Abstract
The polymers of fructose, levan and inulin, as well as sucrose and raffinose, are substrates for the product of the fruA gene of Streptococcus mutans GS-5. The purpose of this study was to characterize the DNA immediately flanking fruA, to explore the regulation of expression of fruA by the carbohydrate source, and to begin to elucidate the molecular basis for differential expression of the gene. Located 3′ to fruA was an open reading frame (ORF) with similarity to β-fructosidases which was cotranscribed with fruA. A transcriptional initiation site, located an appropriate distance from an extended −10-like promoter, was mapped at 165 bp 5′ to the fruA structural gene. By the use of computer algorithms, two overlapping, stable stem-loop sequences with the potential to function as rho-independent terminators were found in the 5′ untranslated region. Catabolite response elements (CREs), which have been shown to govern carbon catabolite repression (CCR) by functioning as negative cis elements in gram-positive bacteria, were located close to the promoter. The levels of production of fruA mRNA and FruA were elevated in cells growing on levan, inulin, or sucrose as the sole carbohydrate source, and repression was observed when cells were grown on readily metabolizable hexoses. Deletion derivatives containing fusions of fruA promoter regions, lacking sequences 5′ or 3′ to the promoter, and a promoterless chloramphenicol acetyltransferase gene were used (i) to demonstrate the functionality of the promoter mapped by primer extension, (ii) to demonstrate that CCR of the fru operon requires the CRE that is located 3′ to the promoter region, and (iii) to provide preliminary evidence that supports the involvement of an antitermination mechanism in fruA induction.
A variety of oral bacteria can utilize sucrose to produce extracellular polysaccharides composed of β2,1- and β2,6-linked fructosyl moieties. For example, Streptococcus salivarius and Actinomyces naeslundii produce fructosyltransferases (FTFs) that synthesize levan-type polymers, which are rich in β2,6 linkages, whereas the FTFs of Streptococcus mutans and Streptococcus sanguis produce inulin-type polymers, composed predominantly of β2,1-linked fructose (4). The ability to synthesize fructan polymers within oral biofilms is believed to allow the organisms to capture a greater proportion of dietary sucrose by converting it to a nondiffusing, extracellular storage polysaccharide which can be catabolized when exogenous sources are lacking (7, 21, 25). Consistent with this model is the observation that organisms which produce fructans usually possess enzymes that are capable of degrading fructan polymers.
S. mutans produces a secreted fructan hydrolase which is encoded by the fruA gene (13). FruA releases fructose from levan, inulin, and raffinose, and it cleaves sucrose into glucose and fructose (13). Analysis of the deduced amino acid sequence of FruA revealed that it is synthesized as a 158-kDa protein, containing a signal sequence typical of gram-positive bacteria, which is presumably cleaved to yield a 155-kDa mature, secreted enzyme (9). The central one-third of the FruA enzyme exhibits similarity to fructan-hydrolyzing enzymes from eubacteria and lower eukaryotes. The N and C termini exhibit no significant similarity to other known proteins, with one exception: an LPXTG anchoring sequence is found at the C terminus, consistent with the finding that the enzyme can be found in significant quantities in association with the cell surface under certain environmental conditions (11). Isogenic mutants were used in a program-fed rat caries model to show that FruA is a virulence determinant that contributes to the progression of dental caries (8).
Results from early studies on the control of fructan hydrolase expression by S. mutans indicated that the activity of this enzyme was increased in supernatant fluids of cell cultures grown on fructans and sucrose (30, 55) compared with that of cells grown on glucose. Cultivation of S. mutans with combinations of both inducing substrates and readily metabolizable hexoses yielded levels of activity similar to those obtained with hexose alone. Also, fructan hydrolase activity was found to be higher when fructose was provided as the limiting carbohydrate in continuous culture than when glucose was the growth carbohydrate (30). More recently, it has been found that although many strains of S. mutans produce only FruA, some strains of S. mutans can produce two fructan-hydrolyzing activities. The major one is FruA, and the second activity is an inulinase with no detectable activity on levans (33). The environmental factors specifically regulating fruA expression and the molecular basis for the apparent differential expression of fructan hydrolase activity remain to be elucidated. In a preliminary report, we noted that fruA expression appeared to be inducible by inulin and was repressible by readily metabolizable hexoses (10). This communication presents the results of a study designed to examine the transcriptional organization of the fruA operon, which includes a second gene with similarity to fructosidase genes, and to uncover the basic molecular mechanisms for control of induction and repression of fruA.
MATERIALS AND METHODS
Bacterial strains and media.
S. mutans strains were maintained on brain heart infusion (BHI) agar (Difco) containing antibiotics when indicated. Genetically competent S. mutans was prepared as previously described (43), and transformants were selected on BHI agar supplemented with tetracycline (5 μg/ml), erythromycin (10 μg/ml), or spectinomycin (250 μg/ml). For induction and repression experiments, and for studies involving chloramphenicol acetyltransferase (CAT) reporter gene fusion measurements, S. mutans was grown to an optical density at 600 nm (OD600) of ca. 0.5 to 0.7 in a tryptone-vitamin base (TV) medium 3.5% tryptone with 0.04 μg of p-aminobenzoic acid/ml, 0.2 μg of thiamine-HCl/ml, 1 μg of nicotinamide/ml, and 0.2 μg of riboflavin/ml) which was supplemented with 0.5% (wt/vol) levan (prepared as detailed below), inulin (from Dahlia tubers; Sigma) or simple sugars, as well appropriate antibiotics when necessary. TV medium was developed because tryptone-yeast extract base medium (58) contained significant amounts of carbohydrate and could support the growth of S. mutans to an OD600 of ca. 0.3 to 0.4. S. mutans was not able to grow in TV medium without carbohydrate supplementation, so experiments designed to look at induction and repression of fruA were not confounded by the presence of significant quantities of uncharacterized sugars. Escherichia coli DH10B was maintained on L agar. E. coli transformants were selected on L medium containing either tetracycline (5 μg/ml), ampicillin (100 μg/ml), spectinomycin (100 μg/ml), chloramphenicol (50 μg/ml), erythromycin (300 μg/ml), or combinations of antibiotics when necessary. Medium components, excluding sugars, were obtained from Difco, and chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Levan was prepared from supernatant fluid of S. salivarius 57.1 cultures (52). The supernate was collected and buffered at pH 6.5 with 10 mM potassium phosphate, and phenylmethylsulfonyl fluoride and sodium azide were added to final concentrations of 1 mM and 0.02%, respectively. Supernatant fluid was concentrated approximately 20-fold at 4°C, using a membrane with a molecular weight cutoff of 30,000. The concentrated material was placed in a dialysis bag with a 6,000-Mr cutoff and dialyzed against 2 liters of 10 mM potassium phosphate, pH 6.5, containing 50 mM raffinose and 0.02% sodium azide for 2 to 3 days at 37°C. Raffinose is a substrate for FTF but not for the glucosyltransferases produced by oral streptococci. Levan that accumulated in the dialysis bag was ethanol precipitated, dissolved in deionized H2O, reprecipitated with ethanol, and lyophilized to dryness. To assess the purity of the levan, the polysaccharide was hydrolyzed in 0.5 M acetic acid for 1 h at 70°C. The ketohexose content was determined by the cysteine-H2SO4 method (20), total reducing sugar was measured by the dinitrosalicylic acid assay (38) with fructose as a standard, and total protein was determined by the method of Bradford (6), using the Bio-Rad protein assay reagent. Levan thus prepared was found to be >99% fructose with <0.5% protein contamination.
DNA manipulations.
Chromosomal DNA was prepared from S. mutans as previously described (13). Small-scale plasmid DNA isolations from S. mutans were performed according to the method of Anderson and McKay (1a), except that mutanolysin was added to a final concentration of 20 U/ml during the lysozyme digestion step. For rapid screening of recombinants, plasmid DNA from E. coli was prepared by an alkaline lysis procedure (5). For transformation of S. mutans, plasmid DNA was prepared from E. coli by a rapid boiling method (39) to minimize nicking of the DNA. For nucleotide sequencing, plasmid DNA was prepared by alkaline lysis followed by polyethylene glycol purification (39).
DNA sequencing was performed by the dideoxynucleotide chain termination method (48). Primers used in sequencing were either M13 forward or reverse primers (U.S. Biochemicals) or custom primers based on derived sequence data. Custom primers either were synthesized with an Applied Biosystems model 391 DNA synthesizer or were purchased from Life Technologies Inc. (LTI; Bethesda, Md.). Sequencing reactions were performed by using Sequenase version 2.0 (U.S. Biochemicals) or a Ladderman sequencing kit (Pan Vera, Madison, Wis.) with Bca thermostable DNA polymerase, as recommended by the manufacturer. Products of sequencing reactions were labeled with [α-35S]dATP, separated by 6% denaturing polyacrylamide gel electrophoresis, and subjected to autoradiography. Sequences were analyzed with MacVector version 4.0.1 and AssemblyLIGN version 1.0.5 from IBI (New Haven, Conn.), with the University of Wisconsin Genetics Computer Group (GCG) Wisconsin Package version 8.1 DNA analysis software and a Vax workstation, and with BLAST search algorithms available through the National Center for Biotechnology Information (Bethesda, Md.).
Constructs containing DNA fragments from the 5′ region of fruA (see Fig. 6) were obtained by the synthesis and use of primers designed to amplify the desired region from plasmid pFRU1 (13). The primers contained mismatched bases such that BamHI and/or PstI sites were generated for subsequent cloning of the PCR products onto pU1, which is pUC18 carrying an E. coli CAT gene (cat) (Pharmacia Biotech, Piscataway, N.J.) in the orientation opposite that of the lac promoter. Positive recombinants were selected for their ability to confer chloramphenicol resistance to E. coli. Sequencing reactions were performed to confirm orientation and error-free PCR amplification. The construct, WHFRU, containing sequences from nucleotides (nt) −564 to +101 with respect to the translation initiation site (see Fig. 6), was cloned into the streptococcal suicide vector pSU20Erm (31). The construct was introduced into S. mutans, and transformants were selected on medium containing erythromycin. Southern hybridization was used to confirm integration of WHFRU at the fruA locus by Campbell insertion. The 5′ deletion constructs were transferred to the E. coli-Streptococcus shuttle vector pDL278 (36), and Spr transformants of S. mutans US100, a recA derivative of S. mutans GS-5 constructed for this study by insertional inactivation of recA with a previously described construct (45), were selected.
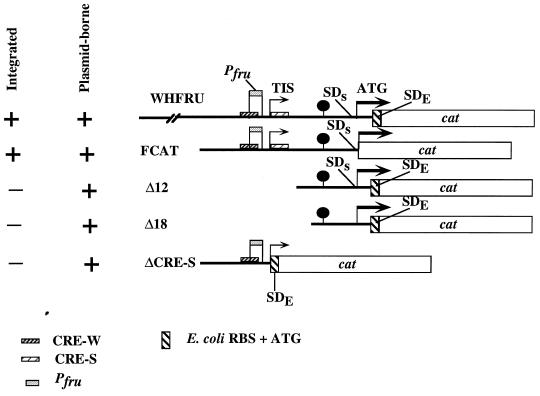
FIG. 6.
Promoter fusions and deletion derivatives. Shown are schematic diagrams of the derivatives of the 5′ region of fruA which were generated by PCRs and fused to an E. coli cat gene. The fruA promoter (Pfru) is indicated with a labeled box. The two CREs are indicated as shaded boxes overlapping the promoter (CRE-W) or beginning at position +2 (CRE-S) in the mRNA. SDs (for Shine-Dalgarno–Streptococcus) indicates the cognate RBS of fruA, and SDE (for Shine-Dalgarno–E. coli) indicates the cognate RBS of cat. (See Materials and Methods for details.) The lollipop-like markings indicate the approximate position of the stem-loop structures in the fruA leader mRNA. The 5′ ends of WHFRU, FCAT, Δ18, and Δ12 promoter fusions described in the text correspond to positions 121, 439, 552, and 581, respectively. The 3′ end point of WHFRU, Δ12, and Δ18 is at position 786, and the 3′ end of FCAT constructs is at position 684 (Fig. 1). All integrated constructs were introduced into wild-type S. mutans GS-5, and all plasmid-borne derivatives were analyzed in US100, a recA derivative of GS-5 constructed for this study. TIS, transcriptional initiation site.
Strain FCAT (see Fig. 6) was constructed by synthesizing primers at positions −254 to −234 and −18 to +10 relative to the fruA translational start site. The primers contained internal mismatched bases such that BamHI sites were generated in the resulting PCR product at the 5′ end and at position −1 relative to the translational start site of fruA. Products, purified as described above, were ligated into pCW24 (16), a pUC-based vector containing a promoterless cat gene. A BamHI site was incorporated at the 5′ end of cat so that PCR products derived from the 5′ end of fruA and containing the cognate ribosome binding site (RBS) of fruA could be correctly spaced from the start codon of the cat gene (see Fig. 6). Positive recombinants were selected for their ability to confer chloramphenicol resistance to E. coli. Sequencing reactions were performed to confirm orientation and sequence identity. The gene fusion cassette was transferred to the suicide vector pSF143 (53). One positive clone, FCAT, was used to transform S. mutans, and Southern hybridizations were performed to confirm integration at the fruA locus by single-crossover insertion.
A 3′ deletion construct was prepared by synthesizing an antisense primer encompassing nt −254 to −156 (ΔCRE) relative to the fruA translational start site; this antisense primer, containing a 5′ overhang with a PstI site, was used in conjunction with the sense primer employed in the construction of FCAT to amplify products by PCR. Products were cloned directly into plasmid pCR2.1 (Invitrogen), sequenced to confirm their identity, and then cloned into pDLCAT, which is pDL278 in which the cat gene from pU1 has been cloned in the orientation opposite that of the lacZ promoter on pDL278, to create pDLΔCRE. Recombinants were selected for their ability to confer both Spr and Cmr to E. coli and used to transform S. mutans US100. Recombinant S. mutans strains were screened by restriction analysis of small-scale preparations of plasmids to ensure plasmid stability.
RNA manipulations.
For preparation of RNA, S. mutans was grown to mid-exponential phase in TV medium supplemented with the desired carbohydrate at 0.5%. Total RNA from S. mutans GS-5 was isolated as previously described (15), using a modification of the protocol of Putzer et al. (44). RNA samples were denatured and transferred to nitrocellulose membranes (Schleicher and Schuell) by using a slot blot apparatus (LTI), and the RNAs were UV cross-linked to the membranes and air dried. Blots were probed with DNA fragments that had been purified after excision from agarose gels (2). The probes consisted of an 834-bp HincII fragment encompassing nt +724 to +1557 of the fruA structural gene (9) or a 1.12-kbp internal NdeI fragment from fruB. Hybridizations and washes were carried out under high-stringency conditions. Signals on autoradiographs were quantified by using an IS1000 digital imaging system from Alpha Innotech Corp. (San Leandro, Calif.).
The transcriptional start site of fruA was mapped by primer extensions with four different primers, and the same result was achieved regardless of the primer used. Primers were end labeled, and primer extension reactions were performed essentially as described previously (2, 31). DNA sequencing reactions were performed with the same primers and pFRU1 (16). Reverse transcriptase (RT) PCR was performed as described previously (16).
Enzyme preparation and assays.
Protein preparations from exponentially growing cultures of S. mutans were assayed for fructan hydrolase activity as previously described (11, 59). One unit of activity was defined as the amount of enzyme needed to release 1 μmol of reducing sugar per h. To prepare samples for CAT assays, cleared lysates were prepared as previously detailed (16) and used directly for determination of CAT activity according to the protocol of Shaw (50). One unit of CAT activity was defined as the amount of enzyme necessary to acetylate 1 nmol of chloramphenicol per min. The values expressed for all enzyme assays are averages of data for three to six independently grown cell cultures, and all assays were performed at least in triplicate.
Nucleotide sequence accession number.
The complete nucleotide sequence of the fruAB genes and flanking regions has been deposited with GenBank and bears accession no. AF093758.
RESULTS
Genetic organization of the fru operon.
The complete nucleotide sequence of the fruA gene of S. mutans GS-5, including 685 nt 5′ of the translational initiation site and approximately 50 nt 3′ of the fruA stop codon, was reported previously (9). At that time, no open reading frames (ORFs) were found 5′ of fruA or immediately following fruA. Also, the small amount of sequence downstream of fruA showed no similarity to known sequences. Preliminary nucleotide sequence analysis of more than 1.5 kbp of additional DNA 5′ of fruA indicated that there were no ORFs encoding proteins potentially involved in fructan metabolism or gene regulation (data not shown).
As reported previously (9), there is a stable stem-loop structure immediately downstream of fruA. An ORF beginning 71 bp from the stop codon of fruA was identified. This gene, designated fruB, consists of 1,560 bp and is preceded by an RBS-like sequence, but there do not appear to be sequences 5′ of fruB with similarity to canonical promoter sequences (Fig. 1). Downstream of fruB is the dnaK operon of S. mutans, which is driven from its own promoter(s) (31), beginning with the hrcA gene (Fig. 1). Consistent with an S. mutans origin, fruB is 36.9% G+C. The deduced amino acid sequence of fruB yields a protein with a molecular weight of 58,612 and a pI of 5.83. FruB has an N-terminal sequence, with characteristics of signal peptides of gram-positive bacteria (56), which could be cleaved to yield a mature polypeptide with a molecular weight of 55,462 and pI of 5.49. Unlike FruA, FruB does not have an LPXTG cell wall-anchoring sequence (49). A BLAST search of FruB against existing databases indicated that the highest degrees of similarities were to fructosidases of bacteria and lower eukaryotes, including levanases, inulinases, and invertases, and that the levels of similarity between FruB and known fructosidases were similar to those generally demonstrated by these proteins. For example, FruB and the Bacillus stearothermophilus levanase SurC (37) exhibit 25% identity (37% similarity; Blast P value, 3.3e−16), and FruB shows 28% identity (36% similarity) to a hypothetical levanase (YveB) identified in Bacillus subtilis (Blast P value, 7.3e−24). Generally, comparisons of fructosidases across genera reveal around 25 to 30% identity and 35 to 50% similarity. Of note, FruB was only 15% identical (29% similar) to FruA, so it seems unlikely that FruB arose from gene duplication. Other notable features of FruB are that the aspartic acid residue (Asp 47 in FruB) that has been shown to be essential for catalytic activity in the yeast invertase (46) was present in the correct relative position in FruB (Fig. 2), as was the cysteine residue (Cys 230, apparently the only cysteine in the mature FruB protein), which has been suggested to be important for activity of sucrases.
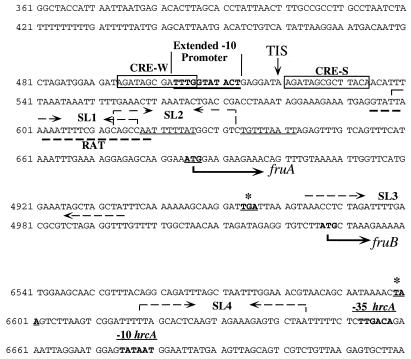
FIG. 1.
Relevant nucleotide sequences and features of the fruAB gene cluster. The nucleotide sequences of the 5′ region of fruA, the fruAB intergenic region, and the 3′ end of fruB are shown. The sequences of the structural genes have been omitted to save space. Sequence numbering corresponds to the original GenBank submission of the fruA sequence (accession no. L03358 and AF093758). The fruA structural gene begins at position 685 and ends at position 4953. The putative RBS of fruB begins at position 5016. The fruB start codon is at position 5027, and the structural gene extends to position 6599. Underlined and in boldface is the extended −10-like promoter element that drives fru transcription. Boxed and labeled as CRE-W (weaker) and CRE-S (strong) are the CREs described in the text. The inverted-repeat structures with characteristics of rho-independent terminators predicted by the Terminator program of the University of Wisconsin GCG software (indicated by opposing dashed arrows above the sequence) are in the 5′ leader mRNA of fruA (SL1, positions 599 to 626; SL2, positions 613 to 633) and after the fruB gene (SL4, positions 6618 to 6644). These three inverted repeats are followed by a T (U)-rich 9-nt sequence. There is also a stable, previously identified inverted repeat (SL3) at the end of fruA (9) indicated by dashed arrows above the sequence, but this is not recognized by computer algorithms as a rho-independent terminator. The sequence with partial similarity to RAT-like sequences (RAT; positions 595 to 619), underlined with a boldfaced dashed line, overlaps with both stem-loop structures in the leader region. The start and stop codons of fruA and fruB are in boldface, and the stop codons are labeled with asterisks. −35 and −10 hrcA indicate the promoter for the dnaK operon (31). TIS, the transcriptional initiation site mapped by primer extension.
FIG. 2.
GAP alignment of S. mutans FruB and B. subtilis YveB. The deduced amino acid sequences of the fruB (top) and yveB genes were aligned by using the GAP program of the University of Wisconsin GCG package. Vertical lines indicate identity, and single and double dots indicate lower and high degrees of amino acid similarity, respectively. The aspartic acid (Asp 47) believed to correspond to that in the yeast invertase which was shown to be involved in catalysis is in boldface, as is a highly conserved cysteine residue (position 230) found in many fructosidases. Overlined with dashed lines are regions in FruB that exhibit some homology to conserved regions in fructosidases or which have some similarity to regions conserved between S. mutans FruA and B. subtilis SacC, which are known levanases. Asterisks indicate a sequence which is very highly conserved among FruB, YveB, and the known levanase of B. stearothermophilus, SurC.
Several attempts have been made to disclose the involvement of FruB, if any, in fructan metabolism. First, a fruA mutant containing a mini-MudE transposon was previously described (59). It is now known, from the results of Northern blotting with a DNA fragment downstream of the insertion site, that the mutation in fruA is not polar (data not shown) and, from the use of a fruB probe, that fruB is transcribed constitutively in the fruA mutant at levels comparable to those observed in the wild type when fruB is fully induced (see below). Still, fructan hydrolase could not be detected in this fruA mutant (59). Second, E. coli carrying the intact fruB gene under the control of the lacZ promoter in pUC-based plasmids had no detectable fructosidase or dextranase activities. Also, FruB lacking the signal sequence and containing an N-terminal 6-His tag has been expressed at high levels in a soluble form in E. coli (data not shown). Screening of several independent clones demonstrated that no levanase, inulinase, sucrase, raffinase, or dextranase activity could be detected in these recombinants, and affinity-purified FruB did not have any of the aforementioned activities. Third, insertional inactivation of fruB in S. mutans with a Tcr determinant had no effect on supernatant or cell-associated fructan hydrolase activity (data not shown). Addition of the purified, histidine-tagged FruB to culture supernates from GS-5 or the fruB mutant, both of which produce FruA, did not alter fructan hydrolase activity appreciably (data not shown). Finally, by measuring total fructan hydrolase activity or by using a cat gene-fruA promoter fusion (FCAT) (see below) in a fruB mutant, it was found that FruB does not affect the regulation of fruA expression detectably (data not shown).
It appears that fruB transcription may not arise from its own promoter. First, the 5′ region of fruB, including roughly 200 bp of the 3′ portion of fruA, was cloned behind a promoterless cat gene and introduced on a shuttle plasmid into S. mutans US100. No evidence suggesting the presence of a functional streptococcal promoter was found (data not shown). Second, primer extension analysis using a primer complementary to fruB mRNA yielded multiple products, typical of results obtained when such experiments are performed with primers internal to bacterial operons. Third, the use of RT PCR revealed the existence of a transcript that contains fruA and fruB sequences. Regardless of whether cells were grown on inulin (Fig. 3A) or glucose (Fig. 3B), a product was observed, indicating that cotranscription was not dependent on growth in medium with inducing substrates. It was also consistently observed that bands produced by RT PCR from inulin-grown cells, as well as from cells grown on levans (data not shown), were slightly smeared and migrated somewhat anomalously compared to the reaction products of cells grown on hexose. This was consistently observed and was possibly due to carryover of some polysaccharides from cells grown on fructans. Finally, insertion of a highly polar kanamycin resistance determinant into fruA ablated fruB transcription (data not shown). This information, coupled with the finding that fruB is coordinately regulated with fruA (see below), indicates that the fruAB genes constitute an operon.
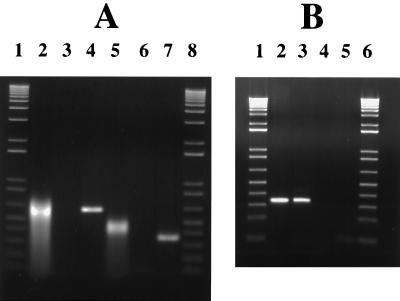
FIG. 3.
RT PCR. Shown are ethidium bromide-stained agarose gels of products obtained by RT PCR with primers spanning the fruA-fruB intergenic region, using RNA from cells grown on inulin (A) or glucose (B). RT PCRs were performed with RNA from cells grown on different carbohydrates to determine whether the carbohydrate source affected transcriptional readthrough and because RT PCR of fructan-grown cells always produced a band that was slightly smeared, perhaps from carryover of polysaccharide. The sense-strand primer corresponded to nt 4794 to 4830, and the antisense primer corresponded to nt 5140 to 5118 for the 0.65-kbp product or to nt 5415 to 5398 for the 0.32-kbp product (nucleotide positions are in reference to those of GenBank accession no. AF093758). (A) Lanes 1 and 8 are DNA size standards (LTI). Products from the PCRs were derived as follows: cDNA prepared from S. mutans RNA with RT (lanes 2 and 5), from S. mutans RNA without RT (lanes 3 and 6), and from GS-5 chromosomal DNA (lanes 4 and 7). (B) Lanes 1 and 6 contain the 1-kbp ladder (LTI). Products from the PCRs were derived as follows: from GS-5 chromosomal DNA (lane 2), from cDNA prepared from S. mutans RNA with RT (lane 3), and from S. mutans RNA without RT (lane 4), Lane 5 contains a no DNA-no RNA-no RT control.
Expression of fru is regulated by the carbohydrate source.
We previously reported that fruA expression was poor in cells grown on BHI medium containing glucose (10, 13). To determine if fruA expression is controlled by the carbohydrate source, cells were cultured in medium containing levan, inulin, sucrose, or fructose. Fructan hydrolase activity in the supernatant (Table 1) and cell-associated (not shown) fractions from mid- to late-exponential-phase cultures (OD600 ≈ 0.6 to 0.7) was measured. FruA activity was found to be optimal when cells were grown on fructan polymers, and activity was also higher when sucrose was provided as the growth carbohydrate. In all cases, levan and inulin were equally efficient at inducing fruA. Conversely, fructan hydrolase activity was low in cells grown on readily metabolizable hexoses. Cells that were grown on mixtures of levan and glucose, or levan and fructose, were found to have levels of FruA activity comparable to those found when only hexoses were used for growth (Table 1). However, when cells were cultured on mixtures of levan and the slowly metabolized sugar alcohol glucitol, induction was seen. Cell-associated activity paralleled that of the supernatant FruA activity in all cases (data not shown). Therefore, fruA expression requires induction and is subject to carbon catabolite repression (CCR).
TABLE 1.
Fructan hydrolase activity in supernates and CAT specific activities of S. mutans GS-5 grown on various carbohydrate sourcesa
| Growth carbohydrateb | Fructan hydrolase activity (U/mg of protein) | CAT activity ofc:
|
|
|---|---|---|---|
| FCAT | WHFRU | ||
| Levan* | 126.9 ± 29.1 | 2.9 ± 0.9 | 2.8 ± 1.1 |
| Inulin* | 132.7 ± 11.9 | 2.6 ± 0.7 | 2.5 ± 0.7 |
| Sucrose* | 64.8 ± 8.1 | 2.3 ± 0.6 | 2.1 ± 0.6 |
| Fructose | 28.4 ± 10.1 | 0.4 ± 0.3 | 0.4 ± 0.2 |
| Glucitol | 29.9 ± 3.7 | 0.3 ± 0.2 | 0.2 ± 0.8 |
| Glucose | NDd | 0.3 ± 0.2 | ND |
| Levan-glucose | 25.5 ± 11.9 | 0.3 ± 0.2 | 0.4 ± 0.2 |
| Levan-fructose | 28.8 ± 2.3 | 0.4 ± 0.1 | 0.2 ± 0.1 |
| Levan-glucitol* | 55 ± 5.78 | 1.9 ± 0.4 | 1.5 ± 0.4 |
Cells were grown in TV medium with the indicated carbohydrates to an OD600 of ca. 0.6 to 0.7. Concentrated supernates from exponentially growing cultures were prepared and assayed for levanase activity as indicated in Materials and Methods. Cell extracts were prepared, and CAT activity was measured as detailed in Materials and Methods. Values represent the averages ± standard deviations for at least three, and in some cases more than six, separate cultures. All assays were performed in triplicate.
An asterisk indicates that the value differed from those of all other groups (P < 0.05) by Tukey-Kramer honestly significant difference analysis, determined by means of the JMP program developed by the SAS Institute (Cary, N.C.).
CAT activity is expressed as units (nanomoles of chloramphenicol acetylated per minute) per milligram of protein.
ND, not determined.
Multiple attempts were made to obtain data on the size of the fru transcript by Northern blotting, but definitive results could not be obtained because of substantial degradation of the RNA, which is typical of preparations from S. mutans. To determine whether the levels of fruA mRNA were higher under induced conditions, RNAs from cells grown in medium with inulin, glucose, or mixtures of the two sugars were subjected to slot blot analysis with a probe derived from an internal portion of the fruA structural gene. The data indicated that expression of fruA was governed in large part at the level of transcription (Fig. 4A). This conclusion was further supported by the results of gene fusion studies (detailed below). To see if transcription of fruB and fruA could be coordinately regulated, RNAs from cells grown on inulin, glucose, or both carbohydrates were probed with an internal fragment of the fruB gene (Fig. 4B). The level of fruB-specific mRNA increased when cells were cultured under conditions that induced fruA expression, and the increase was of a magnitude similar to that of fruA.
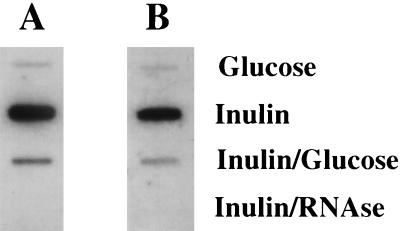
FIG. 4.
Slot blot of total RNA from S. mutans grown under induced and repressed conditions. S. mutans GS-5 was grown in TV medium supplemented with inulin, glucose, or both at concentrations of 0.5%. Total RNA (1 μg) was isolated from exponentially growing S. mutans cells, treated with RNase-free DNase, and transferred to a membrane as detailed in the text. Hybridizations were performed with an internal fragment of either fruA (A) or fruB (B) as indicated in Materials and Methods. Inulin plus RNase samples were treated with RNase prior to application to the membrane as a control to show that the RNAs were free of DNA contamination.
Mapping of the fruA transcription start site and analysis of the 5′ region of the fru operon.
The results of one of four primer extension reactions, all yielding the same conclusion, are shown in Fig. 5. A transcriptional initiation site was identified 165 nt 5′ of the translational start site. Consistent with the results of slot blot analysis, the primer extension signal was markedly stronger in cells grown on fructans than in those grown on glucose. Examination of the sequences flanking the proposed transcriptional start site revealed a −10 sequence of TATACT, which compares well with the consensus TATAAT of ς70-type promoters. However, the sequence (GGATGGAAG) located 10 to 19 bp upstream of position −10 did not closely match the canonical −35 sequence (TTGACA). Further examination revealed that the promoter best resembled extended −10 promoters, which are not unusual in gram-positive bacteria (23) and which have been identified and demonstrated to function in Streptococcus pneumoniae (47). Extended −10 promoters have a conserved extension on the −10 hexanucleotide (TaTGgTATAAT), and some can direct transcription in the absence of a −35 sequence. The proposed extended −10 promoter of fruA matches 5′ extension at all positions except the less highly conserved adenine residue (Fig. 1).
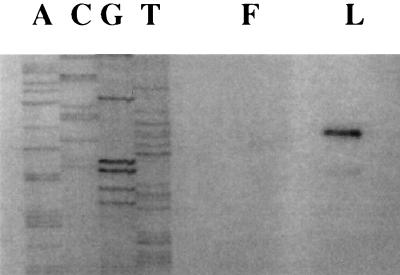
FIG. 5.
Mapping the fruA promoter. This figure shows the results of primer extension reactions used to map the transcriptional initiation site for fruA. RNA was isolated from S. mutans cells growing exponentially with fructose (F) or levan (L) as the carbohydrate source. Reactions were performed as detailed in the text. Adjacent to the primer extensions is a sequencing reaction performed on pFRU1 (13) with the same primer used in the primer extension.
Two potential catabolite response elements (CREs) with strong levels of homology to the consensus sequence for the cis-acting sites controlling catabolite repression in gram-positive bacteria (28) were identified at positions +2 to +15 and positions −27 to −14 relative to the transcriptional start site (Fig. 1). The CRE located at nt +2 of the fru mRNA, which had the sequence AGATAGCGCTTACA, differed from the consensus sequence, TGWNANCGNTNWCA, only at the first position and was designated as CRE-S (with the S being for strong homology). The second CRE (AGATAGCGATTTGG) overlapped the promoter and differed at the 1st, 13th, and 14th positions and thus was designated CRE-W (for weaker homology). No other good matches with CREs were identified in or near the fruAB gene cluster. Two overlapping stable stem-loop structures with the potential to serve as rho-independent terminators, particularly the downstream structure, were identified in the 5′ noncoding region of the fruA mRNA (Fig. 1). The presence of the long leader mRNA and the terminator-like structure suggested the possibility that induction of fruA is regulated by an antitermination mechanism, similar to some saccharolytic operons of B. subtilis (18). Of note, there was a region of dyad symmetry (Fig. 1), overlapping with the potential terminators, with 50% similarity to target sites required for antitermination of saccharolytic operons of B. subtilis, known as ribonucleotide antiterminator (RAT) sequences (3). Interestingly, this sequence is positioned in relation to the potential terminators, particularly SL2, where established RAT sequences are located with respect to known antiterminators in B. subtilis (3).
Construction and regulation of fru::cat transcriptional fusions.
To demonstrate functionality of the fru promoter region and to begin to explore whether transcriptional regulation of fru was exerted through the putative cis-acting elements identified above, the region containing the fru promoter was fused to a promoterless cat gene by strategies detailed in Materials and Methods and outlined in Fig. 6. Briefly, the first set of experiments involved a construct in which the entire promoter region was fused to a cat gene in a manner such that transcription and translation were driven from the cognate streptococcal elements. The fruA::cat cassette was cloned onto the streptococcal suicide vector pSF143 (53), which carries a tetracycline resistance determinant that functions in streptococci. This construct was introduced into S. mutans by transformation of naturally competent cells. Transformants were analyzed by Southern hybridization and PCR to confirm that integration had taken place and that the promoter-reporter construct was intact (data not shown).
The strain chosen for further study, FCAT, was grown in TV medium supplemented with different sugars to ascertain the effects of the carbohydrate source on transcriptional activity from the fruA promoter. As shown in Table 1, CAT activity was six- to sevenfold higher when cells were grown on levan, inulin, or sucrose than when cells were grown on 0.5% fructose, glucose, or glucitol. These findings were consistent with the observations that steady-state levels of fruA mRNA (Fig. 4) and primer extension signals (Fig. 5) were increased to similar degrees under inducing conditions. CAT activity was also measured in cells grown on combinations of sugars. When cells were grown on fructan and either glucose or fructose, CAT activity was comparable to that found with glucose or fructose alone, further demonstrating that fruA expression was sensitive to CCR. Comparison of CAT activity in cells grown on glucitol alone with that of cells grown on levan and glucitol reinforced the concept that expression required an inducing substrate and that repression was occurring when a more quickly metabolized compound, i.e., fructose or glucose, was present at 0.5%. Essentially identical results were obtained with strain WHFRU, which included additional sequences 5′ and 3′ of those in FCAT and in which translation was driven by an E. coli RBS (Table 1). Thus, the use of cat with the E. coli RBS, which differs from the fruA RBS at only one position, would be suitable for 3′ deletion analyses (detailed below).
Deletion analysis of the 5′ region of the fru operon.
To begin to investigate the role of the putative cis elements, identified above, in the regulation of fruA expression, fruA promoter deletion constructs were obtained by PCR (Fig. 6). These deletion constructs were fused to a cat gene containing an E. coli RBS and cloned into pDL278. These constructs were introduced into S. mutans US100 (S. mutans GS-5 recA) by competent transformation. The strains were grown in TV medium with various carbohydrate sources, and CAT activities were measured.
When strains carrying the plasmid-borne, full-length promoter and terminator structures were assayed for CAT activity after growth on a number of different carbon sources, enzyme levels were found to be only marginally above the background obtained with strain DLCAT (31) (data not shown). Interestingly, strains WHFRU and DLFCAT grew very poorly (tg, >4 to 6 h versus 45 to 90 min for the wild type) with fructans as the sole carbon source. Western blot and enzyme-linked immunosorbent assay analyses (data not shown) were performed with a FruA-specific antiserum (13) raised against supernatant proteins prepared from S. mutans US100 or that strain carrying either pDL278 or the full-length promoter-cat fusions. Notably, when the promoter region was present in multiple copies (the copy number of pDL278 has been estimated to be around 20 to 30 [14a]), S. mutans produced barely detectable levels of FruA protein, suggesting the possibility of titration of a trans-acting factor required for efficient induction of fruA. Two 5′ deletion mutants (Fig. 6) in which the putative promoter was deleted but the potential terminators were left intact were created. No CAT activity could be measured in these constructs, the ability to grow well on fructan substrates was restored, and FruA production was detectable at near wild-type levels in these strains (data not shown). These results, supported by additional data detailed below, indicate that the 5′ deletions simply ablated promoter activity.
Specifically, a 3′ deletion mutant was made in which the promoter, and sequences 5′ of it, was left intact and the putative terminator and CRE-S element were deleted, and the construct was fused to cat with an E. coli RBS (Fig. 6). Levels of CAT activity expressed from this construct, pDLΔCRE, were increased dramatically over those of any of the integrated, full-length constructs, and cells grew as well on fructan polymers as did the wild type. Cells grown on fructans exhibited 700 to almost 10,000 times more CAT activity than cells containing either the plasmid-borne or the integrated, full-length promoter fusions, depending on the growth carbohydrate (averages ± standard deviations, 1,919 ± 470, 1,662 ± 98, and 2,170 ± 545 nmol of chloramphenicol acetylated/min/mg of protein for cells grown on inulin, glucose, or a combination thereof, respectively). The presence of the fusion in multiple copies likely results in higher levels than would be seen if a single copy of the construct were present. Importantly, unlike the 6- to 10-fold differences between induced and repressed activities, cells cultivated with glucose as the sole carbohydrate source did not produce levels of CAT statistically different from those of cells grown on inulin. Collectively, these results support the theory that the ascribed promoter (Fig. 1 and 5) is functional and that CRE-S is likely involved in catabolite repression of fruA. The lack of induction by fructans in strains carrying pDLΔCRE strongly suggested that binding of DNA 3′ to the promoter was not responsible for the titration effect noted above, since the 5′ deletions had no effect on FruA synthesis or growth on fructans. Thus, transcription of the 5′ region, including the stem-loop structures, must occur to elicit titration. A recent examination of the sequence data available through the S. mutans genomic sequencing project being conducted at the University of Oklahoma revealed the presence of a LicT/SacY/SacT homologue in S. mutans UA159, supporting the hypothesis that genes for antiterminators are present and could function in the regulation of genes in this organism. Also, it has been determined that the 5′ regions of fruA in GS-5 and UA159 are essentially identical (57a, 59).
Attempts were made to show definitively that the stem-loop structures in the 5′ untranslated region might function as terminators; however, two major technical obstacles were encountered. First, a construct, designated ΔT, which contained the promoter and CREs but harbored a 3′ deletion that eliminated the stem-loop structures could not be stably maintained in any of the gene fusion constructs that were tried, apparently due to intramolecular recombination. Second, as indicated above, the introduction of constructs containing the functional promoter and terminator clearly caused a titration effect, so attempting to analyze the role of the proposed terminator by using promoter fusions on a multicopy plasmid was not appropriate. Efforts to introduce these constructs into the chromosome of S. mutans in single copy are under way, but some problems with stability of the vector and recombinant constructs are evident.
DISCUSSION
For S. mutans, and probably other oral bacteria, fructans appear to serve primarily as storage compounds that can be used when exogenous sources are exhausted (7). Therefore, it should be beneficial to the organisms to separate fructan synthesis and catabolism temporally, postponing breakdown of the polymers until other energy sources are exhausted. S. mutans can enhance expression of FTF when substrate is available, and potentially the bacteria can release the enzyme to where it can access sucrose and convert it to polymer within the biofilm matrix (12, 27, 34, 58). Previously, we demonstrated that low pH favored association of the FruA enzyme with the cell surface, a mechanism that could potentially restrict the access of the hydrolase to fructans when exogenous carbohydrate is present in excess (11). Although it might be possible to optimize fructan utilization simply by producing the hydrolase constitutively, albeit in smaller quantities than the synthase, the data presented here indicate that expression of fruA is tightly regulated.
It appears that fruA is actually the first gene in a two-gene operon, and there appear to be no additional genes in the fru cluster. A fairly thorough analysis of fruB mutants and heterologously expressed FruB proteins has failed to disclose the role of this protein. Of note, our inactivated fruB construct was used to insertionally inactivate that gene in wild-type S. mutans V403, which produces FruA and an inulinase, as well as in a fruA mutant of V403. Strain V403 retained inulinase activity after fruA inactivation (33), but fruB ablation had no discernible effect on fructan metabolism (38a). That observation, coupled with the finding that a full-length FruB protein can be produced in E. coli, indicates that it is unlikely that fruB is a cryptic version of the inulinase gene of V403. Efforts to understand the function of FruB are continuing.
The data presented here show that levans, inulins, and sucrose can induce expression of fruA. Interestingly, biochemical characterization of FruA indicates that this enzyme functions exohydrolytically, releasing only fructose from the polymers, and that digestion of fructans proceeds to completion (13). Since it is unlikely that fructans directly induce expression, and since oligomers of fructans are not produced, it is likely that fructose, at nonrepressing concentrations, is the inducing molecule. In our initial characterization of FruA from S. mutans GS-5 (13), we confirmed results by Jacques et al. (30) which indicated that much higher levels of fructanase production could be achieved in cells in steady-state continuous culture at low growth rates (D = 0.075 h−1; tg = 9.24 h) with fructose as the limiting carbohydrate than could be achieved in batch cultures (13). Consistent with this hypothesis, fructose is the inducer of the B. subtilis levanase, encoded by sacC (17), and high levels of fructose are repressive through CcpA- and PtsI-dependent pathways for CCR exerted through CREs and the transcriptional activator LevR, respectively (41). However, there are significant physiologic differences between S. mutans and B. subtilis that have precluded proving that fru expression is inducible by fructose. Specifically, showing induction of levanase expression by fructose by growing B. subtilis cells on a base medium with minimal amounts of fructose has been fairly straightforward. However, this has not been possible with S. mutans since its growth is entirely dependent on substrate-level phosphorylation and fru transcription is exquisitely sensitive to CCR.
The results of primer extension studies and functional studies using gene fusions are consistent with the ascribed location of the fru promoter. The promoter is most like those of the extended −10 type. Identification of a long leader mRNA containing terminator-like structures which overlap with a possible RAT sequence (3) indicates that fruA expression could be controlled by antitermination, probably in a manner similar to established antitermination models for saccharolytic genes, including the sacPA and sacB genes of B. subtilis (18) and the bgl operon of E. coli (26). Notably, regulation of the levanase of B. subtilis (SacC), which has substantial functional similarity to FruA, is not governed by antitermination; instead, sacC transcription is controlled primarily by the transcriptional activator LevR (19), whose DNA binding activity is modulated via phosphorylation by components of a fructose-specific phosphotransferase system (PTS), encoded by levDEFG (40). Results presented in this communication do not support the theory that transcriptional activation of fru is a control point for induction. Also, S. mutans chromosomal DNA has been probed with levDEFG and levR probes (kindly provided by I. Martin-Verstraete) under low-stringency conditions, and no evidence of sufficiently similar sequences was found (data not shown). Similarly, although weak similarities to the levDEFG genes, which are similar to PTS components, can be found, there is no evidence for a LevR-like protein in the albeit-incomplete genomic sequence. Therefore, the existing data are most consistent with an antitermination mechanism for induction of fru. In B. subtilis, antitermination of certain sac genes is governed by the antiterminator proteins SacY and SacT, whose activities are modified via phosphorylation by PTS components (29, 54). The finding that an ORF with a very high degree of similarity to the SacY and SacT antiterminators is present in S. mutans supports the hypothesis that antitermination is involved in fru induction.
There are two identifiable CREs near the fruA promoter, located at the transcription initiation site and also partially overlapping the promoter. CREs have been shown to be bound by a global regulator of CCR, CcpA, which was originally cloned from B. subtilis (24). CcpA is a repressor of CCR-sensitive genes that appears to be present in many gram-positive bacteria (35) and which has as a corepressor a form of HPr that is phosphorylated at serine residue 46 (32). Clearly, deletion of the more highly conserved of the two CREs, CRE-S, which differs from the consensus sequence at the first position, essentially eliminates CCR. Consistent with this, Weickert and Chambliss (57) have shown that mutation of the first nucleotide in the B. subtilis amylase CRE has little effect on CCR. Notably, mutation of position 13 of the amylase CRE largely alleviated CCR, and thus CRE-W, which differs in the 1st, 13th, and 14th positions, may not be an effective cis element in the absence of CRE-S. Although other catabolite control proteins have been found recently (14, 42), and CCR can occur by direct phosphorylation of specific regulatory elements (41), the findings that sequences near the fruA promoter adhere closely to the consensus for CREs and that deletion of CRE-S alleviates CCR argue that a major control point for CCR of fruA expression could be through binding of a CcpA homologue which is present in S. mutans (1, 51). Interestingly, it is known that Bacillus CcpA can bind in a cooperative fashion to two target CREs located in the xyl operon of Bacillus megaterium (22), so perhaps the two CREs in close proximity to the fru promoter act as target sites for binding and dimerization of CcpA.
In summary, the fruAB gene cluster of S. mutans GS-5 is highly regulated by the carbohydrate source as well as its availability, mediated through CREs known to be bound by CcpA-like proteins, as well as potentially by transcriptional antitermination exerted through elements in the leader region 5′ to the fruA structural gene. On the basis of recent work with B. subtilis (14, 29, 41, 42, 54), it would be surprising if there were not other networks for controlling fruA expression in response to carbohydrate availability. The demonstration of tight transcriptional regulation of fru expression, coupled with our findings on posttranslational control of localization of FruA (11), supports the idea that S. mutans has evolved multiple mechanisms for governing the accumulation and utilization of fructan exopolymers. Future studies oriented toward characterizing the trans-acting factors and other components involved in fru regulation should provide valuable information about carbohydrate and polysaccharide metabolism in the oral streptococci and other lactic acid bacteria.
ACKNOWLEDGMENTS
We thank Don Wexler, Earl Albone, and Christine Hervas for assistance with the generation of promoter constructs and preliminary regulation studies.
This work was supported by grant DE 12236 (to R.A.B.) from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Albone E F, Burne R A. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Regulation of expression of the fruA gene of Streptococcus mutans GS-5, abstr. D-166; p. 237. [Google Scholar]
- 1a.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. [Google Scholar]
- 3.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkhed D, Rosell K-G, Granath K. Structure of extracellular water-soluble polysaccharides synthesized from sucrose by oral strains of Streptococcus mutans, Streptococcus salivarius, Streptococcus sanguis, and Actinomyces viscosus. Arch Oral Biol. 1979;24:53–61. doi: 10.1016/0003-9969(79)90175-4. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid molecules. Nucleic Acids Res. 1979;7:1515–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Burne R A. Oral ecological disasters: the role of short-term extracellular storage polysaccharides. In: Bowen W H, Tabak L A, editors. Cariology for the nineties. Rochester, N.Y: University of Rochester Press; 1991. pp. 351–364. [Google Scholar]
- 8.Burne R A, Chen Y M, Wexler D W, Kuramitsu H, Bowen W H. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific pathogen free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 9.Burne R A, Penders J E C. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-β-d-fructosidase. Infect Immun. 1992;60:4621–4632. doi: 10.1128/iai.60.11.4621-4632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burne R A, Penders J E, Wexler D L, Jayaraman G C, Clancy K A. Regulation of fructan degradation by Streptococcus mutans. Dev Biol Stand. 1995;85:323–331. [PubMed] [Google Scholar]
- 11.Burne R A, Penders J E C. Differential localization of the Streptococcus mutans GS-5 fructan hydrolase enzyme, FruA. FEMS Microbiol Lett. 1994;121:243–250. doi: 10.1111/j.1574-6968.1994.tb07105.x. [DOI] [PubMed] [Google Scholar]
- 12.Burne R A, Penders J E C, Chen Y M. Examination of gene expression in Streptococcus mutans growing in biofilms in vitro. Adv Dent Res. 1997;11:100–109. doi: 10.1177/08959374970110010101. [DOI] [PubMed] [Google Scholar]
- 13.Burne R A, Schilling K, Bowen W H, Yasbin R E. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol. 1987;169:4507–4517. doi: 10.1128/jb.169.10.4507-4517.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Chen, M. Personal communication.
- 15.Chen Y Y, Burne R A. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol Lett. 1996;135:223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y-Y M, Weaver C A, Mendelsohn D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crutz A-M, Steinmetz M, Aymerich S, Richter R, Le Coq D. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J Bacteriol. 1990;172:1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Débarbouillé M, Martin-Verstraete I, Arnaud M, Klier A, Rapoport G. Positive and negative regulation controlling expression of the sac genes in Bacillus subtilis. Res Microbiol. 1991;142:757–764. doi: 10.1016/0923-2508(91)90052-c. [DOI] [PubMed] [Google Scholar]
- 19.Débarbouillé M, Martin-Verstraete I, Klier A, Rapoport G. The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both ς54- and phosphotransferase system-dependent regulators. Proc Natl Acad Sci USA. 1991;88:2212–2216. doi: 10.1073/pnas.88.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dische Z, Devi A. A new colorimetric method for the determination of ketohexoses in presence of aldoses, ketoheptoses and ketopentoses. Biochim Biophys Acta. 1960;29:140–144. doi: 10.1016/0006-3002(60)90129-3. [DOI] [PubMed] [Google Scholar]
- 21.Gold W, Preston F B, Lache M C, Blechman H. Production of levan and dextran in plaque in vivo. J Dent Res. 1974;53:442–449. doi: 10.1177/00220345740530024401. [DOI] [PubMed] [Google Scholar]
- 22.Gosseringer R, Kuster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 23.Graves M C, Rabinowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for “extended” promoter elements in gram-positive bacteria. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 24.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli LacI and GalR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi M, Iwani Y, Yamada T, Araya S. Levan synthesis and accumulation by human dental plaque. Arch Oral Biol. 1970;15:563–567. doi: 10.1016/0003-9969(70)90111-1. [DOI] [PubMed] [Google Scholar]
- 26.Houman F, Diaz-Torres M R, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 27.Hudson M C, Curtiss R., III Regulation of expression of Streptococcus mutans genes important to virulence. Infect Immun. 1990;58:464–470. doi: 10.1128/iai.58.2.464-470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hueck C, Kraus A, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 29.Idelson M, Amster-Choder O. SacY, a transcriptional antiterminator from Bacillus subtilis, is regulated by phosphorylation in vivo. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacques N J, Morrey-Jones J G, Walker G J. Inducible and constitutive production of fructanase in batch and continuous culture of Streptococcus mutans. J Gen Microbiol. 1985;131:1625–1633. doi: 10.1099/00221287-131-7-1625. [DOI] [PubMed] [Google Scholar]
- 31.Jayaraman G C, Penders J E, Burne R A. Transcriptional analysis of the Streptococcus mutans hrcA, grpE, and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- 32.Jones B E, Dossonnet V, Kuster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein, CcpA, to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 33.Kiska D L, Macrina F L. Genetic analysis of fructan-hyperproducing strains of Streptococcus mutans. Infect Immun. 1994;62:2679–2686. doi: 10.1128/iai.62.7.2679-2686.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiska D L, Macrina F L. Genetic regulation of fructosyltransferase in Streptococcus mutans. Infect Immun. 1994;62:1241–1251. doi: 10.1128/iai.62.4.1241-1251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Küster E, Luesink E J, Devos W M, Hillen W. Immunological crossreactivity to the catabolite control protein CcpA from Bacillus megaterium is found in many gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 36.LeBlanc D J, Lee L N, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of streptococcal origin. Plasmid. 1992;28:130–145. doi: 10.1016/0147-619x(92)90044-b. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Triccas J A, Ferenci T. A novel levansucrase-levanase gene cluster in Bacillus stearothermophilus ATCC 12980. Biochim Biophys Acta. 1997;1353:203–208. doi: 10.1016/s0167-4781(97)00103-6. [DOI] [PubMed] [Google Scholar]
- 38.Luchsinger W W, Cornesky R A. Reducing power by the dinitrosalicylic acid method. Anal Biochem. 1962;4:346–347. doi: 10.1016/0003-2697(62)90098-2. [DOI] [PubMed] [Google Scholar]
- 38a.Macrina, F. Personal communication.
- 39.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 40.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990;241:657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulsen I T, Chauvaux S, Choi P, Saier M H., Jr Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel hexose:H+ symporter. J Bacteriol. 1998;180:498–504. doi: 10.1128/jb.180.3.498-504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry D, Kuramitsu H K. Genetic linkage among cloned genes of Streptococcus mutans. Infect Immun. 1989;57:805–809. doi: 10.1128/iai.57.3.805-809.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate control of two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quivey R G, Jr, Faustoferri R C. In vivo inactivation of the Streptococcus mutans recA gene mediated by PCR amplification and cloning of a recA DNA fragment. Gene. 1992;116:35–42. doi: 10.1016/0378-1119(92)90626-z. [DOI] [PubMed] [Google Scholar]
- 46.Reddy V A, Maley F. Identification of an active-site residue in yeast invertase by affinity labeling and site-directed mutagenesis. J Biol Chem. 1990;265:10817–10820. [PubMed] [Google Scholar]
- 47.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 48.Sanger F, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneewind O, Pancholi V, Fischetti V A. Surface proteins of gram-positive cocci have a common motif for membrane anchoring. In: Dunny G M, Cleary P P, McKay L L, editors. Genetics and molecular biology of streptococci, lactococci, and enterococci. Washington, D.C: American Society for Microbiology; 1991. pp. 152–154. [Google Scholar]
- 50.Shaw W V. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 1979;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 51.Simpson C L, Russell R R B. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect Immun. 1998;66:2085–2092. doi: 10.1128/iai.66.5.2085-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sissons C H, Hancock E M, Perinpanayagam H E R, Cutress T W. The bacteria responsible for ureolysis in artificial dental plaque. Arch Oral Biol. 1988;33:727–734. doi: 10.1016/0003-9969(88)90006-4. [DOI] [PubMed] [Google Scholar]
- 53.Tao L, LeBlanc D J, Ferretti J J. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene. 1992;120:105–110. doi: 10.1016/0378-1119(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 54.Tortosa P, Aymerich S, Lindner C, Saier M H, Reizer J, LeCoq D. Multiple phosphorylation of SacY, a Bacillus subtilis transcriptional antiterminator negatively controlled by the phosphotransferase system. J Biol Chem. 1997;272:17230–17237. doi: 10.1074/jbc.272.27.17230. [DOI] [PubMed] [Google Scholar]
- 55.van Houte J, Jansen H M. Levan degradation by streptococci isolated from human dental plaque. Arch Oral Biol. 1968;13:827–830. doi: 10.1016/0003-9969(68)90102-7. [DOI] [PubMed] [Google Scholar]
- 56.von Heinje G, Abrahmsén L. Species-specific variation in signal peptide design: implications for protein secretion in foreign hosts. FEBS Lett. 1989;244:439–466. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]
- 57.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Wexler D L. Ph.D. thesis. Rochester, N.Y: University of Rochester; 1995. [Google Scholar]
- 58.Wexler D L, Hudson M C, Burne R A. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect Immun. 1993;61:1259–1267. doi: 10.1128/iai.61.4.1259-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wexler D L, Penders J E C, Bowen W H, Burne R A. Characteristics and cariogenicity of a fructanase-defective Streptococcus mutans strain. Infect Immun. 1992;60:3673–3681. doi: 10.1128/iai.60.9.3673-3681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]