Abstract
Background/Aims
There are increasing pressures for anonymised datasets from clinical trials to be shared across the scientific community, and differing recommendations exist on how to perform anonymisation prior to sharing. We aimed to systematically identify, describe and synthesise existing recommendations for anonymising clinical trial datasets to prepare for data sharing.
Methods
We systematically searched MEDLINE®, EMBASE and Web of Science from inception to 8 February 2021. We also searched other resources to ensure the comprehensiveness of our search. Any publication reporting recommendations on anonymisation to enable data sharing from clinical trials was included. Two reviewers independently screened titles, abstracts and full text for eligibility. One reviewer extracted data from included papers using thematic synthesis, which then was sense-checked by a second reviewer. Results were summarised by narrative analysis.
Results
Fifty-nine articles (from 43 studies) were eligible for inclusion. Three distinct themes are emerging: anonymisation, de-identification and pseudonymisation. The most commonly used anonymisation techniques are: removal of direct patient identifiers; and careful evaluation and modification of indirect identifiers to minimise the risk of identification. Anonymised datasets joined with controlled access was the preferred method for data sharing.
Conclusions
There is no single standardised set of recommendations on how to anonymise clinical trial datasets for sharing. However, this systematic review shows a developing consensus on techniques used to achieve anonymisation. Researchers in clinical trials still consider that anonymisation techniques by themselves are insufficient to protect patient privacy, and they need to be paired with controlled access.
Keywords: Clinical trials, systematic review, data anonymisation, patient identification systems, personally identifiable information, datasets, data curation, guidelines
Introduction
Clinical trials are complex, time-consuming and costly, and it is wasteful not to use data fully. 1 Therefore, when academic-led clinical trials are completed, their results are usually released to the public and wider scientific community in scientific journals or clinical trials registries. Existing clinical trials’ data can be used to answer novel clinical questions, to reproduce and check analysis, to understand basic science, to investigate new methodologies and for teaching. 2 Also, there are sometimes considerable amounts of data that are not analysed as part of the published results. 3 In addition, trial data are often useful after the end of a trial to perform meta-analyses across several trials and using the individual patient data from each trial adds to the quality of such analyses, 4 for instance, by allowing full investigation of subgroup effects. There is now a drive, particularly from publishers and funders, to encourage the general release of relevant anonymised trial datasets 5 among interested parties.
Clinical trial datasets contain personal health information of the trial participants. It is imperative that data sharing does not disclose personal data to anyone who falls outside the original group to whom the trial participants have provided consent to access their data. Anonymising the trial dataset fulfils this requirement. However, the anonymisation process removes information from the data, and if not done carefully, the original trial analyses could not be reproduced, which in turn will limit the data’s usability for further research. 6
The drive to share data more widely has generated various sets of recommendations to enable sharing.5,7–10 Embedded within these, there is a variety of recommendations on how to anonymise a dataset.
Why it is important to do this review
To our knowledge, there are no reviews of the methods and/or recommendations for the process of generating anonymised clinical trial datasets (a search was executed on the 15 February 2021 on Google Scholar 11 with ‘literature’ ‘review’ ‘anonymization’ ‘methods’ ‘clinical trials’ and also ‘literature’ ‘review’ ‘anonymisation’ ‘methods’ ‘clinical trials’, the first 100 results were screened for each search and relevant results were not found).
To understand and collate the techniques used or recommended for data anonymisation in clinical trials, a systematic scoping review is required.
Objective
To identify, describe and synthesise the existing methods/recommendations to anonymise datasets from clinical trials.
Methods
The Joanna Briggs Institute Reviewers’ Manual: 2015 Methodology for JBI Scoping Reviews 12,13 and the PRISMA Extension for Scoping Reviews (PRISMA-ScR) 14 were followed for the execution of this scoping review.
Types of publications
We included any publications or documentation giving recommendations on anonymising datasets from clinical trials in any therapeutic area. Non-empirical publications, such as editorials, expert views or practice guidelines were also included in this review
Type of outcomes
The primary outcome is the reported methods and/or recommendations for anonymisation of clinical trials datasets.
Search methods for identification of publications
We performed a comprehensive systematic search to identify publications reporting methods or recommendations for anonymising clinical trials datasets. No language restrictions were imposed to attempt worldwide coverage. We did not identify any non-English publications.
Electronic searches
Web of Science (WoS), MEDLINE® (including non-indexed and in-process records) and EMBASE databases were searched from inception to 11 February 2019. The searches were rerun from 1 January 2019 to 8 February 2021 for MEDLINE® and EMBASE. A discrepancy was identified by M.F.D. in the original WoS strategy, so that, we reran the complete search from inception to 8 February 2021.
The search strategy used the following key concept areas, adopting subject headings and keywords as relevant for each database:
(Clinical) and
(trial* or randomi* or research* or control*) and
(principle* or guid* or recomm*) and
(shar* or reus* or re-us* or access* or open) and
(de-identi* or deidenti* or anonym* or privacy or confidential*)
The search was piloted with four indicator papers (Ohmann, 5 Keerie, 9 Tudor-Smith 15 and Hrynaszkie-wicz 16 ) that the searches needed to retrieved to ensure their effectiveness. The resulting detailed electronic search strategies are presented in Appendix 2 in the supplemental materials.
Searching other resources
To ensure the comprehensiveness of our search, we searched the websites of major research governance organisations and public research funding bodies as recommended by the Health Research Authority 17 and the Wellcome Trust, 18 the top 10 wealthiest charities, 19 the top 10 UK charities by brand value 20 and all registered UK academic clinical trials units, 21 to find guidelines published as grey literature from February 2019 until March 2020, so as not to omit documents not published as journal articles and not indexed in the bibliographic databases.
To further supplement our search field, we used citation and reference tracking (backwards and forward citation searching) on the selected articles from the electronic searches in order to identify additional sources. Preliminary results of this project were presented at the Fifth International Clinical Trials Methodology Conference 2019 22 where we requested to be contacted by any author or expert who could assist with the project but we did not receive any replies. During this event, several colleagues suggested publications to include in our grey literature.23,24 Shortly after, the COVID-19 pandemic started and we decided not to burden authors/experts with our requests and to concentrate on getting this project executed with the evidence that we had already collected. All the items included in this review obtained via the search of other resources were re-checked on May 2021 to locate updated versions since the original search.
Data collection and analysis
Records were retrieved and transferred into the reference manager EndNote, 25 which was used for de-duplication and to maintain a master library of the records throughout the review process. Covidence software 26 was used for further de-duplication, screening and full-text review. Two reviewers (A.R. and either C.T. or A.M.) independently screened titles and abstracts for eligibility. Full-text copies of all potentially relevant records were obtained using the reference manager.
Records identified from citation and reference tracking, and major research governance organisations, public research funding bodies and charity websites were collated in MS Excel, 27 for manual de-duplication and title screening. Records selected for full-text review were manually retrieved. Two teams (A.R. and either C.T. or A.M.) independently assessed whether each full-text record met the inclusion criteria. Chosen full-text records were added to the master library in EndNote. 25
Any discrepancies were discussed between the reviewers and if agreement could not be reached then it was arbitrated by a third reviewer (S.C.L., C.J.W. or S.E.).
Publications were excluded if they did not have concrete recommendations/methods of anonymisation, or they were not from a clinical trial framework, or they were focused on omics data or big data.
Data extraction/management and synthesis
A data extraction form to collect relevant data items from eligible sources was developed and piloted in line with Cochrane guidance, 28 this included: publication details (Authors names, Journal, year), country and classification (from electronic search or from other sources).
Data extraction and analysis was undertaken by one reviewer (A.R.) in NVivo®29 using thematic synthesis.30,31 Therefore, the included records were read‘line-by-line’, and when recommendations/methods on anonymisation were found, they were coded to a theme. At this stage, we allowed themes to be free and data-driven (i.e. to emerge from the data), rather than rigidly defining them a priori. It was possible to assign several themes to the same sentence. An independent sense-check was conducted by a second reviewer (A.M.) of the free themes. Any discrepancies were discussed between the reviewers and if an agreement could not be reached then it was resolved by a third reviewer (S.C.L., C.J.W. or S.E.).
The free themes were grouped into broader themes by the study team, this was repeated until we reached a final theme structure. We did not attempt to generate analytical themes 30 as our goal was to only identify the existing recommendations/methods on anonymisation.
Finally, the data from the included publications were summarised in descriptive tables. Themes were summarised by narrative analysis 32 and if applicable descriptive statistics.
Results
We identified 1059 potentially eligible records (Figure 1 in the online supplemental materials). Six hundred thirty-seven records were excluded after title and abstract screening. Three hundred sixty-three records were excluded after full-text review. Fifty-nine records5,9,15,16,23,24,33–86 (representing 43 studies) met the inclusion criteria and were included in the final qualitative synthesis (Appendix 3 has the full list and characteristics of the included records).
Figure 1.
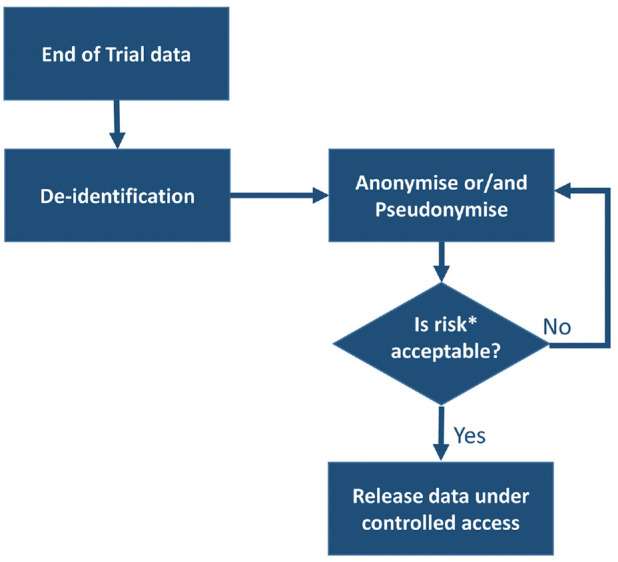
Most suggested method to release anonymised datasets from clinical trials.
Risk of re-identification is a complex variable, which is minimised using controlled access. The description of risk is out of scope for this review. Other processes: five studies described usage of open access instead, one study mentioned both controlled and open access for data release and the remaining two studies did not discussed data release.
Included studies’ characteristics
Table 1 summarises the observed characteristics of the included studies and their associated records, it also shows the included studies by source and country/region and year of publication. Figure 2 in the online supplemental materials shows the included studies over time.
Table 1.
Studies/record characteristics. a
| Parameter | Category | Studies N = 43, n (%) | Records N = 59 n(%) | |
|---|---|---|---|---|
| Source b | Electronic search | 19 (44) | 21 (36) c | |
| Other sources | 24 (56) | 38 (64) d | ||
| Country/region | EU | 12 (28) | 24 (39) | |
| UK | 11 (26) | 14 (23) | ||
| US | 10 (23) | 12 (20) | ||
| Canada | 5 (12) | 5 (8) | ||
| Australia | 2 (5) | 2 (3) | ||
| US–EU–UK | 2 (5) | 3 (5) | ||
| South Korea | 1 (2) | 1 (2) | ||
| Year of publication | 2003–2008 b | 5 (12) | 5 (8) | |
| 2009–2014 b | 15 (35) | 17 (29) | ||
| 2015–2020 b | 23 (53) | 37 (63) | ||
| Studies split by source | ||||
| Parameter | Category | Electronic search N=19, n (%) | Other sources N=24 n(%) | |
| Studies split by country/region | Canada/US | 6 (32) | 9 (37) | |
| EU/UK | 12 (63) | 11 (46) | ||
| Other regions e | 1 (5) | 4 (17) | ||
| Studies split by year of publication | 2003–2008 f | 3 (16) | 2 (8) | |
| 2009–2014 f | 7 (37) | 8 (33) | ||
| 2015–2020 f | 9 (47) | 14 (58) | ||
Therapeutic field was not applicable and it was not recorded.
Where applicable, the oldest record in the included study determined the overall study date.
Consisting of Australia, the United States–EU–the United Kingdom and South Korea.
Where applicable, the oldest record in the included study determined the overall study date.
Deriving the coding themes
A NVivo® exploratory word cloud was generated, it displayed the frequency in which significant words appeared in the included studies from the electronic searches (Figure 3 in the online supplemental materials), and it provided an initial idea of the themes present in the available data.
A.R. started the coding into free themes. As the actual coding progressed, the themes were reviewed and grouped by the study team until its structure was locked on 5 September 2019 by A.R., S.C.L. and C.J.W. The subsequent coding of the studies from other sources did not add any new themes. Eleven themes were identified (see Table 1 in the online supplemental materials).
The body of knowledge after coding themes
The 11 themes were applied to all 43 included studies (see Table 2). The most common theme among the selected studies were the definitions of de-identification (34 studies (79%)), anonymisation (28 studies (65%)), techniques for the manipulations of data (34 studies (79%)) and the implementation of controlled access for data release (38 studies (88%)).
Table 2.
Themes by studies.
| Id/theme | Studies (N = 43) | Associated records | |
|---|---|---|---|
| n | % | ||
| 1. Anonymisation | 28 | 65 | 5, 15, 16, 33, 34, 36, 37, 42, 44–48, 50–54, 56, 57, 63, 65, 66, 68–71,77–80, 82, 83, 85–87 |
| 2. De-identification | 34 | 79 | 5, 15, 16, 33, 34, 36, 37, 42, 44–48, 50–54, 56, 57, 63, 65, 66, 68–71,77–80, 82, 83, 85–87 |
| 2.1. HIPAA identifiers a | 23 | 53 | 5, 16, 24, 33, 34, 36–39, 43, 46, 47, 52–54, 56, 57, 59, 60, 63, 65, 66, 76,77, 80, 82–87 |
| 2.2. Hrynaszkiewicz identifiers b | 12 | 28 | 9, 16, 33, 45–47, 59, 60, 63, 66, 74, 78, 85–87 |
| 3. Pseudonymisation | 23 | 53 | 5, 9, 34, 36, 37, 40–44, 46, 49–51, 55–57, 66, 68, 69, 71, 77, 82 |
| 4. Manipulation of data | 34 | 79 | 9, 15, 16, 24, 33, 36–38, 42–48, 50, 53, 54, 56, 57, 59, 61, 62, 64–66,69, 71, 75, 77, 78, 80–84, 87 |
| 4.1. Perturbation c | 7 | 16 | 9, 36, 55, 66, 67, 77, 84 |
| 4.2. Recalculation c | 12 | 28 | 9, 16, 23, 43, 45, 52, 54, 56, 59, 63, 64, 67, 70, 73, 78, 80, 82, 83 |
| 4.3. Recoding c | 16 | 37 | 9, 33, 35, 43, 51–55, 59, 60, 63, 64, 66, 67, 69, 70, 72, 77–79, 82–84 |
| 4.4. Suppression c | 17 | 39 | 9, 35, 45, 51–54, 56, 57, 59, 60, 62, 63, 65–67, 69, 70, 72, 73, 77,78, 80, 82–84 |
| 4.5. Remove superfluous data c | 2 | 5 | 45, 48 |
| 5. Privacy model | 12 | 28 | 35, 38, 40–42, 46, 55, 57, 66, 69, 84–86 |
| 5.1. K-anonymity c | 7 | 16 | 35, 38, 40, 55, 57, 69, 84 |
| 6. Controlled access | 38 | 88 | 5, 9, 15, 16, 24, 33, 34, 36–39, 44–48, 50, 54, 57, 59, 60, 62–66, 71,72, 75, 77–87 |
| 6.1. Black box c | 3 | 7 | 41, 43, 46 |
| 6.2. Encryption c | 8 | 19 | 36, 39–42, 57, 66, 77 |
| 6.3. Safe haven c | 8 | 19 | 33, 36, 43, 46, 47, 55, 66, 83, 87 |
| 6.4. Split location c | 5 | 12 | 34, 41, 43, 66, 81 |
| 7. Open access | 7 | 16 | 9, 15, 36, 50, 56, 66, 85, 86 |
| 8. Central repositories | 5 | 12 | 16, 33, 46, 62, 66 |
| 9. Expert determination | 12 | 28 | 16, 24, 38, 46, 51, 66, 65, 74, 76, 77, 80, 82–84 |
| 10. Provision of context documents | 12 | 28 | 5, 9, 15, 44, 46–48, 62–64, 66, 75, 79, 82, 83, 87 |
| 11. Risk calculation | 15 | 35 | 16, 33, 35–37, 47–49, 57–59, 66, 69, 72, 78, 81, 82, 84–87 |
HIPAA: Health Insurance Portability and Accountability Act.
HIPAA identifiers refers to the HIPAA Safe Harbor method that requires the removal of 18 items of protected health information. 76
Hrynaszkiewicz identifiers refers to the removal of direct identifiers (information sources such as name and/or address, which on their own canre-identify participants) and the consideration/removal of indirect identifiers (variables that on their own might not represent a risk of re-identification for participants but in combination with of other indirect identifiers might increase the risk of re-identification, e.g. sex combined with age). 16
These are child codes that are included in their parent code.
In general terms, when study authors described anonymisation, de-identification and pseudonymisation, their explanations gravitated around the definitions presented in Table 3.
Table 3.
Most common definitions for anonymisation, de-identification and pseudonymisation.
| Pseudonymisation | De-identification | Anonymisation | ||
|---|---|---|---|---|
| • Attributes are replaced with pseudonyms on a one-to-one
correspondence • It is never an effective means of anonymisation • A security enhancing measure • Pseudonyms bear no relationto the patient details • Preferably reversible |
• Stripping datasets of patients identifying variables as
per either: ○ HIPAA 18 items‘Safe Harbor’ method (US) ○ Hrynaszkiewicz et al.28 items of personal andclinical information (Europe) |
• Any given record lacks any
individuality,distinction or recognisability • Can potentially distort data • The link with the original dataset should be destroyed • Set at a level to reach acceptable risk, but binary in law |
||
| Most common definitions for data manipulation techniquesa | ||||
| Suppression (removal, elimination) | Recoding (grouping, masking, replacement, generalisation, blurring, aggregation) | Recalculation | Perturbation | |
| • Delete outliers • Delete free-text • Delete high-risk variables • Delete high-risk records |
• Keep first three digits of postcode • Categorise age (18–40) and >= 40 |
• Show age instead of DOB • Show study day relativeto randomisationday, instead of date(e.g. day 7) • When dates are important they are presented offset |
• Add randomnoise to variables • Replace data withsimulated randomvalues • Data shuffling • Rounding of variables |
|
HIPAA: Health Insurance Portability and Accountability Act.
aTuck et al. 45 and Tudur-Smith et al. 48 mentioned the removal of superfluous data (e.g. deletion of data, such as audit trails) to supplement data manipulation techniques.
The described aim of data manipulation is to transform variables to reduce detail, without taking away too much data utility. The most common data manipulation methods’ definitions are given in Table 3.
Twelve studies (28%) recommended the use of privacy models (such as k-anonymity, 88 l-diversity 89 and differential privacy 90 ) to further guarantee and assess data anonymity to protect datasets from re-identification attacks.
The theme of controlled access mostly referred to the implementation of data-sharing agreements, the location of data behind a secure access barrier (either physical, virtual or both), the identification and vetoing of secondary research (e.g. checking requesters are bona fide researchers with a valid research question). In contrast, the theme of open access referred to minimal (or non-existent) requirements for allowing access to the data set to secondary researchers.
Central repositories (mentioned by five studies (12%)) were described as destinations where institutions could deposit their datasets to be managed by a third party and accessed by secondary researchers.92–95
The expert determination method for dataset release (12 studies (28%)) was generally described as when an expert (chosen for their knowledge/qualification) could assess the risk of re-identification of clinical trial datasets using ‘generally accepted statistical and scientific principles’, 66 if the risk is low, the data are certified and granted release to a secondary researcher.
Twelve studies (28%) recommended the provision of documental context to avoid erroneous interpretation and use of the anonymised datasets. Suggested documents to be provided included: original study protocol (and applicable amendments), statistical analysis plan, annotated case report forms and a data dictionary.
Finally, 15 studies (35%) highlighted the importance of assessing the risk of the anonymised dataset before making a decision on release, however, only four records35,59,66,69 (three studies) described how the risk could be calculated.
Most suggested processes for sharing anonymised datasets
Thirty-five studies (81%) described that at the end of a clinical trial, data should be de-identified (key items stripped from the dataset). Following this, data manipulation techniques should be used to further anonymise the datasets. Finally, the datasets should be made available under a controlled access approach.
Thirteen of those 35 studies also mentioned a step before release under controlled access in which the risk of re-identification should be assessed. This would start an iterative process, and once the risk is deemed acceptable, the anonymised data set should be made available under controlled access (Figure 1).
Discussion
The EU/UK region provided 53% of the included studies, followed by the US/Canada region with 35%, while the rest (12%) originated from other regions. This result was very similar when studies were split by source. Similarly, 53% of the included studies were published after 2015, 35% of the studies were published from 2009 to 2014 and the rest (12%) of the studies were published from 2003 to 2008. This profile was also observed when the studies were split by source, this shows the greater interest in this topic as time progresses. Overall, the EU/UK region from 2015 to 2020 was the most prolific with 16 studies out of 43 (37%). Where the content in the included studies was congruent regarding the source of the studies, this was noted, while the studies from other sources were coded because there was no need to update the coding themes generated with the studies from the electronic searches. However, a small but crucial difference is that studies from other sources have more detail and examples regarding data manipulations; this is most probably due to the lack of restriction on publication size for this type of source.
Topic 1: The relationship among the themes, pseudonymisation, de-identification and anonymisation, in the context of clinical trials. Anonymisation versus de-identification: they are both described as tools to facilitate data sharing. They rarely appear in isolation in any of the included studies, because they are part of the wider theme of data transparency and patient privacy. In this review, seven records coded to anonymisation, 14 records to de-identification and 28 records coded to both themes.
Anonymisation is presented as an abstract theme with lots of interpretation, mostly shaped by the regional laws where the publications originated (i.e. each researcher would have a theme that they favour which is shaped by their legal framework). These laws could be vague with their definitions and this could explain the existence of multiple concepts. On the other hand, de-identification is a more clear-cut and widely harmonised theme because it is defined in a precise way via Health Insurance Portability and Accountability Act (HIPAA). 96
The themes of anonymisation and de-identification appear to be gradually evolving, for example, older records considered anonymisation and de-identification as equivalent, while newer studies consider de-identification as a mechanical process to remove the identifiers, whereas anonymisation is the next step to prepare data for sharing (via data manipulation and privacy models). In general, most authors adhere to the narrative of further anonymising (via data manipulation and privacy models) the dataset after key variables have been removed, regardless of their previous definition of anonymisation and de-identification. Anonymisation is as a process to balance the minimisation of the probability of re-identification versus the utility of a clinical trial dataset, (e.g. too much anonymisation could render the data unusable).36,44,46,50,66,69,71,86,87 Therefore, data cannot be fully anonymised in the context of clinical trials.
Also, it seems well accepted and understood among authors that some variables in a clinical trial dataset are identifiers and that they can be classified as direct (e.g. name or address) 16 and indirect (also named as quasi-identifiers (e.g. present age instead of date of birth)). 16
Pseudonymisation of data usually occurs in the initial stages of data collection within clinical trials. 5 It also has a regional connotation, bound by the local laws and regulations. Pseudonymisation is declared to carry low risk for re-identification,5,66 however, no authors from the included studies advocated its use in isolation for data sharing. Some authors acknowledge that pseudonymisation alone is not acceptable for data sharing, as the one-to-one correspondence with the original fully identified dataset still exists, which makes it personal information under the EU and UK General Data Protection Regulation (GDPR).34,36,42,57,66
Topic 2: Most common data manipulation techniques to achieve anonymisation. Data manipulation techniques can be applied according to the data holder’s preference and technical capabilities and the intrinsic needs of the clinical trial dataset that is being processed.
Data manipulation techniques have multiple names, but there seems to be a progression towards a concerted set of four tools: perturbation, recalculation, recoding and suppression as presented in Table 3, with suppression, recoding and recalculation being the most talked about techniques. Authors are mostly describing via examples what is available regarding data manipulation techniques without critical judgement of the techniques, however, the majority of authors agree that data manipulation techniques are capable of reducing utility if left unexamined.
Topic 3: The introduction of privacy models. Clinical trials datasets are relatively small when compared to routinely collected data (e.g. medical records) and the implementation of a privacy model (such as differential privacy 90 ) could present challenges, also privacy models could be complicated techniques. This can explain why the uptake of privacy models is modest, despite the fact they come from methodologies that have been tried and tested in big datasets35,97 and they could be applied to clinical trials.35,38,40,55,57 The most common privacy model mentioned is k-anonymity. 88
Topic 4: The importance of controlled access and the tension with open access. The majority of clinical trial researchers strongly advocate for controlled access to the anonymised datasets, stemming from a concern with correct and genuine use of the anonymised data set.87,92
Authors recommend that the secondary researchers should have reasonable research questions and a data-sharing agreement should be put in place, which should include the use of the data for the intended purpose, the implementation of data protection procedures, the prohibition of any patient re-identification, the prohibition of sharing the data with a third party and the acknowledgement of the original authors in the secondary research output.
Regarding the actual sharing of the data, the trend is towards data access (e.g. via a safe haven) instead of data transfer, this means that secondary researchers can see and analyse the dataset but not download it. Here, the central repository plays a key role, because it would prove difficult (when it is necessary), to merge datasets that reside in separate repositories.
It is important to point out that controlled access is not required by laws or regulation, it is something that clinical trials researchers are doing, because it provides better research governance and researchers’ acknowledgement that anonymised datasets are still sensitive. 87 Stripping identifiers from datasets and the use of manipulation techniques are not sufficient on its own to fully anonymise clinical trials datasets and to protect patient privacy. Understandably, researchers do not want to breach patient trust and they want to pre-emptively defend against a potential data breach and its catastrophic consequences (loss of patient trust, hefty legal fines and loss of reputation), 98 but they are generally willing to share. 99
At the other end of the spectrum, open access is a relatively hassle-free release option once the dataset is anonymised, therefore, its existence and practicality is acknowledged, but it is not directly endorsed by any of the included papers as the research governance is very difficult under it. The International Stroke Trial (IST) database91,100 which is often cited as an example of a successful open access dataset by authors,66,87 also drew criticisms from others 86 regarding some of the indirect identifiers left in the dataset. However, IST is yet to report a successful re-identification attack. The limited use of open access causes frustration among secondary researchers who are eager to get fast and easy access to datasets.101,102
Currently, controlled access is still one of the main cornerstones for the release of anonymised clinical trials data and many authors agree that data should only be released if a threshold of acceptable risk is achieved. There are several available methods for calculating risk, but authors of included studies did not explain sufficiently what ‘acceptable’ means, reasonably, this is very difficult to define as it would depend on the context surrounding the release of the anonymised clinical trials datasets and on the datasets own characteristics.
Comparison with existing literature
We identified a similar systematic review by Chevrier, 103 which included all biomedical literature in MEDLINE® between 2007 and 2017. We agreed with them about the existence of multiple interpretations for anonymisation and de-identification and they also discussed the balancing act between the re-identification risk and data manipulation. However, their focus was on electronic health records, and those datasets have different needs and their own challenges when compared with clinical trials datasets.
Strengths and limitations
Strengths to this review are that the electronic databases were searched since inception without any language restrictions and there was a thorough coverage of grey literature. The database searches were complemented by screening of publications on websites of key organisations, and by citation tracking. Despite our extensive search, there might be a lack of representation from other regions outside the United States–Canada, the EU and the United Kingdom. The literature databases used in this review are international in scope, but are published in North America and Europe, and are known to be stronger in coverage of literature from those regions, so that, an unknown quantity of global literature not indexed in those databases was not scrutinised as part of this review.
In the same way, identification of other sources was biased towards websites and funders in the United States–Canada, the EU and the United Kingdom, due to lack of time and funding.
If this review is to be updated, it is possible to only run the electronic searches to obtain a quick actualisation of the recommendations. The records obtained searching other sources have strengthened the evidence found from the electronic searches, contributing more than half of the included studies, but they did not provide brand new information and searching other sources was a manual and time-consuming process. However, it could be worthwhile to directly seek updated records extracted from the Medical Research Council, 71 European Medicines Agency, 57 US Department of Health & Human Services61,76,84 and the Global Healthcare Data Science Community (Pharmaceutical Users Software Exchange – PhUSE).23,24,58–60,67,69,70,72,73
This review is exclusively gathering published recommendations/practices tailored specifically to clinical trials and it could not assess what researchers are actually using (but not reporting) for anonymising clinical trial data for sharing.
As this is a scoping review, there was no assessment of the quality of the evidence, therefore, we did not attempt to either explain how the included studies interpreted their local regulation on anonymisation, or to identify the existence of gaps in current practices from the obtained studies. The coding of the themes was a manual process and therefore subjective, however, a second reviewer sense-checked the coding and disagreements were mediated by a third reviewer which reduced the subjectivity of the findings.
Conclusion
Currently, there is a strong demand for academic researchers to share their data more readily. In clinical trials, data can be shared more widely if they are anonymised, yet, we do not have standardised recommendations on how to do this. As time goes by there seems to be an emerging natural consensus on the definitions of pseudonymisation, de-identification and anonymisation.
The data manipulation techniques currently used are still simple, with an increasing amount of authors recommending a shift towards privacy models, such as k-anonymity. There are other privacy models but they are not routinely used in clinical trials, as they could be complex, time-consuming and not practical for clinical trials datasets (which are relatively small when compared against routine health data).
It is impossible to discuss anonymisation in clinical trials datasets without considering the way in which the data is going to be accessed. Controlled access is still the keystone for the release of clinical trial data.
Finally, an increasing number of authors agree that data should only be released if a threshold of acceptable risk is achieved, but there is not a clear definition of ‘acceptable’ as this is a very complex parameter that not only relies on the dataset but it is also embedded in a wider context out of scope for this review.
The studies identified during this review need to next be critically appraised to identify any gaps in the literature regarding anonymisation methods and data access approaches. Also, clear guidance on methods for quantifying the risk of re-identification need to be developed. This would allow for the creation of standardised worldwide recommendations for data sharing in clinical trials reflecting the growing consensus exhibited in the literature found during this review.
Supplemental Material
Supplemental material, sj-docx-2-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-docx-3-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-docx-4-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-pdf-1-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Footnotes
Author contributions: A.R., S.C.L. and C.J.W. conceived the idea for this work supported by S.E., M.F.D., T.J. and C.T. A.R. wrote the first draft, and all authors contributed to the article.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Ethics and dissemination: This project did not collect any patient data or outcomes; therefore, it was not necessary to seek formal National Health Service Research Ethics Committee’s approval. However, we applied for ethical approval from the Internal Ethics Review Board at The University of Edinburgh’s Usher Institute.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: A.R. has a scholarship from the University of Edinburgh to undertake a PhD with the support from the Asthma UK Centre for Applied Research (AUKCAR) (AUKCAR-17-01a). Neither funder (University of Edinburgh) nor sponsor (AUKCAR) contributed to protocol development. C.J.W. is supported in this work by NHS Lothian via the Edinburgh Clinical Trials Unit. S.C.L. and C.T. are supported in this work by their employment at the Edinburgh Clinical Trials Unit. S.E. is supported in this work by her employment at the Pragmatic Clinical Trials Unit. M.F.D. is supported in this work by their employment at the University of Edinburgh. T.J. is supported by Asthma UK as part of the Asthma UK Centre for Applied Research (grant nos AUK-AC-2012-01 and AUK-AC-2018-01). A.M. is an independent researcher.
Protocol registration: The protocol for this scoping review was not registered with the International Prospective Register of Systematic Reviews (PROSPERO) as the proposed systematic review did not meet the PROSPERO inclusion criteria. However, the final protocol dated 11 January 2019 is attached as Appendix 1 in the supplemental materials.
ORCID iDs: Aryelly Rodriguez  https://orcid.org/0000-0002-1352-3922
https://orcid.org/0000-0002-1352-3922
Tracy Jackson  https://orcid.org/0000-0002-6188-3607
https://orcid.org/0000-0002-6188-3607
Christopher J Weir  https://orcid.org/0000-0002-6494-4903
https://orcid.org/0000-0002-6494-4903
Supplemental material: Supplemental material for this article is available online.
References
- 1. Chan A-W, Song F, Vickers A, et al. Increasing value and reducing waste: addressing inaccessible research. Lancet 2014; 383: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hrynaszkiewicz I, Altman DG. Towards agreement on best practice for publishing raw clinical trial data. Trials 2009; 10: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Song F, Hooper L, Loke Y. Publication bias: what is it? How do we measure it? How do we avoid it? Open Access J Clin Trials 2013; 2013: 71–81. [Google Scholar]
- 4. Berlin JA, Morris S, Rockhold F, et al. Bumps and bridges on the road to responsible sharing of clinical trial data. Clin Trials 2014; 11(1): 7–12. [DOI] [PubMed] [Google Scholar]
- 5. Ohmann C, Banzi R, Canham S, et al. Sharing and reuse of individual participant data from clinical trials: principles and recommendations. BMJ Open 2017; 7: e018647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. El Emam K, Arbuckle L. Anonymizing health data: case studies and methods to get you started. Sebastopol, CA: O’Reilly Media, Inc., 2013. [Google Scholar]
- 7. Hrynaszkiewicz I, Norton ML, Vickers AJ, et al. Preparing raw clinical data for publication trials. Trials 2010; 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Information Commissioner’s Office (ICO). Anonymisation code of practice, 2012, https://ico.org.uk/media/1061/anonymisation-code.pdf [Google Scholar]
- 9. Keerie C, Tuck C, Milne G, et al. Data sharing in clinical trials – practical guidance on anonymising trial datasets. Trials 2018; 19: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tudur Smith C, Hopkins C, Sydes M, et al. Good practice principles for sharing individual participant data from publicly funded clinical trials, 2015, https://www.methodologyhubs.mrc.ac.uk/files/7114/3682/3831/Datasharingguidance2015.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Google. Google Scholar. https://scholar.google.com/ [Google Scholar]
- 12. The Joanna Briggs Institute. Joanna Briggs Institute Reviewers’ manual: 2015 edition / supplement. Methodology for JBI scoping reviews. Adelaide, SA, Australia: The Joanna Briggs Institute, 2015. [Google Scholar]
- 13. Peters MD, Godfrey CM, Khalil H, et al. Guidance for conducting systematic scoping reviews. JBI Evid Implement 2015; 13: 141–146. [DOI] [PubMed] [Google Scholar]
- 14. Tricco AC, Lillie E, Zarin W, et al. Preferred reporting items for systematic review and meta-analysis (PRISMA) extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med 2018; 169: 467–473. [DOI] [PubMed] [Google Scholar]
- 15. Tudur Smith C, Hopkins C, Sydes MR, et al. How should individual participant data (IPD) from publicly funded clinical trials be shared? BMC Med 2015; 13: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hrynaszkiewicz I, Norton ML, Vickers AJ, et al. Preparing raw clinical data for publication: guidance for journal editors, authors, and peer reviewers. Trials 2010; 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Health Research Authority. Funding, 2019, https://www.hra.nhs.uk/planning-and-improving-research/research-planning/funding/
- 18. Wellcome. Clinical trials policy – Grant funding| Wellcome, 2019, https://wellcome.org/grant-funding/guidance/clinical-trials-policy
- 19. Wikipedia. List of wealthiest charitable foundations. Wikipedia, 2019, https://en.wikipedia.org/wiki/List_of_wealthiest_charitable_foundations
- 20. May M. Top 100 UK charities ranked for brand value in new league table. UK Fundraising, 27 November 2018, https://fundraising.co.uk/2018/11/27/top-100-uk-charities-ranked-brand-value-new-league-table/
- 21. UKCRC. Registered CTUs – UKCRC, 2019, https://ukcrc-ctu.org.uk/registered-ctus/. [Google Scholar]
- 22. Meeting abstracts from the 5th International Clinical Trials Methodology Conference (ICTMC 2019). Trials 2019; 20: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. PhUSE. PhUSE DeID Standard – SDTM 3.2 – appendix 1 – date offsetting – v1.91[2]. Kent: PhUSE, 2015. [Google Scholar]
- 24. PhUSE. PhUSE Data De-Identification Standard for SDTM 3.2 – appendix 2-low frequencies-v10-19387. Kent: PhUSE, 2015. [Google Scholar]
- 25. The EndNote Team. EndNote. EndNote X8 ed. Philadelphia, PA: Clarivate Analytics, 2016. [Google Scholar]
- 26. Veritas Health Innovation. Covidence systematic review software. Melbourne, VIC, Australia: Veritas Health Innovation. [Google Scholar]
- 27. Microsoft Corporation. Microsoft Excel. 2016, https://office.microsoft.com/excel [Google Scholar]
- 28. Higgins JPT, Green S. Cochrane handbook for systematic reviews of interventions. London: The Cochrane Collaboration, 2011. [Google Scholar]
- 29. QSR International Pty Ltd. NVivo (Version 11). 2015, https://www.qsrinternational.com/nvivo-qualitative-data-analysis-software/home [Google Scholar]
- 30. Thomas J, Harden A. Methods for the thematic synthesis of qualitative research in systematic reviews. BMC Med Res Methodol 2008; 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gibbs GR. Thematic coding and categorizing. Anal Qual Data 2007; 703: 38–56. [Google Scholar]
- 32. Parcell ES, Baker B. Narrative analysis. In: Allen M. (ed.) The Sage encyclopedia of communication research methods, vol. 3. Thousand Oaks, CA: SAGE, 2017, pp. 1069–1072. [Google Scholar]
- 33. Atzor S, Sorof J, Kelman A, et al. Clinical trial data sharing: from principles to practical implementation – an industry model. Regul Rapp 2014; 11: 4–7. [Google Scholar]
- 34. Demotes-Mainard J, Cornu C, Guerin A, et al. How the new European data protection regulation affects clinical research and recommendations? Therapie 2019; 74: 31–42. [DOI] [PubMed] [Google Scholar]
- 35. El Emam K, Dankar FK. Protecting privacy using k-anonymity. J Am Med Inform Assoc 2008; 15(5): 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. El Emam K, Rodgers S, Malin B. Anonymising and sharing individual patient data. BMJ 2015; 350: h1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J, Jung J, Park P, et al. Design of a human-centric de-identification framework for utilizing various clinical research data. Hum-Centric Comput Inf Sci 2018; 8: 1–12. [Google Scholar]
- 38. Malin B, Karp D, Scheuermann RH. Technical and policy approaches to balancing patient privacy and data sharing in clinical and translational research. J Investig Med 2010; 58(1): 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morse RE, Nadkarni P, Schoenfeld DA, et al. Web-browser encryption of personal health information. BMC Med Inf Decis Mak 2011; 11: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasseh D, Engel J, Mansmann U, et al. Matching study to registry data: maintaining data privacy in a study on family based colorectal cancer. Stud Health Technol Inform 2014; 205: 808–812. [PubMed] [Google Scholar]
- 41. Nitzlnader M, Schreier G. Patient identity management for secondary use of biomedical research data in a distributed computing environment. Stud Health Technol Inform 2014; 198: 211–218. [PubMed] [Google Scholar]
- 42. Noumeir R, Lemay A, Lina JM. Pseudonymization of radiology data for research purposes. J Digit Imaging 2007; 20(3): 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schell SR. Creation of clinical research databases in the 21st century: a practical algorithm for HIPAA Compliance. Surg Infect 2006; 7(1): 37–44. [DOI] [PubMed] [Google Scholar]
- 44. Sudlow R, Branson J, Friede T, et al. EFSPI/PSI working group on data sharing: accessing and working with pharmaceutical clinical trial patient level datasets – a primer for academic researchers. BMC Med Res Methodol 2016; 16: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuck C, Lewis S, Milne G, et al. Data sharing in clinical trials – practical guidance on anonymising trial datasets – oral presentation. Trials 2015; 16. 10.1186/1745-6215-16-S2-O66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tucker K, Branson J, Dilleen M, et al. Protecting patient privacy when sharing patient-level data from clinical trials. BMC Med Res Methodol 2016; 16(Suppl. 1): 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tudur Smith C, Hopkins C, Sydes M, et al. Good practice principles for sharing individual participant data from publicly funded clinical trials. Trials 2015; 16. 10.1186/1745-6215-16-S2-O1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tudur Smith C, Nevitt S, Appelbe D, et al. Resource implications of preparing individual participant data from a clinical trial to share with external researchers. Trials 2017; 18: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wallace SE, Gaye A, Shoush O, et al. Protecting personal data in epidemiological research: DataSHIELD and UK law. Public Health Genomics 2014; 17(3): 149–157. [DOI] [PubMed] [Google Scholar]
- 50. Asthma UK Centre for Applied Research. ASTHMA UK policy data sharing – introduction to sharing individual participant data, version 2. Circa, 2015, p. 2. [Google Scholar]
- 51. Australian National Medical Research Data Storage Facility. Anonymisation. Circa, 2016, https://researchdata.edu.au/meddataeduau/632288 [Google Scholar]
- 52. Clinical Study Data Request (CSDR). CSDR – anonymisation of clinical trial datasets. Circa, 2015, https://www.clinicalstudydatarequest.com/Default.aspx [Google Scholar]
- 53. Clinical Study Data Request (CSDR) and Eisai. CSDR – anonymisation of clinical trial datasets – Eisai circa, 2015, https://www.clinicalstudydatarequest.com/Study-Sponsors/Study-Sponsors-Eisai.aspx [Google Scholar]
- 54. Clinical Study Data Request (CSDR) EL. CSDR – anonymisation of clinical trial datasets – Eli Lilly and Company. Circa, 2015, https://www.clinicalstudydatarequest.com/Documents/Anonymisation_clinicaltrialdata_Lilly_update.pdf
- 55. Ebner H, Hayn D, Falgenhauer M, et al. Piloting the European unified patient identity management (EUPID) concept to facilitate secondary use of neuroblastoma data from Clinical Trials and Biobanking. In: Schreier G, Ammenwerth E, Hörbst A, et al. (eds) Health informatics meets eHealth: predictive modeling in healthcare–from prediction to prevention proceedings of the 10th eHealth2016 conference. Amsterdam: IOS Press, 2016, pp. 31–38. [PubMed] [Google Scholar]
- 56. El Emam K, Malin B. Concepts and methods for de-identifying clinical trial data. Paper commissioned by the Committee on Strategies for Responsible Sharing of Clinical Trial Data, 2014. [Google Scholar]
- 57. European Medicines Agency (EMA). Data anonymisation – a key enabler for clinical data sharing. Workshop Report No. EMA/796532/2018, 4 December 2018. London: European Medicines Agency. [Google Scholar]
- 58. Ferran J-M. PhUSE – De-Identification Standards for CDISC data models – PhUSE, Data Transparency Working Group Lead. In: 4th international clinical trials methodology conference (ICTMC), Liverpool, 7–10 May 2017. [Google Scholar]
- 59. Ferran J-M, El Emam K, Nolan S, et al. PhUSE De-Identification Working Group: providing De-Identification Standards to CDISC data models. Kent: PhUSE, 2015. [Google Scholar]
- 60. Ferran J-M, Lanoue J. PhUSE De-Identification Working Group: providing De-Identification Standards to CDISC data models – DS10, 2015, https://www.pharmasug.org/proceedings/2015/DS/PharmaSUG-2015-DS10.pdf. [Google Scholar]
- 61. Food Drug Administration. HHS – availability of masked and de-identified non-summary safety and efficacy data; request for comments. Federal Register, 2013, https://www.federalregister.gov/articles/2013/06/04/2013-13083/availability-of-masked-and-de-identified-non-summary-safety-and-efficacy-data-request-for-comments
- 62. Hollis S, Fletcher C, Lynn F, et al. Best practice for analysis of shared clinical trial data. BMC Med Res Methodol 2016; 16(Suppl. 1): 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hughes S, Wells K, McSorley P, et al. Preparing individual patient data from clinical trials for sharing: the GlaxoSmithKline approach. Pharm Stat 2014; 13(3): 179–183. [DOI] [PubMed] [Google Scholar]
- 64. Huser V, Shmueli-Blumberg D. Data sharing platforms for de-identified data from human clinical trials. Clin Trials 2018; 15(4): 413–423. [DOI] [PubMed] [Google Scholar]
- 65. International Pharmaceutical Privacy Consortium. IPPC white paper on anonymisation of clinical trial datasets, 2014, https://6a908337-3075-4032-a3f9-cee264a142f8.filesusr.com/ugd/932589_48d9c33238994cdfa5ee4273a29fe444.pdf
- 66. IOM (Institute of Medicine). Sharing clinical trial data: maximizing benefits, minimizing risk. Washington, DC: National Academies Press, 2015. [PubMed] [Google Scholar]
- 67. Iversen JM. PhUSE – Data De-Identification made simple. Ballerup, Denmark: PHUSE – LEO Pharma A/S, 2016. [Google Scholar]
- 68. Jonas S, Siewert S, Spreckelsen C. Privacy-preserving record grouping and consent management based on a public-private key signature scheme: theoretical analysis and feasibility study. J Med Internet Res 2019; 21: e12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kniola L, Hughes A, Paczewska-Sosnowska A, et al. PhUSE – data anonymisation and risk assessment automation. Kent: PhUSE, 2020, p. 10. [Google Scholar]
- 70. Lyathakula S. PhUSE – data anonymization providing clinical trial data to outside researchers. In: NOVARTIS (ed.) PhUSE Single Day Event (SDE). Mumbai, India, 2015. [Google Scholar]
- 71. Medical Research Council (MRC). GDPR guidance note 5: identifiability, anonymisation and pseudonymisation, 2019, https://www.ukri.org/wp-content/uploads/2021/11/MRC-291121-GDPR-Identifiability-Anonymisation-Pseudonymisation.pdf
- 72. Meeh S. PhUSE Data De-Identification Standard for CDSIC SDTM IG 3.2, and EMA Policy 0070. Integrated Data Analytics and Reporting Janssen, 2016, https://slideplayer.com/slide/17911736/
- 73. Meeh S. PhUSE Data De-Identification Standard for CDSIC ADaM 2.1 IG 1.0, and updates for SDTM IG 3.2, 2017, https://slideplayer.com/slide/11742761/ [Google Scholar]
- 74. Miller JD. Sharing clinical research data in the United States under the health insurance portability and accountability act and the privacy rule. Trials 2010; 11: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. National Institutes of Health (NIH). NIH data sharing policy and implementation guidance, 2003, https://grants.nih.gov/grants/policy/data_sharing/data_sharing_guidance.htm
- 76. National Institutes of Health (NIH). HHS – clinical research and the HIPAA privacy rule, 2004, http://privacyruleandresearch.nih.gov/clin_research.asp
- 77. Nelson GS. Practical implications of sharing data: a primer on data privacy, anonymization, and de-identification. In: SAS (ed.) SAS GLOBAL FORUM proceedings 2015, 2015, pp. 1–23, https://www.pharmasug.org/proceedings/2016/IB/PharmaSUG-2016-IB06.pdf [Google Scholar]
- 78. Olesen S. Publishing and sharing sensitive data. Australian National Data Service, 2011, https://www.ands.org.au/guides/sensitivedata
- 79. Pfizer. Clinical trial data access – policy document 01312014, 2014. [Google Scholar]
- 80. Shostak J. De-identification of clinical trials data demystified. SAS Users Group, 2006, https://www.lexjansen.com/pharmasug/2006/PublicHealthResearch/PR02.pdf
- 81. The Expert Panel on Timely Access to Health and Social Data for Health Research and Health System Innovation. Accessing health and health-related data in Canada. Ottawa, ON, Canada: Council of Canadian Academies, 2015. [Google Scholar]
- 82. TransCelerate BioPharma Inc. TransCelerate – data de-identification and anonymization of individual patient data in clinical studies. TransCelerate – Clinical Data Transparency Initiative, 2016, https://www.transceleratebiopharmainc.com/initiatives/clinical-data-transparency/ [Google Scholar]
- 83. TransCelerate BioPharma Inc. TransCelerate – anonymization of individual patient data in clinical studies – a model approach. De-identification, TransCelerate-Data, 2015, https://www.transceleratebiopharmainc.com/wp-content/uploads/2015/04/TransCelerate-De-identification-and-Anonymization-of-Individual-Patient-Data-in-Clinical-Studies-V2.0.pdf
- 84. U.S. Department of Health & Human Services (HHS). HHS – guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. Washington, DC: U.S. Department of Health and Human Services, 2012, p. 26, https://wwwhhsgov/hipaa/for-professionals/privacy/special-topics/de-identification/indexhtml [Google Scholar]
- 85. Walker N. All or nothing: the false promise of anonymity. bioRxiv 2016: 084921, https://www.biorxiv.org/con tent/biorxiv/early/2016/11/03/084921.full.pdf [Google Scholar]
- 86. Walker N. All or nothing: the false promise of anonymity. Data Sci J 2017; 16: 24. [Google Scholar]
- 87. Tudur Smith C, Hopkins C, Sydes MR, et al. Good practice principles for sharing individual participant data from publicly funded clinical trials, version 1. Medical Research Council; Hubs for Trials Methodology Research, 2015, https://www.methodologyhubs.mrc.ac.uk/files/7114/3682/3831/Datasharingguidance2015.pdf
- 88. Sweeney L. K-anonymity: a model for protecting privacy. Int J Uncertain Fuzziness Knowl Based Syst 2002; 10: 557–570. [Google Scholar]
- 89. Machanavajjhala A, Kifer D, Gehrke J, et al. l-diversity; privacy beyond k-anonymity. Acm T Knowl Discov D 2007; 1: 3–es. [Google Scholar]
- 90. Dwork C. Differential privacy: a survey of results, 2008, pp. 1–19, https://web.cs.ucdavis.edu/~franklin/ecs289/2010/dwork_2008.pdf
- 91. Clinical Trials Unit London School of Hygiene & Tropical. freeBIRD (Bank of injury and Emergency Research Data) 2011, https://freebird.lshtm.ac.uk/home/ [Google Scholar]
- 92. Clinical Study Data Request (CSDR). Clinical study data request. https://www.clinicalstudydatarequest.com/ [Google Scholar]
- 93. Project Data Sphere. An independent initiative of the CEO Roundtable on Cancer. https://www.projectdatasphere.org/
- 94. The Yale University. Yale University Open Data Access (YODA) Project, https://yoda.yale.edu/
- 95. Vivli Center for Global Clinical Research Data. A global clinical research data sharing platform, https://vivli.org/home-mar2020/
- 96. Health UDo and Services H. Protecting personal health information in research: understanding the HIPAA privacy rule. Washington, DC: Author, 2003. [Google Scholar]
- 97. Sweeney L. Achieving k-anonymity privacy protection using generalization and suppression. Int J Uncertain Fuzziness Knowl Based Syst 2002; 10: 571–588. [Google Scholar]
- 98. Information Commissioner’s Office (ICO). Anonymisation: managing data protection risk code of practice, 2012, https://ico.org.uk/media/1061/anonymisation-code.pdf
- 99. Rathi V, Dzara K, Gross CP, et al. Sharing of clinical trial data among trialists: a cross sectional survey. BMJ 2012; 345: e7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Roberts I, Shakur H, Coats T, et al. The CRASH-2 trial: a randomised controlled trial and economic evaluation of the effects of tranexamic acid on death, vascular occlusive events and transfusion requirement in bleeding trauma patients – appendix 4 Free Bank of Injury and emergency Research Data – freeBIRD. Health Technol Assess 2013; 17: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Loder E. Data sharing: making good on promises. BMJ 2018; 360: k710. [Google Scholar]
- 102. Dunn AG, Day RO, Mandl KD, et al. Learning from hackers: open-source clinical trials. Sci Transl Med 2012; 4: 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chevrier R, Foufi V, Gaudet-Blavignac C, et al. Use and understanding of anonymization and de-identification in the biomedical literature: scoping review. J Med Internet Res 2019; 21: e13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-2-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-docx-3-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-docx-4-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials
Supplemental material, sj-pdf-1-ctj-10.1177_17407745221087469 for Current recommendations/practices for anonymising data from clinical trials in order to make it available for sharing: A scoping review by Aryelly Rodriguez, Christopher Tuck, Marshall F Dozier, Stephanie C Lewis, Sandra Eldridge, Tracy Jackson, Alastair Murray and Christopher J Weir in Clinical Trials


