after African swine fever virus infection. Transboundary and Emerging Diseases. 2018;65:1318–1328. doi: 10.1111/tbed.12881. [DOI] [PubMed] [Google Scholar]
- Pornthummawat A, Truong QL, Hoa NT, Lan NT, Izzati UZ, Suwanruengsri M, Nueangphuet P, Hirai T, Yamaguchi R. Pathological lesions and presence of viral antigens in four surviving pigs in African swine fever outbreak farms in Vietnam. Journal of Veterinary Medical Science. 2021;83:1653–1660. doi: 10.1292/jvms.21-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, Dwarka R, Onashvili T, Albina E, Dixon LK. African swine fever virus isolate, Georgia, 2007. Emerging Infectious Diseases. 2008;14:1870. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero FJ. Comparative pathology and pathogenesis of African swine fever infection in swine. Frontiers in Veterinary Science. 2020;7:282. doi: 10.3389/fvets.2020.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Cordón P, Montoya M, Reis A, Dixon L. African swine fever: A re-emerging viral disease threatening the global pig industry. The Veterinary Journal. 2018;233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vizcaíno J, Mur L, Martínez-López B. African swine fever: an epidemiological update. Transboundary and Emerging Diseases. 2012;59:27–35. doi: 10.1111/j.1865-1682.2011.01293.x. [DOI] [PubMed] [Google Scholar]
- Schulz K, Schulz J, Staubach C, Blome S, Nurmoja I, Conraths FJ, Sauter-Louis C, Viltrop A. African Swine Fever Re-Emerging in Estonia: The Role of Seropositive Wild Boar from an Epidemiological Perspective. Viruses. 2021;13:2121. doi: 10.3390/v13112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereda A, Kazakova A, Imatdinov A, Kolbasov D. Humoral and cell immune mechanisms under African swine fever. Agricultural Biology. 2015;50:709–718. [Google Scholar]
- Ståhl K, Sternberg-Lewerin S, Blome S, Viltrop A, Penrith M-L, Chenais E. Lack of evidence for long term carriers of African swine fever virus-a systematic review. Virus Research. 2019;272:197725. doi: 10.1016/j.virusres.2019.197725. [DOI] [PubMed] [Google Scholar]
- Tran HTT, Truong AD, Ly DV, Vu TH, Hoang VT, Nguyen TC, Chu TN, Nguyen TH, Pham NT, Nguyen T. Genetic characterisation of African swine fever virus in outbreaks in Ha Nam province, Red River Delta Region of Vietnam, and activity of antimicrobial products against virus infection in contaminated feed. Journal of Veterinary Research. 2020;64:207. doi: 10.2478/jvetres-2020-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HTT, Truong AD, Dang AK, Ly DV, Nguyen CT, Chu NT, Hoang TV, Nguyen HT, Dang HV. Circulation of two different variants of intergenic region (IGR) located between the I73R and I329L genes of African swine fever virus strains in Vietnam. Transboundary and Emerging Diseases. 2021;68:2693–2695. doi: 10.1111/tbed.13996. [DOI] [PubMed] [Google Scholar]
- Van Phan Le DGJ, Yoon S-W, Kwon H-M, Trinh TBN, Nguyen TL, Bui TTN, Oh J, Kim JB, Cheong KM, Van Tuyen N. Outbreak of African swine fever, Vietnam, 2019. Emerging Infectious Diseases. 2019;25:1433. doi: 10.3201/eid2507.190303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak M, Wasiak M, Dudek K, Kycko A, Szacawa E, Olech M, Woźniakowski G, Szczotka-Bochniarz A. Blood Counts, Biochemical Parameters, Inflammatory, and Immune Responses in Pigs Infected Experimentally with the African Swine Fever Virus Isolate Pol18_28298_O111. Viruses. 2021;13:521. doi: 10.3390/v13030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woonwong Y, Do Tien D, Thanawongnuwech R. The future of the pig industry after the introduction of African swine fever into Asia. Animal Frontiers. 2020;10:30–37. doi: 10.1093/af/vfaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The p30, p54, and p72 sequences of two ASFV strains identified in this study have been deposited in GenBank under the accession numbers OM273021–OM273026.