Abstract
Isoaspartyl sites, in which an aspartic acid residue is linked to its C-flanking neighbor via its β-carboxyl side chain, are generally assumed to be an abnormal modification arising as proteins age. The enzyme protein l-isoaspartate methyltransferase (PIMT), present in many bacteria, plants, and animals, catalyzes the conversion of isoaspartate to normal α-linked aspartyl bonds and is thought to serve an important repair function in cells. Having introduced a plasmid into Escherichia coli that allows high-level expression of rat PIMT, we explored the possibility that the rat enzyme reduces isoaspartate levels in E. coli proteins, a result predicted by the repair hypothesis. The present study demonstrates that this is indeed the case; E. coli cells expressing rat PIMT had significantly lower isoaspartate levels than control cells, especially in stationary phase. Moreover, the distribution of isoaspartate-containing proteins in E. coli differed dramatically between logarithmic- and stationary-phase cultures. In stationary-phase cells, a number of proteins in the molecular mass range of 66 to 14 kDa contained isoaspartate, whereas in logarithmic-phase cells, nearly all of the detectable isoaspartate resided in a single 14-kDa protein which we identified as ribosomal protein S11. The near stoichiometric levels of isoaspartate in S11, estimated at 0.5 mol of isoaspartate per mol of S11, suggests that this unusual modification may be important for S11 function.
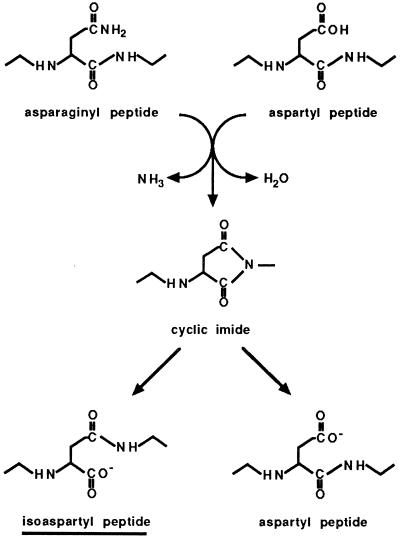
Isoaspartyl residues are generated within labile amino acid sequences by deamidation of asparagine or isomerization of aspartate (10, 13) (Fig. 1) and may result in diminished enzyme activity or other deleterious functional effects (3, 15). Protein l-isoaspartate methyltransferase (PIMT) (EC 2.1.1.77) utilizes the active methyl group of S-adenosyl-l-methionine (AdoMet) to methylate the α-carboxyl group on isoaspartate residues, producing methyl esters. Spontaneous breakdown of the methyl esters results in a mixture of isoaspartate and aspartate in approximately a 3:1 ratio (reference 4 and references therein). Isoaspartate recycling through this pathway converts most of the original isoaspartate to aspartate (16, 26).
FIG. 1.
Formation of isoaspartate by the deamidation of asparagine or the isomerization of aspartate.
Several lines of evidence indicate a role for PIMT in protein repair. In vitro incubation of isoaspartate-containing peptides (9, 16, 26) or proteins (3, 15) with mammalian PIMT and AdoMet results in significant conversion of the damaged sites to normal peptide linkages containing aspartate. Experiments using rat PC12 cells grown in the presence of a methylation inhibitor show a pronounced increase in protein isoaspartate levels (17). Moreover, recent reports have shown that when the mouse gene encoding PIMT is disrupted, cytosolic proteins from the knockout mice have four to eight times more isoaspartyl sites than proteins from the control mice (20, 41). The PIMT-deficient mice grow at a slower rate than the control mice and die from epileptic seizures when they are 26 to 60 days old. PIMT activity has been detected in procaryotic cells (12, 23, 30) and in animal and plant tissues (references 27 and 31 and references therein).
We previously transformed Escherichia coli with a recombinant plasmid (prIM) encoding rat PIMT and demonstrated that the culture produced active rat PIMT at a high level during stationary phase (6). In light of the presumed repair function of mammalian PIMT, we hypothesized that proteins in E. coli transformed with prIM have lower levels of isoaspartate than proteins from control cells. We were also interested in determining whether isoaspartyl sites accumulate in bacterial proteins during stationary phase as previously suggested (24) and whether isoaspartyl sites in E. coli are distributed at low levels among many proteins, as expected, or are present in only a few proteins.
In this study, we show that high-level expression of rat PIMT did indeed result in a significant lowering of isoaspartate in E. coli proteins and that stationary phase was associated with the accumulation of isoaspartate in a number of proteins. In the course of this investigation, we made the unexpected discovery that most of the isoaspartate in logarithmic-phase E. coli apparently resided in a single low-molecular-mass protein which we identified as ribosomal protein S11.
MATERIALS AND METHODS
Materials.
Ampicillin (AMP) (sodium salt), bovine serum albumin, S-adenosyl homocysteine, DNase I, protein A-Sepharose, and bovine gamma globulins were purchased from Sigma. Radiolabeled S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet) was purchased from DuPont NEN. Unlabeled AdoMet was purchased from Sigma and purified on carboxymethyl cellulose (5) before using it to dilute the [3H]AdoMet to the desired specific activity. Rat recombinant PIMT was purified as previously described (6) and had a specific activity of 8,000 to 15,000 U/mg, where 1 U is defined as 1 pmol of methyl transferred to bovine gamma globulins per min at 30°C. Isoaspartyl δ-sleep-inducing peptide (Trp-Ala-Gly-Gly-isoaspartate-Ala-Ser-Gly-Glu) was purchased from BACHEM. Purified E. coli MRE600 30S and 50S ribosomal subunits were donated by Harry Noller, University of California, Santa Cruz. Rabbit polyclonal antibodies to E. coli ribosomal proteins (r-proteins) were donated by Masayasu Nomura, University of California, Irvine.
Transformation of E. coli JM109.
Competent E. coli JM109 cells (Stratagene) were transformed with plasmid DNA, either pΔblue (2) or prIM (6), according to the supplier’s directions. Colonies were picked and grown overnight in Luria-Bertani broth (LB) (34) containing 100 μg of AMP per ml. Cultures were stored in 50% glycerol at −70°C.
Extract preparation with lysozyme.
Cultures of E. coli JM109/pΔblue and JM109/prIM grown overnight (16 h) (optical density at 600 nm [OD600] of 1.98 ± 0.50) were grown in LB containing 100 μg of AMP per ml at 37°C. The starter cultures were centrifuged at 4,350 × g for 10 min and resuspended in LB, and 0.5-ml samples of starter cultures were used to inoculate 500-ml portions of LB containing 200 μg of AMP per ml in 2-liter triple-baffled flasks (Bellco, Vineland, N.J.) to a cell density of approximately 107 cells/ml. Cultures were incubated at 37°C, with shaking at 250 rpm. Samples were removed and centrifuged at 5,500 × g for 10 min at 4°C. The pellets were washed twice in buffer A (50 mM Tris Cl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 10% glycerol, 15 mM 2-mercaptoethanol, 100 μM phenylmethylsulfonyl fluoride [PMSF], and stored at −20°C until used. Pellets were thawed on ice and resuspended in 3.2 ml of buffer A per g of pellet. Cells were lysed by using lysozyme as described previously (6) except RNase was omitted. Lysates were centrifuged at 15,800 × g for 20 min at 4°C. The supernatants were removed and centrifuged at 100,000 × g for 60 min at 4°C. Supernatants were stored at −20°C until used. Lysis yields ranged from 6 to 17 mg of protein/ml.
Methyl-accepting capacity reactions.
Briefly, a 50-μl reaction mixture containing 100 mM sodium phosphate (pH 6.8), 25 to 100 μg of extract protein, 100 μM [3H]AdoMet (specific activity, 500 dpm pmol−1), and 2.7 μM PIMT in a 1.5-ml microcentrifuge tube was incubated at 30°C for 30 min. Reactions were stopped and processed in a methanol diffusion assay as previously described (33).
To determine whether the enzymes used to produce the lysozyme lysis extracts contributed artifactually to the observed isoaspartyl content of the extracts, we assayed samples of lysis buffer containing the same amounts of lysozyme and DNase I used for extract preparation. We detected no significant amounts of isoaspartate in these samples.
Methyltransferase assays.
PIMT activity was measured as previously described (1) by using bovine gamma globulins as the methyl acceptor and [3H]AdoMet as the methyl donor.
Gel electrophoresis.
Samples were methylated for 10 min at 30°C in 100 mM potassium (2-[N-morpholino]ethanesulfonic acid) (potassium MES) (pH 6.2) containing 38 μM [3H]AdoMet (specific activity, 30,000 dpm/pmol) and 2 to 3 μM PIMT. The final volume was 6 μl. Reactions were processed as described previously (32). The acidic pH sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel system of Fairbanks and Avruch was used (7) (10% acrylamide, 0.75-mm-thick gels) followed by fluorography as described previously (17).
Ribosome preparation.
E. coli JM109/pΔblue cells were grown in LB containing 200 μg of AMP per ml at 37°C in 2-liter triple-baffled flasks (500 ml of LB per culture) on a shaker table set at 250 rpm. Logarithmic-phase cells (OD600 of 1.1) were harvested by centrifugation at 4,640 × g for 10 min. The pellets were washed two times in buffer B (10 mM Tris Cl [pH 7.6], 15 mM MgCl2, 500 mM NH4Cl, 1 mM dithiothreitol) and stored at −70°C. After the pellets were resuspended in buffer B, the cells were lysed with a French pressure cell (American Instrument Co.) at 6,000 to 8,000 lb/in2. The lysate was centrifuged at 39,200 × g for 45 min. Ribosomes were pelleted by centrifuging the supernatant at 226,000 × g for 3 h. The pellet was resuspended in a small volume of buffer B and stirred overnight at 4°C. The suspension was centrifuged at 39,200 × g for 45 min, and the pellet was discarded. The supernatant was then subjected to another round of ultracentrifugation and resuspension under the same conditions as described above. The final resuspended material was clarified by centrifugation at 12,100 × g for 15 min, and the supernatant was stored at −70°C until used.
Immunoprecipitation.
70S ribosomes in buffer B were applied to a Pharmacia Micro Spin S-200 HR column equilibrated in a solution containing 50 mM potassium MES (pH 6.2), 200 mM KCl, 1 mM dithiothreitol, 100 μM PMSF. This column was used for buffer exchange by following the manufacturer’s instructions. Next the 70S ribosomes (390 μg of protein) were methylated with 50 μM [3H]AdoMet (specific activity, 29,000 dpm/pmol) and 2 μM PIMT for 15 min at 30°C in a final concentration of 100 mM potassium MES, pH 6.2. The reaction was stopped by the addition of 10 μl of 5 mM S-adenosyl homocysteine, and the reaction mixture was placed on ice. To remove potassium ions, S-adenosyl homocysteine, and unreacted [3H]AdoMet, the reaction mixture was applied to another Micro Spin column equilibrated in a solution of 50 mM sodium MES (pH 6.2), 200 mM NH4Cl, and 100 μM PMSF. To dissociate r-proteins, the mixture was heated at 50°C for 10 min in the presence of 0.5% sodium dodecyl sulfate (SDS). After cooling, modified radioimmunoprecipitation (RIPA) buffer (11) was added to achieve final concentrations of 50 mM sodium MES (pH 6.2), 150 mM NH4Cl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 100 μM PMSF. Twenty-microliter portions of the methylated, dissociated 70S mixture along with 20 μl of modified RIPA buffer were incubated for 1 h at 4°C with 5 μl of polyclonal serum raised against purified r-proteins. Next, 100 μl of protein A-Sepharose (10% suspension in modified RIPA buffer) was added, and the tubes were gently rotated at 4°C for 1 h. After centrifugation at 11,600 × g for 20 s at 4°C, the supernatants were aspirated and discarded, and the pellets were washed two times in modified RIPA buffer, followed by one wash in 50 mM sodium MES (pH 6.2)–200 mM NH4Cl. The pellets were resuspended in 38 mM sodium MES (pH 6.2)–2% SDS–700 mM 2-mercaptoethanol. Samples were heated at 50°C for 10 min and then acidified by the addition of 4 μl of a solution containing 0.5 M sodium phosphate (pH 2.4), 50% glycerol, and 1.5 mg of pyronin Y per ml. After centrifugation, 10-μl portions of the supernatants were loaded onto an acidic pH SDS-PAGE gel as described above.
Other methods.
Protein concentrations were determined by the method of Lowry et al. (25) following precipitation in a final concentration of 5% (wt/vol) trichloroacetic acid. Bovine serum albumin was used as the standard.
RESULTS
Methyltransferase activity and isoaspartate levels in E. coli expressing rat PIMT.
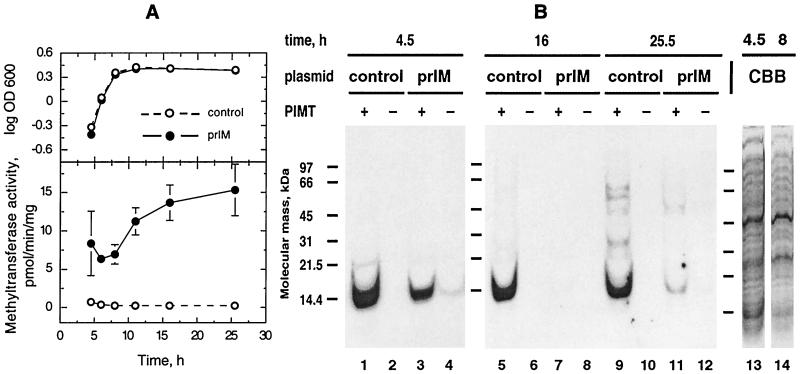
To investigate a possible relationship between PIMT activity and protein isoaspartyl content, we grew E. coli JM109 cells transformed with the rat PIMT-expressing plasmid prIM (prIM cells), and as a control, JM109 cells transformed with the expression vector pΔblue, which lacks the PIMT cDNA. The growth curves of the two cultures were nearly identical as monitored by optical density (Fig. 2A, upper panel). Hence, bacterial expression of the foreign gene was not detrimental to culture growth. As expected, PIMT activity in the overexpressing cells was always higher than in control cells, and the difference was greatest during stationary phase (25.5 h) when PIMT activity in prIM cell extracts was 70 times that in control cells (15.3 ± 3.4 compared to 0.216 ± 0.032 pmol of methyl/min/mg) (Fig. 2A, lower panel).
FIG. 2.
Methyltransferase activity and methyl-accepting capacity of E. coli JM109/pΔblue (control) and JM109/prIM extracts with respect to culture growth phase. (A) Growth curve (with optical density of culture represented by log OD600) (top) and methyltransferase activity of extracts (bottom). Datum points are averages from three separate experiments, and error bars represent standard deviations from the mean. (B) Acidic pH SDS-PAGE analysis of [3H]AdoMet-methylated JM109/pΔblue (control) and JM109/prIM extracts from the same cultures as shown in panel A. Fluorograms of gels are shown for lanes 1 to 12. Purified PIMT was included (+) or was not included (−) in the methylation reaction mixtures. Methylation conditions were as described in Materials and Methods. A 1-week exposure is shown. Lanes 13 and 14 show Coomassie brilliant blue (CBB) staining pattern of JM109/pΔblue (control) extracts. The extract in lane 13 was prepared from cells harvested 4.5 h postinoculation (logarithmic phase), and the extract in lane 14 was prepared from cells harvested 8 h postinoculation (late logarithmic/early stationary phase). All lanes contained 9 μg of extract protein. The figure was prepared with Adobe Photoshop version 5.0 for the Macintosh computer.
We examined the molecular mass distribution of isoaspartate-containing proteins in the extracts by methylating the extracts with [3H]AdoMet and PIMT followed by acidic pH SDS- PAGE and fluorography. We used the pH 2.4 gel system of Fairbanks and Avruch (7) to preserve base-labile methyl esters typically generated by PIMT. Surprisingly, analysis of logarithmic-phase cell extracts revealed only one major methyl-accepting protein band which migrated at about 14 kDa (Fig. 2B, lanes 1, 3, and 4). Methylation was PIMT dependent; no bands were seen in lanes containing control cell extracts subjected to the methylation conditions lacking exogenous PIMT (Fig. 2B, lanes 2, 6, and 10). As the control cell culture aged in stationary phase, several additional bands of higher molecular mass appeared (Fig. 2B, lane 9). The major methyl-accepting 14-kDa band in prIM cell extracts almost disappeared from samples harvested in stationary phase at 16 h postinoculation (Fig. 2B, lane 7), and a diffuse, light pattern of methyl incorporation was seen in the 25.5-h sample (Fig. 2B, lane 11). At the 4.5- and 25.5-h time points, prIM cell extracts had fainter bands migrating at 14 kDa in lanes that had no exogenous PIMT included in the methylation reaction mixtures (Fig. 2B, lanes 4 and 12). These bands were due to methylation catalyzed by the rat PIMT expressed by the cells. In both the prIM and control cell extracts, the heavily methylated 14-kDa protein and the more lightly methylated higher-mass proteins appeared to constitute two distinct classes of methyl acceptors. The overall lower levels of isoaspartate in prIM cell proteins suggested that the highly expressed rat PIMT was retarding the accumulation of isoaspartyl sites.
The Coomassie blue staining patterns of the extracts after SDS-PAGE varied with respect to the sample’s growth phase but not with respect to the culture identity (Fig. 2B, lanes 13 and 14). At the earliest sampling point, 4.5 h postinoculation, there was a darkly stained band that migrated at around 14 kDa, and its position matched that of the major methyl-accepting band seen in the fluorograms. In extracts made from cells in late logarithmic or stationary phase, protein migrating at around 14 kDa did not stain as darkly (Fig. 2B, lane 14), suggesting that the concentration of the methyl acceptor in these extracts was lower than in the logarithmic-phase extracts.
We repeated the methylation reactions, electrophoresis, and fluorography on extracts made from logarithmic- and late stationary-phase E. coli JM109 and DH1 cells to see whether the major methyl-accepting protein(s) was encoded on the plasmid or chromosome. Both JM109 and DH1 extracts showed methylation patterns very similar to that seen with control cell extracts (JM109/pΔblue) (data not shown). These results indicated that the major methyl-accepting protein(s) was encoded by the E. coli chromosome and was not strain specific.
Identity of the 14-kDa isoaspartate-rich protein.
The 14-kDa methyl acceptor appeared to have an overall positive charge. After methylation with PIMT and [3H]AdoMet, extracts made from logarithmic-phase control and prIM cultures were subjected to electrophoresis in a Triton-acetic acid-urea polyacrylamide gel system designed to separate highly basic proteins (39). Fluorography showed a single band in each extract that migrated near calf thymus histone H2B, which is highly basic and has a molecular mass of 13.8 kDa (data not shown). The intensities of the bands on the fluorogram from the Triton-acetic acid-urea-polyacrylamide gel were comparable to those on SDS-PAGE gel fluorograms, suggesting that most of the 14-kDa methyl acceptor seen in Fig. 2B was associated with a highly basic protein.
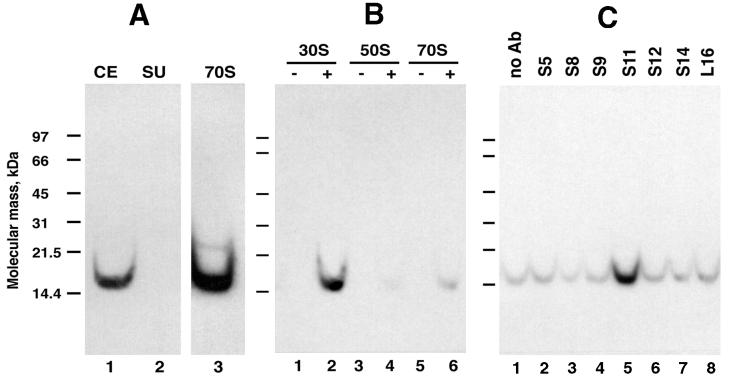
The low-molecular-mass, positive charge, and enrichment in logarithmic-phase cultures led us to suspect that the 14-kDa methyl acceptor may be a r-protein. Most E. coli r-proteins have molecular masses between 7 to 25 kDa, are most abundant in logarithmic phase (28a), and are very basic. To explore the idea that the major methyl acceptor was an r-protein, we prepared crude 70S ribosomes from a logarithmic-phase JM109/pΔblue culture (these cells had previously been used as the control cells in experiments involving cells that overexpress PIMT [see above]). The final fraction containing 70S ribosomes had a methyl-accepting capacity 3.3-fold higher than that of the crude extract. As shown in the Fig. 3A fluorogram, the crude extract contained one major band at around 14 kDa, and this band was enhanced in the 70S ribosome fraction. There was also a faint band in the 70S fraction migrating near the 21.5-kDa molecular mass marker. From these results, we concluded that the 14-kDa substrate was indeed one or more r-proteins.
FIG. 3.
Identification of r-protein S11 as the major methyl acceptor. (A) Major methyl acceptor copurified with 70S ribosomes. Ribosomes were isolated from logarithmic-phase E. coli JM109/pΔblue cultures as described in Materials and Methods. Samples (3.6 μg of protein) were methylated with PIMT and [3H]AdoMet and subjected to acidic pH SDS-PAGE followed by fluorography. Prior to methylation, the extracts were diluted in buffer such that the final magnesium and ammonium ion concentrations were 0.2 and 25 mM, respectively. A 48-h fluorogram is shown. Abbreviations: CE, crude extract; SU, supernatant after centrifugation of CE at 226,000 × g for 3 h; 70S, 70S ribosomal fraction. (B) The major methyl acceptor was in the 30S subunit, and its methylation was blocked by magnesium. Purified 30S (8.5 pmol of subunit, 3.1 μg of protein) and 50S (8.5 pmol of subunit, 4.4 μg of protein) subunits and 70S ribosomes (1.7 pmol of ribosome, 3 μg of protein) were methylated with PIMT and [3H]AdoMet and subjected to acidic pH SDS-PAGE followed by fluorography. The presence (+) or absence (−) of EDTA is indicated above the lanes. EDTA, when present in the reaction mixtures (lanes 2, 4, and 6), was at a final concentration of 15 mM. A 7-h fluorogram is shown. (C) Immunoprecipitation of the major methyl acceptor by antibodies (Ab) raised against E. coli r-protein S11. Experimental details are described in Materials and Methods. Antibodies directed against r-protein L16 (lane 8) were included in this experiment as a negative control. A 47-h fluorogram is shown. The brightness and contrast of each fluorogram were optimized, and the figure was prepared with Adobe Photoshop version 5.0 for the Macintosh computer.
In the course of our experiments with 70S ribosomes and ribosomal subunits (see below), we found that the 14-kDa substrate could not be methylated when magnesium and ammonium ions (components of the buffer used in ribosome purification) were present at concentrations of 5.0 mM and 170 mM, respectively, but could be methylated when the concentrations of these ions in the reaction mixtures were lowered to 0.26 and 27 mM, respectively. To see whether the effect was substrate or enzyme specific, we tested the ability of PIMT to methylate a synthetic isoaspartyl-containing peptide at various concentrations of magnesium and ammonium ions (Table 1). Methylation of the peptide was not inhibited even at the highest concentration of magnesium tested, 22.5 mM, a concentration 4.5-fold higher than was present during methylation of extracts and r-proteins prior to SDS-PAGE analysis. Ammonium ion had a slight inhibitory effect on PIMT methylation when it was present at 200 mM, and no inhibition was seen when it was present at 25 mM. When the highest tested concentrations of magnesium and ammonium ions were included together in the reaction mixture, PIMT methylation of the peptide was inhibited by 34% compared to the control. We conclude that the inability of PIMT to methylate the 14-kDa substrate in the presence of magnesium and ammonium ions was due to an interaction of these cations with the ribosome or with the individual substrate itself and not with the methyltransferase.
TABLE 1.
Effects of magnesium and ammonium ions on the ability of PIMT to methylate isoaspartyl δ-sleep-inducing peptidea
| Mg2+ concn (mM) | NH4+ concn (mM) | Methylation (% control) |
|---|---|---|
| 0 | 0 | 100 |
| 4.5 | 0 | 102 |
| 22.5 | 0 | 98 |
| 0 | 25 | 96 |
| 0 | 200 | 82 |
| 22.5 | 200 | 66 |
See Materials and Methods for a description of the methyl-accepting capacity assay. Final concentrations of isoaspartyl δ-sleep-inducing peptide and PIMT were 1 and 2 μM, respectively. Reactions were done in duplicate, and the average result is reported.
To determine whether the 14-kDa substrate was in the 30S, 50S, or both of the ribosomal subunits, we methylated purified subunits with [3H]AdoMet and PIMT and subjected them to SDS-PAGE followed by fluorography. Figure 3B shows that the 14-kDa substrate was solely a component of the 30S subunit. No methylation occurred in the absence of EDTA due to the presence of 5 mM magnesium in the subunit storage buffer (compare lanes 1 and 2 in Fig. 3B). The ammonium ion concentration in the methylation reaction mixtures was 50 mM. The results in Fig. 3B also demonstrate that the 14-kDa substrate was present in another strain of E. coli, namely, MRE600.
We next used an immunoprecipitation technique to identify which r-protein in the 30S subunit was the 14-kDa substrate. We tested antibodies made against purified r-proteins that were basic and that had molecular masses which fell within 15% of 14 kDa. Our screening included antibodies directed towards S5, S8, S9, S10, S11, S12, S13, and S14. The results shown in Fig. 3C are from a typical experiment and indicate that r-protein S11 was the only significant methyl acceptor. Radiolabel precipitated by antibodies other than anti-S11 was no greater than in the control (lane 1). S11 has a molecular mass of 13,728 Da (40), and its isoelectric point is greater than 12 (19).
DISCUSSION
Isoaspartate in E. coli.
It appears that isoaspartate in E. coli has two sources. One source, dominant in logarithmic-phase cells, resides almost exclusively in the 14-kDa protein we identified as r-protein S11. The second source is a group of higher-mass proteins that slowly accumulate isoaspartate during stationary phase. A rise in isoaspartate during stationary phase was anticipated previously by Li and Clarke, who suggested that isoaspartate accumulation might contribute to the loss of viability that occurs in stationary phase (24). More recently, Visick et al. provided the first direct evidence that isoaspartate does indeed accumulate during stationary phase (38). In that study, however, no comparisons were made between isoaspartate levels in early logarithmic and stationary phases and no information on the nature of the methyl-accepting species was reported.
PIMT from a mammalian source is likely to be better than a bacterial PIMT in detecting isoaspartate in proteins because it has a higher specific activity and recognizes isoaspartate in a wide variety of proteins and peptides. PIMT from E. coli has low activity toward proteins (23) and moderate activity toward an isoaspartyl-containing nonapeptide (8).
Isoaspartate in r-protein S11.
Twenty-one years ago, well before it was known that PIMT is selective for isoaspartate, Kim et al. (21) reported base-labile methylation of 30S E. coli r-proteins S3 and S9, and, possibly, S6 and S11. r-protein S3 has a molecular mass of 25,852 Da (40), which makes it an unlikely candidate for the 14-kDa methyl acceptor we have observed; however, S3 could be the less-pronounced methyl acceptor that migrated above the 21.5-kDa marker (Fig. 3A, lane 3). r-protein S6 has a molecular mass of 15,704 Da, but it is acidic (isoelectric point of 4.9). It would not have migrated into the Triton-acetic acid-urea-polyacrylamide gels, so although S6 may have a small amount of isoaspartate, we eliminated it as a candidate for the major methyl acceptor.
Our immunological results indicate that r-protein S11 was the major methyl-accepting protein in E. coli extracts. S11 is located on the platform region in the small (30S) subunit (29, 35). This platform region faces the large (50S) subunit (22) and is believed to be involved with codon-anticodon recognition (see reference 29 and references therein).
Both magnesium and ammonium ions appeared to inhibit the in vitro methylation of S11 catalyzed by PIMT. Magnesium has been shown to stabilize 70S ribosomes and subunits in vitro (36). At low (<2 mM) concentrations of magnesium, the 30S subunit becomes inactive with regard to the binding of Phe-tRNA (42). This inactivity has been attributed to a magnesium-dependent conformational change. A decrease in the ammonium ion concentration also leads to the inactivation of the 30S subunit. Additionally, incubation temperature affects the activity state of subunits. 30S subunits that have been inactivated by decreasing the concentration of magnesium or ammonium ions can be reactivated by replenishing the missing cation and heating the mixture. Perhaps these changes in 30S subunit conformation are intimately involved in changes in the tertiary structure of S11, leading to different states of the protein which have various degrees of accessibility to PIMT. Our observation that E. coli cultures overproducing PIMT had significantly lower amounts of isoaspartate in S11 argues that the PIMT-accessible form of S11 does exist in vivo.
The mole fraction of isoaspartate in S11 is at least 30-fold higher than in the average mammalian or E. coli cytosolic protein. By measuring the picomoles of [3H]methyl incorporated per microgram of ribosome protein and accounting for the known stoichiometry of S11 within the ribosome, we calculated that S11 contains approximately 0.5 mol of isoaspartate per mol of protein. The unusually high level of isoaspartate in S11 suggests that isoaspartate may be involved in the function of S11.
Do the E. coli and rat forms of PIMT have similar functions?
One of the original goals of this study was to test the putative repair function of mammalian PIMT by observing the relationship between isoaspartate and PIMT levels in E. coli transformed with prIM or a control plasmid. The high PIMT activity afforded by expression of the rat enzyme in prIM cells led to a significant reduction in isoaspartate in S11 as well as in the higher-mass proteins that accumulated isoaspartate during stationary phase. The rat PIMT apparently recognizes and methylates the S11 isoaspartyl site even though the E. coli homologue may not. These findings lend strong support to the idea that mammalian PIMT converts atypical isoaspartyl peptide bonds back to normal linkages.
It is unclear at present whether the E. coli enzyme serves a similar repair function. Although inactivation of the PIMT gene in mice leads to premature death in conjunction with high isoaspartate levels (20, 41), disruption of the E. coli PIMT gene alone has no marked effect on prolonged stationary-phase survival (37) nor does it lead to an increase in isoaspartate levels (38). As noted by Visick et al. (38), this unexpected result does not rule out a repair function for E. coli PIMT because other enzymes, such as proteases, may compensate for the loss of PIMT by rapidly degrading the abnormal isoaspartate-containing proteins. On the other hand, there is additional evidence that PIMT in E. coli differs functionally from its mammalian counterpart. First, as shown by Fu et al. (8), the E. coli and mammalian forms of PIMT differ dramatically in their abilities to methylate ovalbumin relative to a synthetic isoaspartate-containing peptide, suggesting that the E. coli enzyme recognizes a more restricted range of sequences than does the mammalian enzyme. Second, PIMT is apparently not required for all life forms, since it appears to be absent from several bacteria, including all gram-positive species tested (23), and from the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae (18). Because isoaspartate repair enzymes are apparently not essential for all bacteria, it follows that the presence of the isoaspartate-modifying methyltransferase in E. coli could, in principle, have a function other than protein repair. Isoaspartyl sites are characterized by an atypical α-carboxyl group that resembles the α-carboxyl normally found only at the C terminus of a polypeptide (14, 28). It seems possible that the E. coli PIMT is designed to modify isoaspartate-like carboxyl groups in a context unrelated to protein damage. Further studies on the substrate specificity of E. coli PIMT may provide important new insights as to its function and mechanism of action.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant NS17269.
We thank Harry Noller and Masayasu Nomura for providing materials and Lynn Young for technical assistance in figure preparation.
REFERENCES
- 1.Aswad D W, Deight E A. Purification and characterization of two distinct isozymes of protein carboxymethylase from bovine brain. J Neurochem. 1983;40:1718–1726. doi: 10.1111/j.1471-4159.1983.tb08147.x. [DOI] [PubMed] [Google Scholar]
- 2.Brandt M E, Vickery L E. Expression and characterization of human mitochondrial ferredoxin reductase in Escherichia coli. Arch Biochem Biophys. 1992;294:735–740. doi: 10.1016/0003-9861(92)90749-m. [DOI] [PubMed] [Google Scholar]
- 3.Brennan T V, Anderson J W, Jia Z, Waygood E B, Clarke S. Repair of spontaneously deamidated HPr phosphocarrier protein catalyzed by the L-isoaspartate-(D-aspartate) O-methyltransferase. J Biol Chem. 1994;269:24586–24595. [PubMed] [Google Scholar]
- 4.Brennan T V, Clarke S. Deamidation and isoaspartate formation in model synthetic peptides: the effects of sequence and solution environment. In: Aswad D W, editor. Deamidation and isoaspartate formation in peptides and proteins. Boca Raton, Fla: CRC Press; 1995. pp. 65–90. [Google Scholar]
- 5.Chirpich T P. Ph.D. dissertation. Berkeley: University of California; 1968. [Google Scholar]
- 6.David C L, Aswad D W. Cloning, expression, and purification of rat brain protein L-isoaspartyl methyltransferase. Protein Expr Purif. 1995;6:312–318. doi: 10.1006/prep.1995.1041. [DOI] [PubMed] [Google Scholar]
- 7.Fairbanks G, Avruch J. Four gel systems for electrophoretic fractionation of membrane proteins using ionic detergents. J Supramol Struct. 1972;1:66–75. doi: 10.1002/jss.400010110. [DOI] [PubMed] [Google Scholar]
- 8.Fu J C, Ding L, Clarke S. Purification, gene cloning, and sequence analysis of L-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem. 1991;266:14562–14572. [PubMed] [Google Scholar]
- 9.Galletti P, Ciardiello A, Ingrosso D, Di Donato A. Repair of isopeptide bonds by protein carboxyl O-methyltransferase: seminal ribonuclease as a model system. Biochemistry. 1988;27:1752–1757. doi: 10.1021/bi00405a055. [DOI] [PubMed] [Google Scholar]
- 10.Geiger T, Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. J Biol Chem. 1987;262:785–794. [PubMed] [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 12.Ichikawa J K, Clarke S. A highly active protein repair enzyme from an extreme thermophile: the L-isoaspartyl methyltransferase from Thermotoga maritima. Arch Biochem Biophys. 1998;358:222–231. doi: 10.1006/abbi.1998.0830. [DOI] [PubMed] [Google Scholar]
- 13.Johnson B A, Aswad D W. Enzymatic protein carboxyl methylation at physiological pH: cyclic imide formation explains rapid methyl turnover. Biochemistry. 1985;24:2581–2586. doi: 10.1021/bi00331a028. [DOI] [PubMed] [Google Scholar]
- 14.Johnson B A, Aswad D W. Fragmentation of isoaspartyl peptides and proteins by carboxypeptidase Y: release of isoaspartyl dipeptides as a result of internal and external cleavage. Biochemistry. 1990;29:4373–4380. doi: 10.1021/bi00470a017. [DOI] [PubMed] [Google Scholar]
- 15.Johnson B A, Langmack E L, Aswad D W. Partial repair of deamidation-damaged calmodulin by protein carboxyl methyltransferase. J Biol Chem. 1987;262:12283–12287. [PubMed] [Google Scholar]
- 16.Johnson B A, Murray E D, Jr, Clarke S, Glass D B, Aswad D W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J Biol Chem. 1987;262:5622–5629. [PubMed] [Google Scholar]
- 17.Johnson B A, Najbauer J, Aswad D W. Accumulation of substrates for protein-L-isoaspartyl methyltransferase in adenosine dialdehyde-treated PC12 cells. J Biol Chem. 1993;268:6174–6181. [PubMed] [Google Scholar]
- 18.Kagan R M, McFadden H J, McFadden P N, O’Connor C, Clarke S. Molecular phylogenetics of a protein repair methyltransferase. Comp Biochem Physiol B. 1997;117:379–385. doi: 10.1016/s0305-0491(96)00333-1. [DOI] [PubMed] [Google Scholar]
- 19.Kaltschmidt E. Ribosomal proteins. XIV. Isoelectric points of ribosomal proteins of E. coli as determined by two-dimensional polyacrylamide gel electrophoresis. Anal Biochem. 1971;43:25–31. doi: 10.1016/0003-2697(71)90103-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim E, Lowenson J D, MacLaren D C, Clarke S, Young S G. Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc Natl Acad Sci USA. 1997;94:6132–6137. doi: 10.1073/pnas.94.12.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Lew B, Chang F N. Enzymatic methyl esterification of Escherichia coli ribosomal proteins. J Bacteriol. 1977;130:839–845. doi: 10.1128/jb.130.2.839-845.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lake J A. Ribosomal subunit orientations determined in the monomeric ribosome by single and by double-labeling immune electron microscopy. J Mol Biol. 1982;161:89–106. doi: 10.1016/0022-2836(82)90280-7. [DOI] [PubMed] [Google Scholar]
- 23.Li C, Clarke S. Distribution of an l-isoaspartyl protein methyltransferase in eubacteria. J Bacteriol. 1992;174:355–361. doi: 10.1128/jb.174.2.355-361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Clarke S. A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci USA. 1992;89:9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.McFadden P N, Clarke S. Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins. Proc Natl Acad Sci USA. 1987;84:2595–2599. doi: 10.1073/pnas.84.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mudgett M B, Lowenson J D, Clarke S. Protein repair L-isoaspartyl methyltransferase in plants. Plant Physiol. 1997;115:1481–1489. doi: 10.1104/pp.115.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray E D, Jr, Clarke S. Synthetic peptide substrates for the erythrocyte protein carboxyl methyltransferase. J Biol Chem. 1984;259:10722–10732. [PubMed] [Google Scholar]
- 28a.Neidhardt F C, Ingraham J L, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinaver Associates, Inc.; 1990. pp. 420–434. [Google Scholar]
- 29.Oakes M, Henderson E, Scheinman A, Clarke M, Lake J A. Ribosome structure, function, and evolution: mapping ribosomal RNA, proteins, and functional sites in three dimensions. In: Hardesty B, Kramer G, editors. Structure, function, and genetics of ribosomes. New York, N.Y: Springer-Verlag; 1985. pp. 47–67. [Google Scholar]
- 30.O’Connor C M, Clarke S. Specific recognition of altered polypeptides by widely distributed methyltransferases. Biochem Biophys Res Commun. 1985;132:1144–1150. doi: 10.1016/0006-291x(85)91926-6. [DOI] [PubMed] [Google Scholar]
- 31.O’Connor M B, Galus A, Hartenstine M, Magee M, Jackson F R, O’Connor C M. Structural organization and developmental expression of the protein isoaspartyl methyltransferase gene from Drosophila melanogaster. Insect Biochem Mol Biol. 1997;27:49–54. doi: 10.1016/s0965-1748(96)00071-9. [DOI] [PubMed] [Google Scholar]
- 32.Orpiszewski J, Aswad D W. High mass methyl-accepting protein (HMAP), a highly effective endogenous substrate for protein L-isoaspartyl methyltransferase in mammalian brain. J Biol Chem. 1996;271:22965–22968. doi: 10.1074/jbc.271.38.22965. [DOI] [PubMed] [Google Scholar]
- 33.Potter S M, Henzel W J, Aswad D W. In vitro aging of calmodulin generates isoaspartate at multiple Asn-Gly and Asp-Gly sites in calcium-binding domains II, III, and IV. Protein Sci. 1993;2:1648–1663. doi: 10.1002/pro.5560021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Stöffler G, Stöffler-Meilicke M. Immuno electron microscopy on Escherichia coli ribosomes. In: Hardesty B, Kramer G, editors. Structure, function, and genetics of ribosomes. New York, N.Y: Springer-Verlag; 1985. pp. 28–46. [Google Scholar]
- 36.Tissiéres A, Watson J D, Schlessinger D, Hollingworth B R. Ribonucleoprotein particles from Escherichia coli. J Mol Biol. 1959;1:221–233. [Google Scholar]
- 37.Visick J E, Cai H, Clarke S. The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol. 1998;180:2623–2629. doi: 10.1128/jb.180.10.2623-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visick J E, Ichikawa J K, Clarke S. Mutations in the Escherichia coli surE gene increase isoaspartyl accumulation in a strain lacking the pcm repair methyltransferase but suppress stress-survival phenotypes. FEMS Microbiol Lett. 1998;167:19–25. doi: 10.1111/j.1574-6968.1998.tb13202.x. [DOI] [PubMed] [Google Scholar]
- 39.Wiekowski M, DePamphilis M L. Gel analysis of histone synthesis. Methods Enzymol. 1993;225:489–501. doi: 10.1016/0076-6879(93)25033-x. [DOI] [PubMed] [Google Scholar]
- 40.Wittmann H G. Components of bacteria ribosomes. Annu Rev Biochem. 1982;51:155–183. doi: 10.1146/annurev.bi.51.070182.001103. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto A, Takagi H, Kitamura D, Tatsuoka H, Nakano H, Kawano H, Kuroyanagi H, Yahagi Y, Kobayashi S, Koizumi K, Sakai T, Saito K, Chiba T, Kawamura K, Suzuki K, Watanabe T, Mori H, Shirasawa T. Deficiency in protein L-isoaspartyl methyltransferase results in a fatal progressive epilepsy. J Neurosci. 1998;18:2063–2074. doi: 10.1523/JNEUROSCI.18-06-02063.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamir A, Miskin R, Elson D. Inactivation and reactivation of ribosomal subunits: amino acyl-transfer RNA binding activity of the 30 s subunit of Escherichia coli. J Mol Biol. 1971;60:347–364. doi: 10.1016/0022-2836(71)90299-3. [DOI] [PubMed] [Google Scholar]