Abstract

Managed honey bee colonies used for crop pollination are fed artificial diets to offset nutritional deficiencies related to land-use intensification and climate change. In this study, we formulated novel microalgae diets using Chlorella vulgaris and Arthrospira platensis (spirulina) biomass and fed them to young adult honey bee workers. Diet-induced changes in bee metabolite profiles were studied relative to a natural pollen diet using liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) metabolomics. Untargeted analyses of pollen- and microalgae-fed bees revealed significant overlap, with 248 shared features determined by LC-MS and 87 shared features determined by GC-MS. Further metabolomic commonalities were evident upon subtraction of unique diet features. Twenty-five identified metabolites were influenced by diet, which included complex lipids, essential fatty acids, vitamins, and phytochemicals. The metabolomics results are useful to understand mechanisms underlying favorable growth performance as well as increased antioxidant and heat shock protein gene expression in bees fed the microalgae diets. We conclude that the tested microalgae have potential as sustainable feed additives and as a source of bee health-modulating natural products. Metabolomics-guided diet development could eventually help tailor feed interventions to achieve precision nutrition in honey bees and other livestock animals.
Keywords: honey bee, Apis mellifera, microalgae, pollen substitute, artificial diet, nutrition, Chlorella vulgaris, spirulina, Arthrospira platensis, antioxidant, metabolomics, gene expression
1. Introduction
The honey bee (Apis mellifera) is the world’s primary managed pollinator. In the United States alone, honey bee pollination services contribute ∼$20 billion per year to the value of crop production.1 Nevertheless, commercial beekeepers are experiencing annual colony losses that are on average twice as high as historical records, jeopardizing pollination services and human food security.2 Honey bee colony losses are attributed to multiple stressors, notably parasites and pathogens. However, many lines of evidence indicate that malnutrition is a major factor underlying colony mortality.3−5 Abundant floral resources are required for honey bee colony growth, immune function, and stress responses.6−10 Nectar provides energy in the form of carbohydrates, while pollen is the main source of proteins, lipids, and micronutrients.7 Under ideal conditions, varied flower sources are necessary to meet bee nutritional requirements since the composition of pollen varies by plant species.11,12 Unfortunately, modern intensive agriculture is associated with reduced flower diversity and, hence, lower nutritional value.13−15 Plant responses to climate change, such as altered flower, nectar, and pollen production, will alter the landscape of floral resource availability.16−18 These conditions may further exacerbate the challenges of honey bee nutrition and health, especially within a managed setting.
Managed bee colonies used for crop pollination are routinely fed artificial “pollen substitute” diets to compensate for a lack of pollen forage in the environment and to prevent nutritional deficiencies. Various diet formulations have been used as a substitute for natural pollen, and these often incorporate protein-rich ingredients, such as soy, corn gluten, yeast, casein, and egg, as a source of essential amino acids.7 However, diet comparisons suggest the existence of potentially overlooked nutritional factors or other pollen components that might improve artificial diet effectiveness (i.e., providing phytochemicals that might stimulate bee immunity or improve stress resistance).19,20 In addition to protein content, pollen contains a variety of necessary lipids, essential fatty acids,21−23 and a broad diversity of bee health-modulating bioactive compounds, such as vitamins and phenolic acids.24,25 Thus, there are opportunities to enhance feed to more closely mimic the chemical composition of pollen, especially to serve the growing demands of a majority of US beekeepers who feed supplemental nutrition to their colonies.20 Importantly, given the challenges of feeding the world’s human population, sustainable ingredients that do not compete with human food production are good candidates to address this crucial need of modern beekeeping.
Microalgae are nutritious and sustainable feed ingredients that have been used in a variety of livestock,26 including recent applications in managed honey bees.27 Notably, eukaryotic microalgae in the genus Chlorella and prokaryotic cyanobacteria (blue-green microalgae) in the genus Arthrospira (commonly called spirulina) are excellent sources of protein, fatty acids, sterols, and other bioactive compounds with nutraceutical potential. These microalgae are digestible by honey bees and appear to reproduce the growth characteristics of a natural pollen diet;28,29 however, little is known about the metabolic mechanisms underlying their impact on bee health.
Mass spectrometry-based metabolomics enables comprehensive and systematic analyses of all metabolites in an organism,30 and it has emerged as a powerful tool in nutrition and food sciences.31 Metabolomics-guided diet development could enable precision nutrition and an improved understanding of the mechanisms underlying the effects of feed.32 Using a mass spectrometry-based metabolomics approach, the objective of this study was to investigate diet-induced changes in honey bees, so as to better understand the nutritional and metabolic effects of microalgae relative to a natural pollen diet (Figure 1).
Figure 1.
Schematic overview of metabolite extraction and analyses. Honey bees were fed four different diets: sugar, pollen, Chlorella (Chlorella vulgaris), and spirulina (Arthrospira platensis). Bee abdomens were harvested, extracted, and examined through untargeted and targeted liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) to analyze their metabolomic compositions.
2. Materials and Methods
2.1. Honey Bee (Apis mellifera L.) Experimental Design
Experiments were conducted in the summer of 2021 at the USDA-ARS Honey Bee Breeding Genetics and Physiology Laboratory in Baton Rouge, Louisiana. Newly emerged worker bees were obtained by incubating sealed brood frames at 35 °C and 50% relative humidity overnight. Three brood frames were obtained from a healthy, vigorous colony that was treated for Varroa mites and had no visible signs of disease. Bees (<24 h old) were collected into a container and then randomly assigned to diet treatment groups (50 bees/cage). Four cages were established for each diet (16 cages total). Thus, each diet had four biological replicates. After 8 days of ad libitum feeding, bees were separately collected from each cage, frozen on dry ice, and then stored at −80 °C.
2.2. Diet Preparation and Consumption Measures
The different diet groups consisted of sugar, bee-collected pollen, Chlorella (C. vulgaris), and spirulina (A. platensis). All diets were mixed with 1:1 (v/v) sucrose syrup/honey to achieve a paste consistency. Thus, the formulated diet pastes were approximately two parts of dry ingredients to one part of sucrose/honey syrup. Diet pastes were loaded into modified microcentrifuge tubes (see experimental design schematic, Figure S1) and stored at −20 °C until use. There were four cage replicates per diet treatment (i.e., biological replicates). For the pollen diet, mixed corbicular pollen pellets were collected using entrance-mounted pollen traps during the late fall of 2020 (thus, predominantly Solidago spp. in floral composition) from a USDA-ARS apiary in Baton Rouge, Louisiana, and frozen at −80 °C until needed. The Chlorella diet consisted of organic, powdered, cracked cell wall C. vulgaris biomass (Micro Ingredients, California). The spirulina diet consisted of organic, powdered A. platensis biomass (Micro Ingredients, California). Approximately, 1.25 g of formulated diet paste was provided to each cage. The pollen and spirulina diets used in this study were characterized for amino acid content and protein bioavailability previously.29,33 The amount of diet consumed by each cage was recorded on day 4, then the diet was refreshed with approximately 1.25 g of new diet paste, and consumption was measured again on day 8. As a control, diet samples were placed in cages without bees and weight loss was measured to determine the evaporation rate for each diet type. Diet consumption in each cage was adjusted for moisture loss (<1.5%) and recalculated to give the total diet consumed over the 8-day period
2.3. Honey Bee Dissection
Frozen bees were dissected on dry ice into three parts: head, thorax (excluding legs and wings), and abdomens with guts intact. Then, dissected parts were collected into pools of eight parts per cage. Two separate pools of eight abdomens were made for each cage. One abdomen pool was used for RNA extraction and gene expression, while the other abdomen pool was used for metabolite extraction and metabolomic analyses. The bee abdomen was chosen for gene expression and metabolite analyses since it contains the fat body, a tissue with central nutrient storage, and metabolic functions, as well as the entire digestive tract.
2.4. Honey Bee Physiological Measures and Gene Expression Analyses
The average head and thorax weights per bee cage were determined by drying pools of eight heads or thoraces to a constant weight (60 °C for 48 h) and recording to the nearest 0.1 mg. For gene expression analyses, pools of eight frozen bee abdomens per cage were subjected to RNA extraction with a Monarch total RNA miniprep kit (New England BioLabs) according to the manufacturer’s protocol. cDNA synthesis was carried out using 1 μg of DNAase-treated RNA and a LunaScript RT SuperMix Kit (New England BioLabs) according to the manufacturer’s protocol. Quantitative polymerase chain reaction (qPCR) was performed in triplicate to quantify expression levels of vitellogenin (vg), superoxide dismutase (CuZn SOD), catalase, heat shock protein 70 (hsp70), and heat shock protein 90 (hsp90). All qPCR reactions were performed as follows: initial denaturation at 95 °C for 5 min; 40 cycles with denaturation at 95 °C for 15 s; and a primer-pair-specific annealing and extension temperature (Table S4) for 30 s. The reactions were carried out using SsoAdvanced Universal SYBR Green Supermix (Biorad) in triplicate on a CFX96 Real-Time PCR Detection System (Biorad). To confirm the absence of contaminating genomic DNA and primer dimers, amplification and melting curves were tested in negative control reactions containing only DNase-treated total RNA. Relative transcript levels were determined based on standardized Ct values (ΔCt) using β-actin for normalization.
2.5. Honey Bee Metabolite Extractions
Each bee sample represented one biological replicate that contained eight honey bee abdomens that were fed a specific diet (i.e., sugar, pollen,Chlorella, or spirulina). First, abdomens were ground with a mortar and pestle using liquid nitrogen (Figure S2). Then, the crushed abdomens were transferred to a scintillation vial, submerged with 5 mL of acetone, and were shaken for 16 h. The solvent was transferred to an Eppendorf tube and centrifuged. The supernatant (i.e., acetone layer) was retained. The residual solids were resuspended in 1:1 MeOH/CHCl3 (5 mL) and sonicated for 30 min. The solvent layer was transferred to an Eppendorf tube and centrifuged, where the supernatant (i.e., MeOH/CHCl3 layer) was saved and combined with the acetone layer described above. The combined layers were dried under a nitrogen atmosphere to produce a bee extract fed on a diet, and each of these consisted of four biological replicates (Table S1). Standards of pollen, Chlorella (C. vulgaris), and spirulina (A. platensis) were treated and extracted in a manner identical to the above. Extraction solvents were chosen based on their widespread use in the processing of biological materials.34,35
2.6. Mass Spectrometry Analysis
2.6.1. LC-MS Analysis
The extracts were examined at 0.2 mg/mL dissolved in MeOH. An Acquity ultraperformance liquid chromatography system (UPLC, Waters Corp.) coupled with a Thermo Q Exactive Plus MS (Thermo Fisher) was used for the analysis. MeOH was used as a blank sample. The flow rate of the UPLC was set to 0.3 mL/min using a BEH C18 (2.1 mm × 50 mm × 1.7 μm) column equilibrated at 40 °C. The mobile phase consisted of Fisher Optima LC-MS grade CH3CN–H2O (with 0.1% formic acid added). The analysis started at 15% CH3CN and increased linearly to 100% CH3CN over 8 min; it was then held at 100% CH3CN over 1.5 min before returning to the starting conditions over 0.5 min, making the total run time 10 min. Photodiode-array (PDA) detection was used to acquire data from 200 to 500 nm with a resolution of 4 nm. The Q Exactive Plus with electrospray ionization (ESI) was used to collect high-resolution accurate mass measurements and fragmentations of the detected ions. The initial data were collected from m/z 135 to 2000 at a resolving power of 70,000 for both positive and negative modes; however, only positive mode data were used for these studies. The spray voltage was set to either 3000 V (+) or 3000 V (−). Sheath gas was 47.50, aux gas was set to 11.25, spare gas 2.25, the heater temperature was 350.0 °C, the capillary temperature was 256.25 °C, and s-lens was 50.0. The acquired LC-MS data were analyzed using Xcalibur (Thermo Scientific).
2.6.2. GC-MS Analysis
The extracts were analyzed with a GC-MS-QP2010S (Shimadzu). The samples were prepared at 1 mg/mL dissolved in CHCl3. An AOC-20i/s autosampler was used for injection of the samples, with the injection temperature at 270 °C and split mode used (10.0 ratio), all via an Agilent DB-1HT (30 mm × 0.10 μm × 0.25 mm) column. The analysis started at 50 °C, where it was held for 5 min, then increased to 350 °C at a rate of 15 °C/min, where it was held for 20 min. GC-MS solution Version 4.20 (Shimadzu) was used to process the results and to apply the similarity search to a NIST library (2011).
2.7. Statistical Analysis
Data generated by LC-MS and GC-MS were processed through MZmine 2.5336 using the tabulated parameters (Table S2–S3). To filter and clean the LC-MS data for principal component analyses (PCA) and Volcano plots, a 1 × 104 blank cutoff was used (i.e., to remove background noise from signals with peak areas under 1 × 104), and the mass spectrometry data were filtered between m/z 135 and m/z 2000 with a retention time window of 0 to 10 min. Four technical injections per biological replicate were used and averaged, using a relative standard deviation (RSD) threshold value below 0.4 (i.e., to remove system variance).37 To filter the GC-MS data, the blank cutoff was 1 × 102, and the mass spectrometry data were filtered between m/z 19 and m/z 350 with a retention time window of 10.00 to 22.75 min with a 0.4 RSD cutoff. For generation of the PCA plots, Jupyter lab (Python) was used. Volcano plots were made by VolcaNoseR.38 Venn diagrams were made by using an available webTool (https://bioinformatics.psb.ugent.be/webtools/Venn/). To generate the PCA and volcano plots that display only the unique and/or upregulated features, the data acquired from MZmine were further filtered.36 The features (peak areas and appropriate m/z over retention time values) from bees fed sugar, as well as features from the specific diet samples (i.e., pollen, Chlorella, or spirulina), were all subtracted from the feature list acquired from bees fed the respective diets. Thus, the generated filtered feature list only contained the unique and or upregulated metabolites when the bees were fed on the pollen, Chlorella, or spirulina diets (Figure S3).
3. Results and Discussion
3.1. Diet Consumption and Honey Bee Growth Performance
Diet consumption was measured in caged honey bees fed sugar, pollen, Chlorella, and spirulina diets after 8 days of ad libitum feeding. Consumption is an important metric in feed comparison studies since the amount of diet consumed determines the pool of available nutrients. Of the protein-containing diets, consumption was highest for the pollen diet and lowest for the Chlorella diet (P < 0.0001). Overall diet consumption was as follows: sugar > pollen > spirulina > Chlorella (Figure 2). These results are consistent with our previous observations that bees consume less microalgae than pollen in similar experimental designs.29,33,39 Bees fed sugar had the lowest head weights, but there were no differences between bees fed pollen and microalgae (P = 0.0013). Similarly, the sugar diet produced the lowest thorax weights (P < 0.0001), but there were no differences among pollen-,Chlorella-, or spirulina-fed bees (Figure 2). Increases in head and thorax weights, respectively reflect head gland and flight muscle development, attributes that are central to honey bee colony fitness and productivity.8,40,41 The protein content of spirulina biomass is 60–66% and the protein content of Chlorella ranges from 38 to 48%.26 Further, the lipid content of spirulina ranges from 2 to 7%, whereas Chlorella ranges from 13 to 21%. Protein and lipid contents reported for a variety of pollens ranged from 2–60 and 2–20%, respectively.11,12 Taken together, the high macronutrient content and bioavailability of the microalgae diets may explain how reduced consumption led to bee growth characteristics that were similar to those produced by a natural pollen diet. Since honey bees do not appear to consume pollen based on its nutritional quality,42 it can be postulated that non-nutrient components might underlie pollen’s attractiveness. Indeed, pollen phagostimulants are solvent-extractable in sufficient quantities to increase the consumption of artificial diets by honey bees, although the specific compounds involved are largely unknown.43
Figure 2.
Effects of pollen and microalgae diets on honey bee feed consumption and growth performance after 8 days. (A) Diet consumption. (B) Average head weight. (C) Average thorax weight. Error bars represent standard error (SE). Columns with different letters are significantly different at α = 0.05.
3.2. Nutritionally Regulated Gene Expression
Nutritional genomics approaches can indicate how nutrients impact gene expression as well as measure an organism’s response to changes in feed composition.44 Nutritionally regulated gene expression was measured in bees fed the different diets. In honey bees, Vitellogenin (Vg) is a central storage and regulatory protein that has been used as a nutritional biomarker since protein and mRNA titers are linked to diet quality.10,45,46vg mRNA expression was highest in pollen- and Chlorella-fed bees and was lowest in sugar-fed bees (P = 0.0010). Overall vg expression was as follows: pollen = Chlorella > spirulina > sugar, with Chlorella-fed bees trending toward higher vg levels than pollen-fed bees (Figure 3). Antioxidant enzyme gene expression is associated with longevity in honey bees47 and is nutritionally regulated.48 Bees fed spirulina had significantly higher transcript levels of the antioxidant genes catalase (P < 0.0001) and superoxide dismutase (P < 0.0001). Heat shock proteins are highly conserved and have important roles in protecting cells from thermal-induced (including cold) and oxidative stresses,49 as well as innate immune functions.50 Bees fed sugar and spirulina had higher levels of heat shock protein 70 (hsp70) (P = 0.0073). Spirulina-fed bees had the highest levels of heat shock protein 90 (hsp90) (P < 0.0001) (Figure 3).
Figure 3.
Gene expression profiles of honey bees fed pollen and microalgae diets. General nutrition status was assessed by quantifying mRNA transcript levels of vitellogenin (vg), a vital nutritional storage protein. Stress response potential was measured by quantifying transcript levels of the antioxidant genes catalase and superoxide dismutase, as well as heat shock proteins 70 (hsp70) and 90 (hsp90). For each gene, columns with different letters are significantly different at α = 0.05.
3.3. Untargeted Metabolomics Via LC-MS and GC-MS
Untargeted metabolomics has proven useful to identify bee metabolites,51 and thus, a combination of both LC-MS and GC-MS was used to study the effects of different diets on the bee metabolome. These techniques generate mass-to-charge ratio (m/z) and retention time (RT) pairs, hereafter referred to as features.52 The generated feature lists acquired by LC-MS and GC-MS were compared using a suite of computational tools and plotting techniques to analyze and explore honey bee metabolome compositions.
LC-MS analyses revealed 248 features that were shared among bees fed the different diets, and GC-MS analyses revealed 87 shared features (Figure 4). In the case of the GC-MS data, a peak area threshold of 500 was applied to each feature. For both instruments, features were determined prior to applying a subtractive metabolomics approach.
Figure 4.
Venn diagrams showing feature distributions of honey bee metabolites from LC-MS and GC-MS analyses. Diagrams represent the number of features belonging to bees fed the different diets. Each feature is defined as an m/z value and retention time pair. These orthogonal approaches revealed a high number of shared features across bees fed the diets (248 and 87, respectively).
To explore bee metabolomes among diet treatment groups, principal component analyses (PCA) were performed. We also used a subtractive approach to represent unique and/or upregulated features of bee metabolomes that responded to the pollen, Chlorella, or spirulina diets. To accomplish this, features of sugar-fed bees and features from the respective diet extracts were subtracted from the total feature list (Figure S3) While there are pros and cons to this approach (Table S8), it is a pragmatic way to focus attention on certain features uniquely impacted in the experimental bees. For example, unique and/or upregulated features of pollen-fed bees were determined by subtracting sugar-fed bee features and features that were specific to the pollen extract itself (Figure S3). As displayed in the PCA plots (Figures 5 and 6), metabolomes of bees fed pollen and either Chlorella or spirulina had a distinct separation regardless of using an LC-MS or GC-MS approach. Interestingly, bees fed Chlorella and spirulina exhibited some overlap despite the taxonomic divergence of the microalgae biomass used for the diets. Chlorella is a eukaryotic microalga and spirulina is derived from Arthrospira, a genus of prokaryotic cyanobacteria (blue-green algae). Consistent with our PCA results, a large-scale analysis of mass spectrometry data from diverse algae samples revealed similar clustering patterns among marine and freshwater algae specimens when compared to groups of marine and terrestrial actinobacteria and lichens.53
Figure 5.
Principal component analysis (PCA) plots of untargeted honey bee metabolites acquired through LC-MS and GC-MS. Each data point represents a biological replicate (i.e., eight bees pooled from an independent cage with each sample analyzed in four technical replicates). Bees fed pollen, Chlorella, and spirulina had distinct separations that were evident by both the LC-MS and GC-MS. Chlorella- and spirulina-fed bees exhibited overlapping metabolome profiles and were more similar to each other than to the metabolomes of bees fed pollen.
Figure 6.
Principal component analysis (PCA) plots of untargeted honey bee metabolites acquired through LC-MS (scores plots (A) and (C)) and GC-MS (scores plots (B) and (D)) after applying a subtractive metabolomics approach. To better evaluate bee metabolomic responses, the features originating from extracts of pollen, Chlorella, or spirulina diets were subtracted from the respective feature lists of the bees fed those diets (scores plots (A) and (B)). Further, features of sugar-fed bees were separately subtracted (scores plots (C) and (D)). Bees fed Chlorella and spirulina exhibited similarities to pollen-fed bees, but their metabolomes were more similar to each other than to pollen.
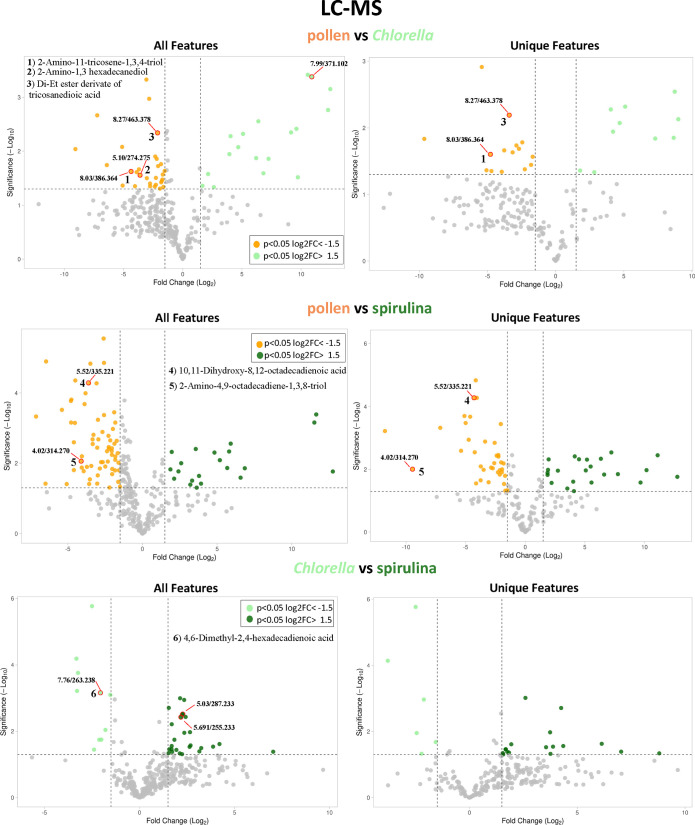
Volcano plots were generated based on LC-MS and GC-MS data (Figures 7 and S4, respectively). Each dot on the plots represents a feature (i.e., RT/m/z value). All features that exhibited > 1.5-fold change and P < 0.05 were considered statistically significant. Subtractive analyses (Figure S3) removed features associated with diet extracts and indicated bee metabolites that were differentially expressed upon consumption of the different diets (Figure 7, Unique Features panel). In general, LC-MS and GC-MS volcano plots revealed high similarities among bees that were fed pollen, Chlorella, and spirulina. A nonsubtractive approach across LC and GC-MS volcano plot data showed that the two algae species are highly similar due to a majority of the features being shared and/or nonsignificant (Figure 7). Although pollen-fed bees exhibited features that were significantly different from microalgae-fed bees, many features were shared among the diets. In addition, when the subtractive approach was utilized, even fewer features remained, which indicated that some differences originated from the diets and not from the metabolomic response of the bee. When the two algae species were compared after subtraction, only a few features remained, indicating that metabolomes of bees fed Chlorella are highly similar to bees fed spirulina. Applying the subtractive approach to LC-MS data, we identified selected features that may have contributed to diet effects on bee growth performance and gene expression (Figure 7 and Table S5). Putative identification was based on accurate mass searches in the Dictionary of Natural Products54 and aimed to highlight the potential of volcano plot data for addressing diet deficiencies. For example, the feature 8.27/463.378 (RT/m/z) was putatively identified as a di-Et ester derivate of tricosanedioic acid; a fatty acid that was upregulated in bees fed pollen but not in bees fed Chlorella when all features were examined (Figure 7, pollen vs Chlorella, All features plot). Upon subtraction of diet extract features, this feature remained, suggesting a potentially important role in the nutritional value of the pollen diet (Figure 7, pollen vs Chlorella, Unique features plot). Similarly, the feature 5.52/335.221 (RT/m/z) was putatively identified as 10,11-dihydroxy-8,12-octadecadienoic acid, a fatty acid that was upregulated in bees fed pollen but not in bees fed spirulina after subtractive analyses (Figure 7, pollen vs Chlorella, Unique features plot). Another use case for the subtractive approach is the feature 5.10/274.275 (RT/m/z) found in bees fed the pollen diet, putatively identified as 2-amino-1,3 hexadecanediol. Upon subtraction of the diet features, this putatively identified compound was not statistically different, suggesting that its signal primarily originated from the diet (Figure 7, pollen vs Chlorella, Unique features plot). These results highlight the potential of untargeted metabolomics for artificial diet development in honey bees. By comparison to pollen, the bee’s natural source of macro- and micronutrients, feed could ultimately be tailored to reproduce the metabolomes of pollen-fed bees. This approach could further be applied to optimize feed ingredients that support seasonal and regional nutritional requirements of honey bees, which vary based on interactions between available pollen forage and seasonal colony demography.55
Figure 7.
Volcano plots of honey bees fed different diets generated using untargeted LC-MS metabolomics. “All features” plots represent all RT/m/z values processed. The “Unique features” plots only show uniquely expressed and/or upregulated features produced by the bees. On these plots, a subtractive approach was used, thus the features that came from the bees fed only sugar and the features originating in the pollen and microalgae diet extracts were subtracted. A selection of RT/m/z pairs (highlighted dots) that may contribute to diet effects was putatively identified. These features are further discussed in Table S5. Overall, the LC-MS volcano plots revealed high similarities among the metabolomes of bees that were fed pollen and microalgae.
Unique features of Chlorella- and/or spirulina-fed bee metabolomes warrant further investigation to better understand the effects of specific algal metabolites on bee physiology, particularly since such compounds are not naturally encountered. Some metabolites derived from algae appear to have ecological roles as allelochemicals, including compounds that may inhibit competing microorganisms. These allelochemicals may also serve as protection against aquatic invertebrates and their larvae.56 Commercially grown strains of Chlorella and spirulina are generally recognized as safe for human and animal consumption. However, mass produced strains can coexist in the same habitats as potentially toxic algae, and if so, such biomass can become contaminated with toxins produced by other microorganisms.57 Therefore, future work could incorporate screening for known algal toxins and their metabolites in bees, especially when testing novel strains and wild-harvested biomass. On the other hand, microalgae are a rich source of natural products with unique structures that also have potential as therapeutic agents.53 Notably, a sulphated polysaccharide derived from the red alga, Porphyridium spp., led to decreased parasite loads and decreased honey bee mortality due to infection by the gut parasite Nosema ceranae.58 In our study, microalgae diets led to increased mRNA levels of antioxidant enzymes and heat shock proteins. These genes apparently respond to diet quality in honey bees and may be differentially regulated by certain algal metabolites. Consistent with our results, dietary spirulina supplementation led to increased antioxidant gene expression and total antioxidant capacity in rainbow trout.59 It remains to be determined if prolonged upregulation of antioxidant gene pathways is beneficial to bees or if it presents a metabolic cost. Nevertheless, further studies could lead to the identification of potentially health-modulating metabolites for therapeutic development in honey bees.
3.4. Comparative Quantification of Select Metabolites by LC-MS and GC-MS
Where pure reference standards were available, select metabolites were identified and quantified by LC-MS and GC-MS. Specifically, LC-MS data were used to identify metabolites through comparisons to available standard materials. Extracted ion chromatograms (XIC) of each compound were examined (Figure S5–S11). The relative abundances of the compounds were calculated based on their average peak area (Table S6). Accurate mass measurements, retention time, and UV absorptions were used to confirm identification60 of the following compounds: linoleic acid, α-linolenic acid, zeaxanthin, lutein, quinic acid, and α-tocopherol (Figure S12). Due to the low ionization of β-carotene standard under the same experimental parameters using ESI, the identification of β-carotene was recorded as “putative,” as opposed to the previously mentioned compounds, which were all first level compound identifications.60 Linoleic acid and α-linolenic acid are two polyunsaturated fatty acids that are considered essential for honey bees.21−23 The Chlorella diet led to the highest levels of linoleic acid, and the levels of α-linolenic acid were comparable to pollen-fed bees (Figure 8). Spirulina-fed bees accumulated the lowest levels of both essential fatty acids (Figure 8). The abundance of pollen-derived polyunsaturated fatty acids is positively correlated with abdominal vg expression.21−23 Consistent with linoleic and α-linolenic acid levels, bees fed pollen and Chlorella had significantly higher abdominal vg mRNA levels than spirulina-fed bees (Figure 3). Lipid accumulation in green algae, such as Chlorella, is well known to exceed that of spirulina, which is renowned for its protein content.26 Based on fatty acid composition and vitellogenin expression, our results suggest that Chlorella and related eukaryotic algae are promising lipid sources for bee diet development. Nevertheless, spirulina is a natural source of many phytochemicals that occur in pollen, including carotenoids, which are potent antioxidants and vitamin precursors that modulate gene activity in a variety of animals.61 For instance, diets containing the carotenoid, β-carotene, extracted from spirulina led to increased expression of superoxide dismutase and catalase as well as increased total antioxidant capacity in Nile tilapia.62 In our study, spirulina-fed bees accumulated significantly higher levels of β-carotene (Figure 8), which may explain the observed increases in catalase, superoxide dismutase, and heat shock protein 90 (Figure 3). Other carotenoids, lutein and zeaxanthin, were only identified in microalgae-fed bees (Figure 8). Most carotenoids occurring in higher plants, plus a variety of additional carotenoid structures, are produced by microalgae (e.g., lutein, zeaxanthin, astaxanthin, and fucoxanthin).61 Our results indicate that microalgae are promising natural sources of carotenoids for use in bee feed. Overall, the targeted LC-MS results demonstrate the utility of metabolomics to quantify and compare the relative abundances of beneficial compounds among bees fed different diet formulations.
Figure 8.
Targeted LC-MS metabolite analyses of bees fed pollen and microalgae diets. For each compound, columns with different letters are significantly different at α = 0.05.
For metabolite identification through GC-MS, total ion chromatograms (TICs) were used, and the peaks were compared to a NIST 2011 library for similarity match based on their ion fragmentation. The putatively identified metabolites all possessed higher than 90% similarity scores (Figure 9). The prominent compounds identified by this technique were mainly fatty alcohols and hydrocarbons (Figure S12). Insects rely on blends of waxy hydrocarbons as pheromones for mating and nestmate recognition.63 Insect hydrocarbons are influenced by nutrition, as experimentally demonstrated using different host plants and artificial diets.64 In honey bees, currently unknown factors in the colony environment may contribute directly or indirectly to molecular processes regulating pheromone synthesis.65 Thus, it is plausible that local nutrition could contribute to development of similar pheromone profiles among individuals. To test this hypothesis within the context of our study, relative metabolite abundances were calculated based on average peak areas of compounds detected by GC-MS (Table S7). The abundances of 1-heneicosanol, n-nonadecanol-1, n-tetracosanol, and docosane were significantly impacted by diet. Additionally, two fatty acids, n-hexadecanoic (i.e., palmitic) acid and erucic acid, were significantly impacted by diet. Overall, the GC-MS results suggest that nutrition can influence honey bee hydrocarbon and fatty acid profiles, which may have future utility as dietary or health biomarkers.
Figure 9.

Putatively identified metabolites from GC-MS analyses of bees fed pollen and microalgae diets. A comparison to the NIST 2011 compound library was used for metabolite identifications.
4. Conclusions
Malnutrition is a serious threat to managed honey bees that is exacerbated by landscape agricultural intensification and climate change. As beekeeper reliance on artificial diets increases, there is a growing need for efficacious and sustainable feed that can support bee nutrition across seasons and diverse management conditions. Current methods for honey bee diet development involve measuring a few pre-selected biochemical and/or physiological parameters to test the effects of diet formulations on growth performance. However, higher-resolution methods that can directly target diet deficiencies are necessary. Here, we applied mass spectrometry-based metabolomics to better understand the nutritional and metabolic impacts of microalgae-based artificial diets relative to a natural pollen diet. The use of both LC-MS and GC-MS methods provided coverage across a broad range of metabolite groups and overcame the individual limitations associated with both approaches. Pollen and microalgae diets had similar nutritional and metabolomic impacts in bees, especially after subtraction of unique diet features. Chlorella provided more essential fatty acids than spirulina, which likely contributed to its enhanced nutritional value. Nevertheless, spirulina is a promising source of bioavailable protein and phytochemicals, notably carotenoids, that may augment stress response pathways in bees. We conclude that the tested microalgae have potential as sustainable bee feed additives and health-modulating natural products. Finally, this study showed that metabolomics approaches have significant potential to achieve precision nutrition in honey bees as well as identify beneficial attributes of natural and artificial diets.
Acknowledgments
This work was funded by Agriculture and Food Research Initiative grant no. 2021-67013-33556 from the USDA National Institute of Food and Agriculture and Project Apis m. grant no. 58-6050-1-003.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.2c02583.
Overview of the feeding experiment; honey bee metabolite extraction workflow; process of feature filtering; volcano plots of GC-MS data; extracted ion chromatograms of zeaxanthin, lutein, linolenic acid, quinic acid, α-tocopherol, β-carotene, and linoleic acid; structures of all identified compounds; table of extraction; MZmine processing parameters for LC-MS and GC-MS; primers used in this study; identified features from LC-MS volcano plot analysis; relative abundance of compounds identified through LC-MS and GC-MS; and benefits and limitations of the subtractive approach (PDF)
Author Contributions
§ V.A.R. and K.B.C. contributed equally to this study.
The authors declare no competing financial interest.
Supplementary Material
References
- Chopra S. S.; Bakshi B. R.; Khanna V. Economic Dependence of U.S. Industrial Sectors on Animal-Mediated Pollination Service. Environ. Sci. Technol. 2015, 49, 14441–51. 10.1021/acs.est.5b03788. [DOI] [PubMed] [Google Scholar]
- Kulhanek K.; Steinhauer N.; Rennich K.; Caron D. M.; Sagili R. R.; Pettis J. S.; Ellis J. D.; Wilson M. E.; Wilkes J. T.; Tarpy D. R.; Rose R.; Lee K.; Rangel J.; vanEngelsdorp D. A national survey of managed honey bee 2015–2016 annual colony losses in the USA. J. Apic. Res. 2017, 56, 328–340. 10.1080/00218839.2017.1344496. [DOI] [Google Scholar]
- Dolezal A. G.; Toth A. L. Feedbacks between nutrition and disease in honey bee health. Curr. Opin. Insect Sci. 2018, 26, 114–119. 10.1016/j.cois.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Dolezal A. G.; Carrillo-Tripp J.; Judd T. M.; Allen Miller W.; Bonning B. C.; Toth A. L. Interacting stressors matter: diet quality and virus infection in honeybee health. R. Soc. Open Sci. 2019, 6, 181803. 10.1098/rsos.181803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrandi-Hoffman G.; Chen Y. Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci. 2015, 10, 170–176. 10.1016/j.cois.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Di Pasquale G.; Alaux C.; Le Conte Y.; Odoux J.-F.; Pioz M.; Vaissière B. E.; Belzunces L. P.; Decourtye A. Variations in the Availability of Pollen Resources Affect Honey Bee Health. PLoS one 2016, 11, e0162818 10.1371/journal.pone.0162818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodschneider R.; Crailsheim K. Nutrition and health in honey bees. Apidologie 2010, 41, 278–294. 10.1051/apido/2010012. [DOI] [Google Scholar]
- Crailsheim K. The protein balance of the honey bee worker. Apidologie 1990, 21, 417–429. 10.1051/apido:19900504. [DOI] [Google Scholar]
- Huang Z. Pollen nutrition affects honey bee stress resistance. Terr. Arthropod Rev. 2012, 5, 175–189. 10.1163/187498312X639568. [DOI] [Google Scholar]
- Alaux C.; Dantec C.; Parrinello H.; Le Conte Y. Nutrigenomics in honey bees: digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genom. 2011, 12, 496 10.1186/1471-2164-12-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulston T. H.; Cane J. H.; Buchmann S. L. What governs protein content of pollen: pollinator preferences, pollen–pistil interactions, or phylogeny?. Ecol. Monogr. 2000, 70, 617–643. 10.2307/2657188. [DOI] [Google Scholar]
- Roulston T. H.; Cane J. H. Pollen nutritional content and digestibility for animals. Plant Syst. Evol. 2000, 222, 187–209. 10.1007/BF00984102. [DOI] [Google Scholar]
- Donkersley P.; Rhodes G.; Pickup R. W.; Jones K. C.; Wilson K. Honeybee nutrition is linked to landscape composition. Ecol. Evol. 2014, 4, 4195–206. 10.1002/ece3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponsler D. B.; Johnson R. M. Honey bee success predicted by landscape composition in Ohio, USA. PeerJ 2015, 3, e838 10.7717/peerj.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal A. G.; Carrillo-Tripp J.; Miller W. A.; Bonning B. C.; Toth A. L. Intensively Cultivated Landscape and Varroa Mite Infestation Are Associated with Reduced Honey Bee Nutritional State. PLoS one 2016, 11, e0153531 10.1371/journal.pone.0153531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbergen A. J.; Initiative tI. P. Threats to an ecosystem service: pressures on pollinators. Front. Ecol. Environ. 2013, 11, 251–259. 10.1890/120126. [DOI] [Google Scholar]
- Bartomeus I.; Ascher J. S.; Wagner D.; Danforth B. N.; Colla S.; Kornbluth S.; Winfree R. Climate-associated phenological advances in bee pollinators and bee-pollinated plants. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 20645–20649. 10.1073/pnas.1115559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settele J.; Bishop J.; Potts S. G. Climate change impacts on pollination. Nat. Plants 2016, 2, 16092 10.1038/nplants.2016.92. [DOI] [PubMed] [Google Scholar]
- Ricigliano V. A.; W S.; Oliver R. Effects of different artificial diets on commercial honey bee colony performance, health biomarkers, and gut microbiota. BMC Vet. Res. 2022, 18, 52 10.1186/s12917-022-03151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordyke E. R.; Ellis J. D. Reviewing the Efficacy of Pollen Substitutes as a Management Tool for Improving the Health and Productivity of Western Honey Bee (Apis mellifera) Colonies. Front. Sustainable Food Syst. 2021, 5, 772897 10.3389/fsufs.2021.772897. [DOI] [Google Scholar]
- Avni D.; Hendriksma H. P.; Dag A.; Uni Z.; Shafir S. Nutritional aspects of honey bee-collected pollen and constraints on colony development in the eastern Mediterranean. J. Insect Physiol. 2014, 69, 65–73. 10.1016/j.jinsphys.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Arien Y.; Dag A.; Zarchin S.; Masci T.; Shafir S. Omega-3 deficiency impairs honey bee learning. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 15761. 10.1073/pnas.1517375112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener J.; Jakop U.; Schiller J.; Müller K. The membrane phospholipid composition of honeybee (Apis mellifera) workers reflects their nutrition, fertility, and vitellogenin stores. Insectes Soc. 2018, 65, 381–391. 10.1007/s00040-018-0623-x. [DOI] [Google Scholar]
- Liao L.-H.; Pearlstein D. J.; Wu W.-Y.; Kelley A. G.; Montag W. M.; Hsieh E. M.; Berenbaum M. R. Increase in longevity and amelioration of pesticide toxicity by natural levels of dietary phytochemicals in the honey bee, Apis mellifera. PLoS one 2020, 15, e0243364 10.1371/journal.pone.0243364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylanc V.; Falcão S. I.; Ertosun S.; Vilas-Boas M. From the hive to the table: Nutrition value, digestibility and bioavailability of the dietary phytochemicals present in the bee pollen and bee bread. Trends Food Sci. Technol. 2021, 109, 464–481. 10.1016/j.tifs.2021.01.042. [DOI] [Google Scholar]
- Madeira M. S.; Cardoso C.; Lopes P. A.; Coelho D.; Afonso C.; Bandarra N. M.; Prates J. A. M. Microalgae as feed ingredients for livestock production and meat quality: A review. Livest. Sci. 2017, 205, 111–121. 10.1016/j.livsci.2017.09.020. [DOI] [Google Scholar]
- Ricigliano V. A. Microalgae as a promising and sustainable nutrition source for managed honey bees. Arch. Insect Biochem. Physiol. 2020, 104, e21658 10.1002/arch.21658. [DOI] [PubMed] [Google Scholar]
- Jehlík T.; Kodrík D.; Krištůfek V.; Koubová J.; Sábová M.; Danihlík J.; Tomčala A.; Čapková Frydrychová R. Effects of Chlorella sp. on biological characteristics of the honey bee Apis mellifera. Apidologie 2019, 50, 564–577. 10.1007/s13592-019-00670-3. [DOI] [Google Scholar]
- Ricigliano V. A.; Simone-Finstrom M. Nutritional and prebiotic efficacy of the microalga Arthrospira platensis (spirulina) in honey bees. Apidologie 2020, 51, 898–910. 10.1007/s13592-020-00770-5. [DOI] [Google Scholar]
- Dettmer K.; Aronov P. A.; Hammock B. D. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.; Xu G. Mass-spectrometry-based metabolomics analysis for foodomics. TrAC, Trends Anal. Chem. 2013, 52, 36–46. 10.1016/j.trac.2013.09.005. [DOI] [Google Scholar]
- Chakrabarti P.; Morré J. T.; Lucas H. M.; Maier C. S.; Sagili R. R. The omics approach to bee nutritional landscape. Metabolomics 2019, 15, 127 10.1007/s11306-019-1590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricigliano V. A.; Dong C.; Richardson L. T.; Donnarumma F.; Williams S. T.; Solouki T.; Murray K. K. Honey Bee Proteome Responses to Plant and Cyanobacteria (blue-green algae) Diets. ACS Food Sci. Technol. 2021, 1, 17–26. 10.1021/acsfoodscitech.0c00001. [DOI] [Google Scholar]
- Flores-Bocanegra L.; Augustinović M.; Raja H. A.; Kurina S. J.; Maldonado A. C.; Burdette J. E.; Falkinham J. O. 3rd; Pearce C. J.; Oberlies N. H. Cytotoxic and antimicrobial drimane meroterpenoids from a fungus of the Stictidaceae (Ostropales, Ascomycota). Tetrahedron Lett. 2021, 68, 152896 10.1016/j.tetlet.2021.152896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T.; Raja H. A.; Day C. S.; Chen W.-L.; Swanson S. M.; Oberlies N. H. Greensporones: Resorcylic Acid Lactones from an Aquatic Halenospora sp. J. Nat. Prod. 2014, 77, 2088–2098. 10.1021/np500497r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T.; Castillo S.; Villar-Briones A.; Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar L. K.; Kvalheim O. M.; Cech N. B. Hierarchical cluster analysis of technical replicates to identify interferents in untargeted mass spectrometry metabolomics. Anal. Chim. Acta 2018, 1021, 69–77. 10.1016/j.aca.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J.; Luijsterburg M. S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560 10.1038/s41598-020-76603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricigliano V. A.; Ihle K. E.; Williams S. T. Nutrigenetic comparison of two Varroa-resistant honey bee stocks fed pollen and spirulina microalgae. Apidologie 2021, 52, 873–886. 10.1007/s13592-021-00877-3. [DOI] [Google Scholar]
- DeGrandi-Hoffman G.; Chen Y.; Huang E.; Huang M. H. The effect of diet on protein concentration, hypopharyngeal gland development and virus load in worker honey bees (Apis mellifera L.). J. Insect Physiol. 2010, 56, 1184–1191. 10.1016/j.jinsphys.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Hendriksma H. P.; Pachow C. D.; Nieh J. C. Effects of essential amino acid supplementation to promote honey bee gland and muscle development in cages and colonies. J. Insect Physiol. 2019, 117, 103906 10.1016/j.jinsphys.2019.103906. [DOI] [PubMed] [Google Scholar]
- Corby-Harris V.; Snyder L.; Meador C.; Ayotte T. Honey bee (Apis mellifera) nurses do not consume pollens based on their nutritional quality. PLoS one 2018, 13, e0191050 10.1371/journal.pone.0191050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgett R. J.; Kirk W. D. J.; Drijfhout F. P. The use of a within-hive replication bioassay method to investigate the phagostimulatory effects of pollen, bee bread and pollen extracts, on free-flying honey bee colonies. Apidologie 2015, 46, 315–325. 10.1007/s13592-014-0324-z. [DOI] [Google Scholar]
- Nowacka-Woszuk J. Nutrigenomics in livestock—recent advances. J. Appl. Genet. 2020, 61, 93–103. 10.1007/s13353-019-00522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corby-Harris V.; Jones B. M.; Walton A.; Schwan M. R.; Anderson K. E. Transcriptional markers of sub-optimal nutrition in developing Apis mellifera nurse workers. BMC Genom. 2014, 15, 134 10.1186/1471-2164-15-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricigliano V. A.; Mott B. M.; Maes P. W.; Floyd A. S.; Fitz W.; Copeland D. C.; Meikle W. G.; Anderson K. E. Honey bee colony performance and health are enhanced by apiary proximity to US Conservation Reserve Program (CRP) lands. Sci. Rep. 2019, 9, 4894 10.1038/s41598-019-41281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona M.; Hughes K. A.; Weaver D. B.; Robinson G. E. Gene expression patterns associated with queen honey bee longevity. Mech. Ageing Dev. 2005, 126, 1230–1238. 10.1016/j.mad.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Li C.; Xu B.; Wang Y.; Yang Z.; Yang W. Protein content in larval diet affects adult longevity and antioxidant gene expression in honey bee workers. Entomol. Exp. Appl. 2014, 151, 19–26. 10.1111/eea.12167. [DOI] [Google Scholar]
- Moura C. S.; Lollo P. C. B.; Morato P. N.; Amaya-Farfan J. Dietary Nutrients and Bioactive Substances Modulate Heat Shock Protein (HSP) Expression: A Review. Nutrients 2018, 10, 683. 10.3390/nu10060683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsan M. F.; Gao B. Heat shock protein and innate immunity. Cell. Mol. Immunol. 2004, 1, 274–279. [PubMed] [Google Scholar]
- Kešnerová L.; Mars R. A. T.; Ellegaard K. M.; Troilo M.; Sauer U.; Engel P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467 10.1371/journal.pbio.2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinaixa M.; Schymanski E. L.; Neumann S.; Navarro M.; Salek R. M.; Yanes O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. TrAC, Trends Anal. Chem. 2016, 78, 23–35. 10.1016/j.trac.2015.09.005. [DOI] [Google Scholar]
- Luzzatto-Knaan T.; Garg N.; Wang M.; Glukhov E.; Peng Y.; Ackermann G.; Amir A.; Duggan B. M.; Ryazanov S.; Gerwick L.; Knight R.; Alexandrov T.; Bandeira N.; Gerwick W. H.; Dorrestein P. C. Digitizing mass spectrometry data to explore the chemical diversity and distribution of marine cyanobacteria and algae. eLife 2017, 6, e24214 10.7554/eLife.24214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictionary of Natural Products, Buckingham J., Eds.; Taylor & Francis Group: London, 2021. [Google Scholar]
- DeGrandi-Hoffman G.; Gage S. L.; Corby-Harris V.; Carroll M.; Chambers M.; Graham H.; Watkins deJong E.; Hidalgo G.; Calle S.; Azzouz-Olden F.; Meador C.; Snyder L.; Ziolkowski N. Connecting the nutrient composition of seasonal pollens with changing nutritional needs of honey bee (Apis mellifera L.) colonies. J. Insect Physiol. 2018, 109, 114–124. 10.1016/j.jinsphys.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Berry J. P.; Gantar M.; Perez M. H.; Berry G.; Noriega F. G. Cyanobacterial toxins as allelochemicals with potential applications as algaecides, herbicides and insecticides. Mar. Drugs. 2008, 6, 117–146. 10.3390/md6020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshagauer S.; Kraemer K.; Somoza V. The True Value of Spirulina. J. Agric. Food Chem. 2020, 68, 4109–4115. 10.1021/acs.jafc.9b08251. [DOI] [PubMed] [Google Scholar]
- Roussel M.; Villay A.; Delbac F.; Michaud P.; Laroche C.; Roriz D.; El Alaoui H.; Diogon M. Antimicrosporidian activity of sulphated polysaccharides from algae and their potential to control honeybee nosemosis. Carbohydr. Polym. 2015, 133, 213–220. 10.1016/j.carbpol.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Teimouri M.; Yeganeh S.; Mianji G. R.; Najafi M.; Mahjoub S. The effect of Spirulina platensis meal on antioxidant gene expression, total antioxidant capacity, and lipid peroxidation of rainbow trout (Oncorhynchus mykiss). Fish. Physiol. Biochem. 2019, 45, 977–986. 10.1007/s10695-019-0608-3. [DOI] [PubMed] [Google Scholar]
- Sumner L. W.; Amberg A.; Barrett D.; Beale M. H.; Beger R.; Daykin C. A.; Fan T. W. M.; Fiehn O.; Goodacre R.; Griffin J. L.; Hankemeier T.; Hardy N.; Harnly J.; Higashi R.; Kopka J.; Lane A. N.; Lindon J. C.; Marriott P.; Nicholls A. W.; Reily M. D.; Thaden J. J.; Viant M. R. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes A. C.; Amaro H. M.; Malcata F. X. Microalgae as sources of carotenoids. Mar. Drugs 2011, 9, 625–644. 10.3390/md9040625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaan M. S.; Mohammady E. Y.; Soaudy M. R.; Sabae S. A.; Mahmoud A. M. A.; El-Haroun E. R. Comparative study on the effect of dietary β-carotene and phycocyanin extracted from Spirulina platensis on immune-oxidative stress biomarkers, genes expression and intestinal enzymes, serum biochemical in Nile tilapia, Oreochromis niloticus. Fish Shellfish Immunol. 2021, 108, 63–72. 10.1016/j.fsi.2020.11.012. [DOI] [PubMed] [Google Scholar]
- Chung H.; Carroll S. B. Wax, sex and the origin of species: Dual roles of insect cuticular hydrocarbons in adaptation and mating. BioEssays 2015, 37, 822–830. 10.1002/bies.201500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingleby F. C. Insect Cuticular Hydrocarbons as Dynamic Traits in Sexual Communication. Insects 2015, 6, 732–742. 10.3390/insects6030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernier C. L.; Krupp J. J.; Marcus K.; Hefetz A.; Levine J. D.; Ben-Shahar Y. The cuticular hydrocarbon profiles of honey bee workers develop via a socially-modulated innate process. eLife 2019, 8, e41855 10.7554/eLife.41855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.