SUMMARY
Emerging evidence suggests that G protein-coupled receptor (GPCR) kinases (GRKs) are associated with the pathophysiology of Alzheimer’s disease (AD). However, GRKs have not been directly implicated in regulation of the amyloid-β (Aβ) pathogenic cascade in AD. Here, we determine that GRKs phosphorylate a non-canonical substrate, anterior pharynx-defective 1A (APH1A), an integral component of the γ-secretase complex. Significantly, we show that GRKs generate distinct phosphorylation barcodes in intracellular loop 2 (ICL2) and the C terminus of APH1A, which differentially regulate recruitment of the scaffolding protein β-arrestin 2 (βarr2) to APH1A and γ-secretase-mediated Aβ generation. Further molecular dynamics simulation studies reveal an interaction between the βarr2 finger loop domain and ICL2 and ICL3 of APH1A, similar to a GPCR-β-arrestin complex, which regulates γ-secretase activity. Collectively, these studies provide insight into the molecular and structural determinants of the APH1A-βarr2 interaction that critically regulate Aβ generation.
Graphical Abstract

In brief
GRKs phosphorylate a growing list of non-GPCR substrates to regulate GPCR-independent signaling cascades. Todd et al. show that GRKs regulate phosphorylation of the 7 TMD subunit of the γ-secretase complex APH1A, which differentially affects recruitment of the scaffolding protein βarr2 to APH1A and modulation of γ-secretase activity and Aβ generation.
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized pathologically by the deposition of extracellular plaques composed of the amyloid-β (Aβ) peptide and intracellular inclusions of the misfolded and post-translationally modified microtubule-associated protein tau (Hyman et al., 2012; Serrano-Pozo et al., 2011). Sequential cleavage of the amyloid precursor protein (APP) by the β-secretase and γ-secretase yields Aβ peptides, which range in length from 37 to 46 amino acids (Zhang et al., 2011). γ-Secretase is a 4-subunit complex consisting of nicastrin (NCT), anterior pharynx-defective (APH) 1A or APH1B, presenilin-enhancer 2 (PEN2), and presenilin 1 (PS1) or presenilin 2 (PS2), the catalytic core of the complex (Hur et al., 2008; De Strooper, 2003; Vetrivel et al., 2004). APH1 is required for γ-secretase complex assembly, stability, and catalytic activity (Serneels et al., 2005; De Strooper, 2003). In humans, two genes (APH1A and APH1B) encode for the two APH1 proteins, APH1A and APH1B, respectively. We previously determined that β-arrestin 2 (βarr2), a multifunctional G protein-coupled receptor (GPCR) scaffolding protein, interacts with APH1A and stabilizes localization of the γ-secretase complex in lipid raft, or detergent-resistant membrane (DRM), domains where it is more catalytically active (Thathiah et al., 2013).
The β-arrestin family of proteins (βarr1 and βarr2) recognizes and binds to phosphorylated serine or threonine residues on the carboxy terminus (C terminus) or intracellular cytoplasmic loops (ICLs) of GPCRs following receptor activation (DeWire et al., 2007; Lefkowitz and Shenoy, 2005). GPCR kinases (GRKs) are serine/threonine kinases that are primarily responsible for GPCR phosphorylation (Gurevich et al., 2012). Evidence within the past decade suggests that multiple GRKs can phosphorylate a single receptor at distinct sites. The specific phosphorylation pattern or barcode generated by GRKs leads to differential β-arrestin binding conformations and downstream signaling events (Choi et al., 2018; Latorraca et al., 2020; Nobles et al., 2011; Ren et al., 2005; Yang et al., 2017; Zhou et al., 2017). Additionally, recent structural studies suggest multiple functional binding conformations of β-arrestins with a GPCR. Specifically, the N-terminal domain of β-arrestins can interact with the phosphorylated C terminus of a GPCR, or alternatively, a small region on β-arrestin known as the finger loop domain can engage with the ICLs of a GPCR transmembrane (TM) core (Chen et al., 2017; Latorraca et al., 2018; Ranjan et al., 2017; Shukla et al., 2014). These conformationally distinct complexes differentially regulate downstream signaling pathways and are regulated by GRK-mediated phosphorylation barcodes (He et al., 2021; Kumari et al., 2016). Furthermore, GRKs can phosphorylate a growing list of non-GPCR substrates to regulate GPCR-independent signaling cascades (Gurevich et al., 2012; Ribas et al., 2007). Accordingly, we hypothesized that GRK phosphorylation of APH1A regulates βarr2 recruitment to APH1A and catalytic activity of the γ-secretase complex.
Here, we use a combination of label-free quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS), in vitro biochemical assays, structural modeling, and molecular dynamics (MD) simulations to investigate the molecular determinants that regulate βarr2 interaction with APH1A and how regulation of this critical protein-protein interaction mediates Aβ generation. We determine that GRKs 2, 3, 5, and 6 impart distinct APH1A phosphorylation patterns within the second intracellular loop (ICL2) and C terminus of APH1A to differentially regulate γ-secretase activity and Aβ generation. Furthermore, our MD simulation studies reveal that the βarr2 finger loop region engages with ICL2 and ICL3 of APH1A to facilitate the interaction between βarr2 and APH1A. Our structural analysis of βarr2 binding to APH1A suggests a conformation that closely resembles a fully engaged GPCR-β-arrestin complex. We demonstrate that mutagenesis of specific residues in the parr2 finger loop region or ICL3 of APH1A significantly reduces binding and Aβ generation, thus confirming a critical role of this interaction in regulating γ-secretase activity. Overall, we propose that GRK-mediated phosphorylation barcodes on the APH1A subunit of the γ-secretase complex lead to conformationally distinct APH1A-βarr2 complexes, which differentially affect γ-secretase activity and Aβ generation.
RESULTS
APH1A is phosphorylated in the second intracellular loop and C terminus
β-Arrestins are canonically involved in GPCR desensitization and internalization via recognition and binding to phosphorylated serine and threonine residues in the ICLs and/or C terminus of GPCRs (DeWire et al., 2007; Gurevich and Gurevich, 2019). We previously demonstrated that APH1A interacts with βarr2 in cells and mouse brain tissue (Thathiah et al., 2013). Similar to GPCRs, APH1A contains putative phosphorylation sites in ICL2 and the C terminus on the basis of a phosphorylation-site prediction algorithm (Blom et al., 1999). To determine whether APH1A is indeed phosphorylated, we expressed APH1A in HEK293 cells and performed label-free LC-MS/MS analysis on phosphopeptide-enriched trypsin digests. LC-MS/MS analysis confirms that APH1A is phosphorylated at S103 and S110 in ICL2 and S251 and S257 in the C terminus (Figures 1 and S1).
Figure 1. The APH1A subunit of the γ-secretase complex is phosphorylated in ICL2 and the C terminus.
(A) Table of identified phosphorylated peptides on APH1A determined by label-free LC-MS/MS on trypsin-digested and phosphorylation-enriched HEK293 cells following expression of APH1A.
(B) Snake diagram of APH1A. The phosphorylated amino acids in ICL2 and the C terminus are indicated in yellow.
Chemical inhibition of GRKs affects βarr2 recruitment and Aβ generation in a physiological model of AD
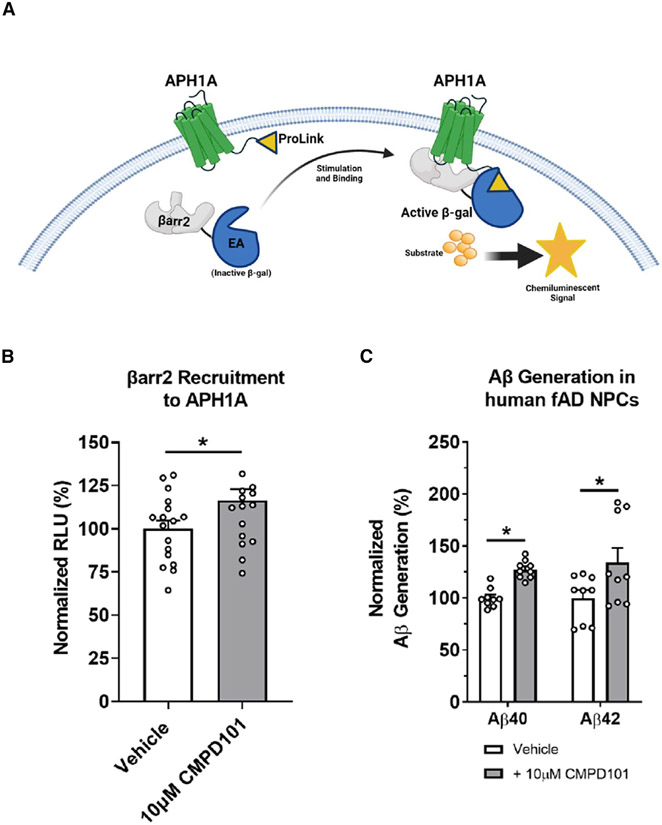
GRKs are serine/threonine kinases primarily responsible for phosphorylating the ICLs and C termini of GPCRs to initiate β-arrestin recruitment. GRKs are also capable of phosphorylating non-GPCR substrates (Gurevich et al., 2012; Watari et al., 2014). GRK2, GRK3, GRK5, and GRK6 are ubiquitously expressed in tissues throughout the body and in the brain (Gurevich et al., 2012; Komolov and Benovic, 2018). We hypothesized that GRK activity is involved in mediating the βarr2 interaction with APH1A. To initially test this hypothesis and gain preliminary insight into whether GRKs regulate βarr2 recruitment to APH1A and γ-secretase activity, we used the commercially available Takeda compound 101 (CMPD101), which inhibits the kinase activity of both GRK2 and GRK3 (Thal et al., 2011). To measure βarr2 recruitment to APH1A, we used a PathHunter βarr2 recruitment assay (Figure 2A). Surprisingly, we determined that treatment of cells that express APH1A with 10 μM CMPD101 results in a significant increase in βarr2 recruitment to APH1A in comparison with vehicle-treated cells (Figure 2B). To determine whether GRK2 and GRK3 are also involved in regulating Aβ generation, we turned to a more biologically- and AD-relevant system. We treated a human neural progenitor cell (NPC) line, ReN, which harbors familial AD (fAD) mutations in APP (Choi et al., 2014; Kim et al., 2015), with 10 μM CMPD101. Significantly, CMPD101 treatment leads to an increase in both Aβ40 and Aβ42 generation in the human fAD NPCs (Figure 2C). Collectively, our data demonstrate that chemical inhibition of GRK2/3 enhances βarr2 interaction with APH1A and Aβ generation. Additionally, the studies with the human fAD NPCs suggest a putative pathogenic role of the GRK family of kinases in AD.
Figure 2. Chemical inhibition of GRK2 and GRK3 increases βarr2 recruitment to APH1A and Aβ generation.
(A) Schematic diagram depicting the principle of the PathHunter assay used to detect the βarr2 interaction with APH1A. The CHO-βarr2 cells, which stably express βarr2 covalently attached to a portion of β-galactosidase (enzyme acceptor [EA]), were transfected with ProLink (PK)-tagged APH1A (APH1A-PK). Upon βarr2 binding to APH1A, the EA and PK form an active β-galactosidase enzyme. Upon the addition of a β-galactosidase substrate, the interaction between βarr2 and APH1A is detected by a chemiluminescent signal.
(B) βarr2 recruitment to APH1AWT in the CHO-βarr2 cell line following treatment with 10 μM CMPD101 relative to vehicle (0.1% DMSO) for 30 min as measured using the PathHunter assay. **p < 0.01 by unpaired t test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in quadruplicate. Individual technical replicates across all independent experiments are indicated (white circles).
(C) Aβ40 and Aβ42 generation in human fAD neural progenitor cells (NPCs) that stably express APPSwe/Ind. *p < 0.05 by ANOVA with Sidak’s post-test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate. Individual technical replicates across all independent experiments are indicated (white circles).
GRKs differentially regulate γ-secretase activity and Aβ generation
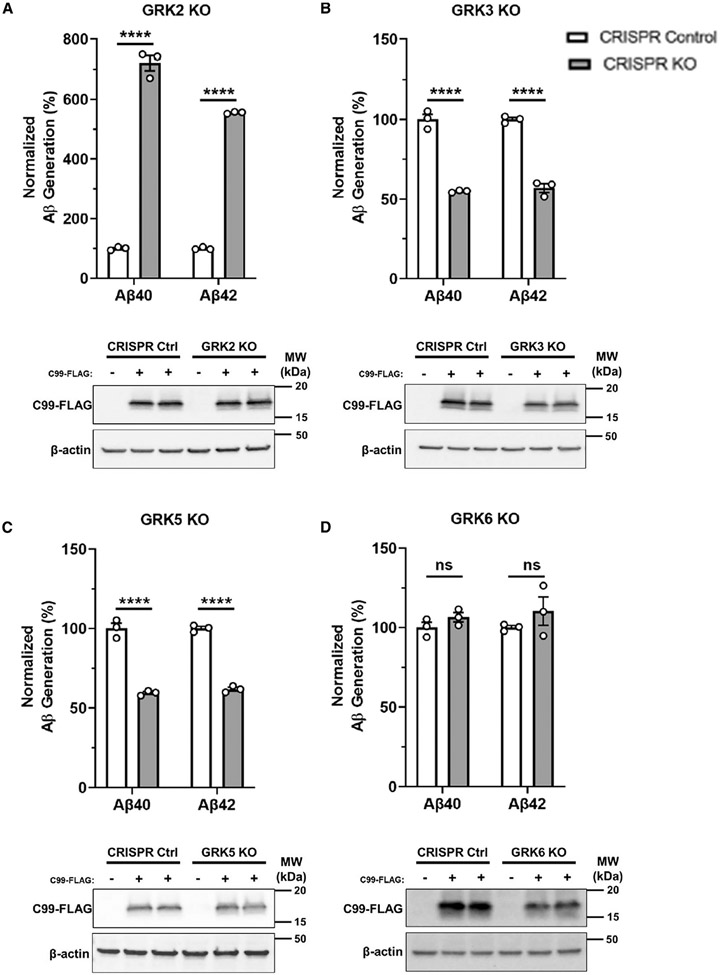
We next sought to comprehensively investigate the putative involvement of each ubiquitously expressed GRK in mediating APH1A phosphorylation and γ-secretase activity. To accomplish this, we used a CRISPR-Cas9 genome-editing strategy (Ran et al., 2013) to genetically delete Adrbk1, Adrbk2, Grk5, or Grk6 in HEK293 cells and generate monoclonal GRK knockout (KO) cell lines, herein referred to as GRK2 KO, GRK3 KO, GRK5 KO, or GRK6 KO cells, respectively. Successful genetic deletion of each Grk was confirmed using western blot analysis (Figures S2A-S2D). Notably, genetic deletion of each Grk did not affect the protein expression of the other GRKs. Furthermore, we did not detect changes in γ-secretase subunit expression in the GRK KO cell lines (Figures S2E-S2H).
We then sought to determine whether genetic deletion of each Grk mediates distinct functional outcomes, specifically in the regulation of γ-secretase activity and Aβ generation. We transiently expressed APP-C99, a direct substrate of γ-secretase, in each GRK KO and an unedited CRISPR control cell line and determined the levels of Aβ40 and Aβ42 generation by enzyme-linked immunosorbent assay (ELISA). Surprisingly, we detect a 6- to 7-fold increase in Aβ40 and Aβ42 generation in GRK2 KO cells in comparison with control cells (Figure 3A). These results are consistent with CMPD101 treatment in human fAD NPCs (Figure 2B). In contrast, we detect a ~50% reduction in Aβ40 and Aβ42 generation GRK3 KO and GRK5 KO cells (Figures 3B and 3C) and no change in Aβ40 or Aβ42 generation in GRK6 KO cells relative to control cells (Figure 3D). Similar to the results in the CRISPR-Cas9-generated GRK2 KO cell line, small interfering RNA (siRNA)-mediated knockdown of GRK2 (~70%) leads to an increase in Aβ generation (Figures S3A and S3B). Interestingly, the magnitude of the effect on Aβ generation is similar to the effect observed with CMPD101 treatment (Figure 2B). In contrast, we do not observe a significant change in Aβ levels following siRNA knockdown of GRK6 (Figures S3A and S3B), which is also consistent with the results observed in the GRK6 KO cell line (Figure 3D). Although we observe a 50% reduction in Aβ levels in the GRK3 and GRK5 KO cell lines (Figures 3B and 3C), we do not observe a change in Aβ levels following siRNA knockdown of GRK3 and GRK5 (Figures S3A and S3B). The remaining GRK3 and GRK5 expression (~20%) may be sufficient to maintain kinase activity, which may preclude the potential detection of appreciable changes in Aβ levels. Collectively, these results establish a role for the GRKs in differential regulation of γ-secretase activity and Aβ generation.
Figure 3. Genetic deletion of each GRK differentially regulates γ-secretase activity and Aβ generation.
(A–D) Aβ40 and Aβ42 generation and representative western blot analysis of HEK293 cells following expression of APP-C99 in CRISPR control cells (white bars) and each GRK KO cell line (gray bars): (A) Adrbk1 (GRK2), (B) Adrbk2 (GRK3), (C) Grk5 (GRK5), and (D) Grk6 (GRK6). ****p < 0.0001 by ANOVA with Tukey’s post-test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate. The means from each experiment are indicated (white circles).
GRKs differentially regulate APH1A phosphorylation
Given the identification of phosphorylation sites in ICL2 and the C terminus of APH1A and the differential effects of Grk genetic deletion on Aβ generation, we hypothesized that the GRKs generate distinct phosphorylation patterns in APH1A, which differentially affect γ-secretase activity and Aβ levels. To test this hypothesis, we first determined the phosphorylation pattern of APH1A in each GRK KO cell line. We transiently expressed equivalent amounts of APH1A in each GRK KO and control cell line and performed label-free quantitative LC-MS/MS analysis. We detect different APH1A phosphorylation patterns in the GRK2 KO and GRK6 KO cell lines and a similar APH1A phosphorylation pattern in the GRK3 KO and GRK5 KO lines. Interestingly, we observe the most striking differential phosphorylation changes in ICL2. We identify a phosphorylation site at S105 and an increase in phosphorylation at S110 in the GRK2 KO line (Figures 4A and S4A). Genetic deletion of Grk6 also leads to a substantial increase in APH1A phosphorylation at S110. In contrast, phosphorylation at S103 is almost completely abolished in the GRK2, GRK3, and GRK5 KO cell lines. Genetic deletion of each Grk reduces, but does not eliminate, S251 and S257 C-terminal phosphorylation. We observe a greater loss of phosphorylation at S257 relative to S251 in each GRK KO cell line, suggesting that phosphorylation at S251 may affect or facilitate phosphorylation at S257. Collectively, MS analysis of APH1A phosphorylation in the GRK KO cell lines highlights distinct ICL2 and C-terminal phosphorylation patterns, with ICL2 displaying the primary differential sites of phosphorylation in APH1A (Figures 4A and 4B).
Figure 4. Differential APH1A phosphorylation is mediated by GRKs and regulates interaction with βarr2 and γ-secretase activity.
(A) Heatmap of the fold change in APH1A phosphorylation following expression in each GRK KO cell line relative to control cells as measured by label-free LC-MS/MS in 3 independent experiments. The “x” in specific squares indicates that phosphorylation was not detected.
(B) Snake diagram of APH1A. The serine residues that display increased (red) or decreased (blue) levels of phosphorylation in each GRK KO cell line are indicated.
(C) βarr2 recruitment to APH1A in the CHO-βarr2 cell line following expression of APH1AWT or the APH1A ICL2 and C-terminal phosphorylation-deficient or phosphorylation-mimetic mutants. *p < 0.05, **p < 0.01, and ****p < 0.0001 by two-way ANOVA with Tukey’s post-test.
(D) Aβ40 generation in cells following expression of APP-C99 and APH1AWT, APH1A phosphorylation-deficient, or phosphorylation-mimetic mutants. *p < 0.05 and ****p < 0.0001 by two-way ANOVA with Tukey’s post-test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate. Individual technical replicates across all independent experiments are indicated (white circles).
(E) Aβ42 generation in cells following expression of APP-C99 and APH1AWT, APH1A phosphorylation-deficient, or phosphorylation-mimetic mutants. **p < 0.01 and ****p < 0.0001 by two-way ANOVA with Tukey’s post-test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate. Individual technical replicates across all independent experiments are indicated (white circles).
Site-specific APH1A phosphorylation mediates the interaction with βarr2 and γ-secretase activity
Our lab previously demonstrated that βarr2 interacts with APH1A to stabilize the γ-secretase complex in lipid-raft or DRM domains where the complex is more enzymatically active and can process substrates, including APP-C99 (Thathiah et al., 2013). As such, we hypothesized that APH1A phosphorylation patterns differentially regulate βarr2 recruitment to APH1A and consequent γ-secretase activity and Aβ generation. To test this hypothesis and establish the presence of an APH1A phosphorylation barcode, we generated individual phosphorylation-deficient (APH1AS103A, APH1AS110A, APH1AS251A, and APH1AS257A) and phosphorylation-mimetic (APH1AS105D and APH1AS110D) mutants, which correspond to phosphorylation changes observed in the GRK KO cell lines relative to control cells (Figures 4A and 4B). In addition, to gain insight into a putative multi-site phosphorylation barcode on APH1A, we generated double-phosphorylation mutants in ICL2 representative of phosphorylation changes in the GRK3/GRK5 KO (APH1AS103A/S110A) and GRK2 KO (APH1AS105D/S110D) cell lines and in the C terminus (APH1AS251A/S257A), as observed in the GRK2/GRK3/GRK5 KO cell lines. We then expressed APH1AWT or the phosphorylation mutants along with APP-C99 in CHO-βarr2 cells (Figures S4B and S4C) and used the PathHunter assay to measure βarr2 recruitment to APH1A (Figure 4C) and ELISAs to measure Aβ40 and Aβ42 generation (Figures 4D and 4E). Following expression of the individual phosphorylation-deficient APH1AS103A or APH1AS110A mutants, βarr2 recruitment is reduced or unaffected, respectively, relative to cells that express APH1AWT. Neither the APH1AS103A nor APH1AS110A mutation affects γ-secretase activity. However, expression of the APH1AS103A/S110A double ICL2 phosphorylation-deficient mutant leads to a significant reduction (~60%) in βarr2 recruitment to APH1A and Aβ generation compared with APH1AWT. As complementary validation for the PathHunter assay, we also performed co-immunoprecipitation (coIP) experiments to determine the effect of the APH1AS103A/S110A double ICL2 phosphorylation-deficient mutation on the interaction between βarr2 and APH1A (Figures S5A and S5B). Consistent with the PathHunter results, the APH1AS103A/S110A mutant displays a reduced interaction with βarr2. These results suggest that phosphorylation at both S103 and S110 within ICL2 regulates both βarr2 recruitment and γ-secretase activity and that loss of phosphorylation at both sites, as observed in both the GRK3 KO and GRK5 KO cells (Figure 4A), significantly hinders βarr2 recruitment and Aβ generation.
In the GRK2 KO cell line, MS analysis identified a phosphorylation site at S105 and an increase in phosphorylation at S110 (Figure 4A). Interestingly, expression of the individual APH1AS105D and APH1AS110D phosphorylation-mimetic mutants reduces βarr2 recruitment and Aβ generation relative to APH1AWT. However, expression of the APH1AS105D/S110D double phosphorylation-mimetic mutant results in a significant increase in both βarr2 recruitment to APH1A and Aβ40 and Aβ42 generation (Figures 4C-4E). These results are consistent with the effect we observe in the GRK2 KO cell line (Figure 3A) and with CMPD101 treatment in CHO-βarr2 cells (Figure 2B) and human fAD NPCs (Figure 2C). Moreover, sucrose density fraction studies indicate that the γ-secretase complex subunits NCT and APH1A are enriched in DRM domains in GRK2 KO cells relative to control cells (Figures S5C-S5H). Taken together, these data highlight the importance of phosphorylation at multiple ICL2 sites (i.e., a phosphorylation barcode) to mediate βarr2 recruitment to APH1A and direct the functional outcome of γ-secretase activity.
MS analysis of the GRK KO cell lines reveals a decrease in C-terminal phosphorylation at S251 and S257 in the GRK2, GRK3, and GRK5 KO lines and only a decrease in phosphorylation at S257 in the GRK6 KO line (Figure 4A). Expression of the individual APH1AS251A and APH1AS257A or double APH1AS251A/S257A phosphorylation mutant reduces βarr2 recruitment to APH1A relative to APH1AWT. Surprisingly, despite a reduction in βarr2 recruitment, the APH1AS251A mutant exhibits increased Aβ generation. In contrast, the APH1AS257A and double APH1AS251A/S257A mutant exhibit a decrease in βarr2 recruitment and Aβ generation relative to APH1AWT. Consistent with the PathHunter results, coIP experiments also indicate that the APH1AS251A/S257A mutant displays a reduced interaction with βarr2 (Figures S5A and S5B). Given that we observe reduced C-terminal phosphorylation in each GRK KO cell line and still detect increased Aβ generation in GRK2 KO cells or equivalent Aβ generation in GRK6 KO cells relative to control cells (GRK6 KO), we can conclude that APH1A phosphorylation changes by GRKs on the C terminus function in conjunction with ICL2 phosphorylation to dictate overall βarr2 binding and downstream functional consequences on γ-secretase activity.
Structural modeling and MD simulations reveal βarr2 finger loop domain engagement with APH1A cytoplasmic loops
After establishing the differential effects of the ubiquitously expressed GRKs in mediating an APH1A phosphorylation barcode and a role of each specific APH1A ICL2 and C-terminal phosphorylation site in mediating βarr2 binding and γ-secretase activation, we sought to gain additional structural insights into the mechanism of the βarr2-APH1A interaction. To this aim, we first generated a structural model for the complex formed between APH1A and βarr2 using the X-ray structure resolved for constitutively active rhodopsin (Kang et al., 2015) in the presence of visual arrestin (Figure S6A). Although the relative orientations of the TM helices exhibited some differences between APH1A and rhodopsin (Figure S6B), TM6, TM7, and helix 8 were closely superposable, which allowed the construction of a structural model for the βarr2-APH1A complex by optimally aligning the APH1A and βarr2 molecules onto their counterparts in the resolved rhodopsin-arrestin structure and refining the generated model using MD simulations (Figure S6C). In addition to this so-called alignment model, we independently generated two additional models termed DOCK1 (Figures S6D and S6E) and DOCK2 (Figure 5), by docking simulations followed by MD refinement, to explore the possible occurrence of alternative binding poses and the robustness of interfacial contacts. Figure 5 illustrates the results for the DOCK2 model (Figures 5A and 5B) and displays the time evolution of the corresponding interfacial contacts observed during two independent runs of 40 ns carried out for this model (Figure 5C). Equivalent results for the other two models are presented in Figure S6F.
Figure 5. Structural modeling of the interaction between APH1A and βarr2 and the time evolution of the most stable interfacial contacts.
(A) Structural model DOCK2 generated by docking simulations followed by MD refinement carried out at full atomic scale in explicit membrane (gray sticks) and water. The simulations were performed for the intact γ-secretase complex that displays tight interaction between APH1A (magenta) and βarr2 (salmon) and transient involvement of other γ-secretase complex subunits such as the PS1 (cyan) and NCT (green).
(B) Detailed view of interfacial interactions robustly observed at the interface. Residues engaged in persistent interfacial contacts are shown as sticks and labeled.
(C) Interactions between APH1A and βarr2 for two independent runs (40 ns each) were carried out for this model (the first two columns) and corresponding time evolution of interactions for both runs. The last column displays the cumulative fractional time during which those pairs made contacts. Here, interactions are defined when any pairs of heavy atoms belonging to the two respective proteins are separated by less than 5 Å. Equivalent results for two other models, termed Alignment and DOCK1, are presented in Figure S6, supporting the robustness of the regions (ICL2 and ICL3 in APH1A and finger loop and C terminus in βarr2) and residues engaged in interfacial association, despite minor redistributions of the specific pairs of residues.
The three models consistently exhibited interfacial interactions at APH1A ICL2 and ICL3 and βarr2 finger loop, despite some local differences in specific residue pairs. The C terminus of APH1A was also engaged in close contact with βarr2 in the two docking models (Figures 5B and S6E) enclosed in the respective yellow (ICL2/3 interactions) and blue (C terminus interactions) circles. Contacts validated by at least two independent models include the three main groups of interactions: E183 or R184 (ICL3) with G73, L74, and/or S75 (finger loop); I114-R115 (ICL2) with V71 -L72 finger loop; and E83-R184 (ICL3) with G73 and S75. Contacts observed between the C terminus of APH1A and βarr2 in the DOCK1 and DOCK2 models involved APH1A residues R241, C245, Q248, and E249 (C terminus) making contacts with K153, E156, E157, and R52 on βarr2. As shown in Figures 5C and S6F, these interactions were stably maintained during extended portions of the MD runs.
βarr2 finger loop engagement with APH1A ICL2 and ICL3 facilitates binding and γ-secretase activity
Our computational docking and MD simulation studies indicate a putative model of βarr2 binding to APH1A that resembles a fully engaged GPCR-arrestin complex (Ranjan et al., 2017; Shukla et al., 2014; Yin et al., 2019). As shown in Figure 5, MD simulations indicate that the βarr2 finger loop domain engages in hydrophobic and polar interactions with ICL2 and ICL3 of the APH1A cytoplasmic core. We sought to experimentally validate these interactions and to further determine whether specific residues in these regions on βarr2 and APH1A are critical for engagement and downstream functional effects on γ-secretase activity. Guided by computational predictions, we generated a βarr2 mutant containing the substitution S75R at the primary site of interaction. Importantly, S75 was consistently observed to engage in interfacial associations in all three models (Figures 5 and S6D). We also generated the βarr2 L72E mutation at a secondary site of interaction shared by DOCK1 and DOCK2 (Figures 5 and S6E). In HEK293 cells expressing APH1AWT and βarr2WT, βarr2L72E, or βarr2S75R, we used a coIP assay to determine the effect of mutagenesis of L72 and S75 on the interaction between βarr2 and APH1A. Our data demonstrate that mutagenesis of the βarr2 finger loop region at L72 and S75 reduces binding to APH1A (Figure 6A), indicating that the βarr2 finger loop domain is critical for interaction with APH1A.
Figure 6. βarr2 finger loop engagement with APH1A ICL3 and ICL2 facilitates binding and γ-secretase activity.
(A) Co-immunoprecipitation experiments in HEK293 cells expressing APH1A and βarr2WT or the βarr2 finger loop mutants (βarr2L72E, βarr2S75R).
(B–D) Representative western blot expression of C99-FLAG and βarr2WT, βarr2L72E, or βarr2S75R (B). (C) Aβ40 and (D) Aβ42 generation in HEK293 cells following expression of APP-C99 and empty-vector, βarr2WT, or βarr2 finger loop mutants. **p < 0.01 by ANOVA with Tukey’s post-test (comparison with empty). #p < 0.05 and ###p < 0.001 by ANOVA with Tukey’s post-test (comparison with βarr2WT). Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate or quadruplicate. Individual technical replicates across all independent experiments are indicated (white circles).
(E–G) Representative western blot expression of C99-FLAG and APH1AWT-PK or APH1AR184D-PK (E). (F) βarr2 recruitment to APH1AWT or APH1AR184D mutant in the CHO-βarr2 cell line. **p < 0.01 by unpaired t test. (G) Aβ40 and Aβ42 generation in the CHO-βarr2 cell line following expression of C99-FLAG and APH1AWT or APH1AR184D. ****p < 0.0001 by ANOVA with Sidak’s post-test. Bars denote mean ± SEM of 3 independent experiments (n = 3) performed in triplicate or quadruplicate. Individual technical replicates across all independent experiments are indicated (white circles).
(H–J) Results for the βarr2 mutants (H) L72E and (I) S75R and (J) for the APH1A mutant R184D are presented in the respective panels using the structural model DOCK1. In each case, a representative snapshot for the complex with the mutant (top), and the histograms of the intermolecular Cα-Cα distances between the indicated residue pairs of APH1A and βarr2 are shown (bottom, solid curves) on the basis of duplicate runs (total of 80 ns) conducted for the indicated mutant. Dashed curves indicate the counterparts obtained with the βarr2WT or APH1AWT proteins.
Given the involvement of L72 and S75 in the βarr2 finger loop domain in interaction with APH1A, we then determined the effect of L72E and S75R mutagenesis on γ-secretase activity and Aβ generation. We expressed APP-C99 in HEK293 cells along with an empty vector, βarr2WT, βarr2L72E, or βarr2S75R (Figure 6B) and used ELISA to measure Aβ40 and Aβ42 generation (Figures 6C and 6D). As expected, expression of βarr2WT increased Aβ generation in comparison with vector control samples. Significantly, the βarr2L72E and βarr2S75R finger loop mutants reduced Aβ generation to control conditions, indicating that engagement of the βarr2 finger loop domain with APH1A is necessary for both the βarr2 interaction with APH1A and γ-secretase activity.
To further investigate the putative involvement of specific residues in ICL3 of APH1A identified by MD simulations in the interaction with βarr2, we generated an APH1A ICL3 mutant (APH1AR184D) to disrupt interaction with the primary interaction site at S75 on βarr2. In our CHO-βarr2 cell line, we expressed APP-C99 and APH1AWT or the APH1AR184D mutant (Figure 6E) and used the PathHunter assay and ELISA to determine the effect of APH1A ICL3 mutagenesis on βarr2 recruitment and the downstream functional effect on γ-secretase activity. Expression of the APH1AR184D mutant significantly reduced both βarr2 recruitment to APH1A (Figure 6F) and Aβ40 and Aβ42 generation (Figure 6G) compared with cells expressing APH1AWT. These data provide evidence of critical βarr2 finger loop domain interactions with the APH1A cytoplasmic core to facilitate γ-secretase activation and Aβ generation.
Additional MD simulations for the L72E and S75R mutants corroborate the decreased binding to APH1A. As illustrated in Figure 6H, the L72 of βarr2WT makes contacts with I114, R115, D180, and E183 of APH1A within a Cα-Cα distance range of 4–10 Å (dashed curves), while intermolecular distances in the presence of the E72 mutant (βarr2L72E) are shifted to longer distances (solid curves), indicating that the interaction becomes weaker upon substitution of L72 with E72. On the other hand, the R75 mutant (βarr2S75R) shows slightly weaker interactions compared with βarr2WT, with a bimodal distribution of inter-residue distances (Figure 6I). As such, one might anticipate a smaller reduction in binding affinity. However, we observed that L72, at the middle of the finger loop, is entropically amenable to a diversity of contacts (including those with ICL2 D184), which is impaired by the perturbations caused by the S75R mutation. The introduction of a charged amino acid R75 in the immediate neighborhood of the hydrophobic residues, L74, and highly flexible G73, further destabilize the interfacial contacts in this region, as evidenced by the distance changes presented in Figure 6I. Furthermore, our MD simulations conducted with APH1AR184D also clearly show a weakening in the interaction of D184 with finger loop residues L74 and S75 (Figure 6J). Cα-Cα distances of R184-L74 and R184-S75 increase by about 2 Å compared with those achieved in APH1AWT, demonstrating the occurrence of a looser binding. These structural and dynamic characteristics consistently support the experimentally observed decrease in binding affinity for both mutants.
We finally sought to determine whether the phosphorylation status of APH1A would modulate the structure and dynamics of βarr2. For these studies, we selected the APH1AS103A/S110A phosphorylation-deficient mutant, which leads to a drastic reduction in βarr2 recruitment to APH1A and Aβ levels (Figures 4C-4E), and the APH1AS105D/S110D phosphorylation-mimetic mutant, which leads to an increase in βarr2 recruitment to APH1A and Aβ levels (Figures 4C-4E). The APH1AS105D/S110D mutant shows a stronger interface interaction with a larger deformation of βarr2, and the APH1AS103A/S110A mutant shows a weaker interface interaction with a slight deformation of βarr2 compared with APH1AWT (Figures S7A-S7C). Intriguingly, the APH1A mutations also lead to mutation-specific conformational changes in βarr2. The D105/D110 mutants show conformational changes in the three regions around P134, Y250, and K285 compared with APH1AWT (Figures S7A and S7C), whereas the A103/A110 mutants show minor changes in the loop with P134 (Figures S7A and S7B). These results indicate that the stronger interface interaction of the D105/D110 mutants induce a larger deformation of βarr2. The contact duration was also generated by analyzing the most important residues (D107 and R109) in APH1A and their closest residues in βarr2. The D105/D110 mutants exhibit a stronger contact duration whereas the A103/A110 mutants show a weaker contact duration relative to APH1AWT (Figure S7D). Collectively, these data suggest that changes in the APH1A phosphorylation status affect both the extent of the direct interaction with βarr2 as well as the specific βarr2 conformation upon interaction. Overall, our data suggest a model in which the GRK-mediated APH1A phosphorylation barcodes regulate βarr2 finger loop domain interactions with the cytoplasmic TM core of APH1A to differentially regulate γ-secretase activity and proteolytic cleavage of APP-C99 (Figure 7).
Figure 7. GRKs differentially regulate γ-secretase activity by mediating an APH1A phosphorylation barcode.
(A) GRKs mediate APH1A phosphorylation within ICL2 and the C terminus of APH1A to regulate βarr2 recruitment via interactions between the βarr2 finger loop domain and the cytoplasmic TM core (ICL2/3) of APH1A. In turn, βarr2 recruitment to APH1A leads to γ-secretase cleavage of APP-C99 and the generation of Aβ peptides.
(B) In GRK2 KO cells, APH1A ICL2 phosphorylation at both S105 and S110 is elevated relative to control cells. The γ-secretase complex exhibits increased activity and cleavage of APP-C99.
(C) In GRK3 KO and GRK5 KO cells, APH1A displays reduced levels of ICL2 phosphorylation at S103 and S110 relative to control cells. The γ-secretase complex exhibits decreased activity and cleavage of APP-C99.
(D) In GRK6 KO cells, APH1A phosphorylation at S110 alone is elevated relative to control cells. The γ-secretase activity and Aβ generation are unaffected relative to control cells. We hypothesize that changes in the phosphorylation barcode affect both the extent of βarr2 recruitment to APH1A and the APH1A-βarr2 conformation following binding. We predict the positioning of H171 and H197 in APH1A is altered by βarr2 engagement, which affects recognition and cleavage of APP-C99. The phosphorylation sites highlighted in red and blue indicate elevated or reduced levels of phosphorylation, respectively, relative to control (or basal) levels of phosphorylation at the sites highlighted in yellow. The figure was created with BioRender.com.
DISCUSSION
GRKs are a family of proteins canonically involved in GPCR phosphorylation that regulate receptor desensitization and internalization. GRK phosphorylation of GPCRs also leads to the activation of intracellular signaling cascades via G proteins and β-arrestins (DeWire et al., 2007; Gurevich et al., 2012; Lefkowitz and Shenoy, 2005). Different ligands or stimuli initiate distinct GRK phosphorylation patterns or barcodes on GPCRs to differentially regulate cellular signaling (Drube et al., 2022; Nobles et al., 2011). Here, we uncover a role for the four ubiquitously expressed GRKs in phosphorylation of a non-canonical substrate, APH1A, an integral component of the γ-secretase complex. Quantitative LC-MS/MS analysis indicates that APH1A is phosphorylated at S103 and S110 within ICL2 and at S251 and S257 on the C terminus. Further investigation reveals that chemical inhibition of GRK2/3 increases the direct interaction between βarr2 and APH1A as well as γ-secretase activity and Aβ generation in a human fAD NPC model. Genetic deletion of each ubiquitously expressed Grk, namely GRK2, GRK3, GRK5, and GRK6, generates distinct phosphorylation patterns in ICL2 and the C terminus of APH1A and differentially affects γ-secretase activity and Aβ generation. Intriguingly, we determine that APH1A phosphorylation barcodes differentially regulate interaction with the GPCR scaffolding protein βarr2. Further computational modeling and MD simulation studies reveal that a conserved region in βarr2, known as the finger loop, interacts with the cytoplasmic TM core (ICL2/3) of APH1A. Experimental validation of the computational modeling studies indicates that the βarr2 finger loop domain and APH1A TM core interaction regulates γ-secretase catalytic activity. Collectively, these studies suggest a model whereby APH1A phosphorylation barcodes dictate different βarr2-APH1A binding conformations that affect γ-secretase substrate recognition of APP-C99 and consequent proteolytic cleavage and Aβ generation (Figure 7).
In this study, we uncover a functional role for the ubiquitously expressed GRKs in differential modulation of γ-secretase activity. In the GRK2 KO cells, we observe a substantial increase in γ-secretase activity and Aβ generation (Figure 3A). Surprisingly, we observe an increase in ICL2 phosphorylation at S105 and S110 in the GRK2 KO cells and S110 in the GRK6 KO cells, suggesting potential compensatory phosphorylation at S105 and S110 by other kinases in these cell lines and hierarchical phosphorylation of APH1A by GRKs. Indeed, GRKs are known to act hierarchically to mediate ICL and C-terminal phosphorylation of many GPCRs to regulate downstream functional signaling outcomes (Gurevich and Gurevich, 2020; Kim et al., 2004; Kouhen et al., 2000; Palmer and Stiles, 2000). We also detect an increase in Aβ generation following chemical inhibition of GRK2/3 in human fAD NPCs (Figure 2C). These results suggest that GRK2-mediated APH1A phosphorylation precludes pathogenic phosphorylation at S105 and S110 in ICL2 by other GRKs. In agreement with these data, we observe a decrease in constitutive γ-secretase activity and Aβ generation in both the GRK3 KO and GRK5 KO cells in comparison with control cells, which suggests that GRK3 and GRK5 activity contributes to increased γ-secretase activity and Aβ generation. Interestingly, our lab recently determined that both GRK3 and GRK5 are abundantly localized around amyloid plaques in human AD brains (Guimarães et al., 2021). Together, these data support the hypothesis that GRK3 and GRK5 activity may contribute to pathogenic Aβ generation while GRK2 activity may act in a protective manner to regulate γ-secretase activity.
Genetic deletion of each ubiquitously expressed Grk reduces APH1A C-terminal phosphorylation. The APH1A phosphorylation patterns in the GRK KO cell lines also suggest hierarchical GRK phosphorylation at the C terminus. In each GRK KO line, we detect a greater loss of phosphorylation at S257 than at S251. Accordingly, phosphorylation at S251 may dictate the level of phosphorylation at S257 by GRKs. In fact, GRK2 has been demonstrated to preferentially phosphorylate serine and threonine residues that are preceded by negatively charged amino acids (Onorato et al., 1991), which may explain how the loss of phosphorylation at S251 could reduce S257 phosphorylation to an even greater extent.
Although individual APH1A ICL2 and C-terminal serine mutants can independently alter the interaction with βarr2 and/or γ-secretase activity, the double serine-mutant results suggest that distinct, multi-site APH1A phosphorylation patterns determine differential functional outcomes with regard to regulating βarr2 interaction and γ-secretase cleavage of APP-C99. Furthermore, ICL2 and C-terminal phosphorylation of APH1A appears to act in conjunction to modulate the overall outcome of βarr2 recruitment and γ-secretase activity. For example, we measure an increase in γ-secretase activity in the GRK2 KO cells (Figure 3A), in which levels of APH1A ICL2 phosphorylation at S103 and S105 are elevated in comparison with a reduction in C-terminal S251 and S257 phosphorylation (Figure 4A). Given that loss of S251 and S257 C-terminal phosphorylation alone (APH1AS251A/S257A) reduces βarr2 recruitment to APH1A and γ-secretase activity (Figures 4C-4E), our GRK2 KO data suggest an important role of ICL2 phosphorylation in determining the functional γ-secretase outcomes. Likewise, our MD simulation data reveal a critical role of APH1A ICL2 and regulating engagement with βarr2 (Figure 5) which would be altered by the addition or removal of negatively charged phosphates. Overall, we establish the existence of an APH1A phosphorylation barcode in ICL2 and the C terminus that regulates βarr2 recruitment and γ-secretase activity.
Our previous work demonstrated that overexpression of βarr2 stabilizes localization of the γ-secretase subunits in DRMs where the complex is catalytically active (Thathiah et al., 2013). In support of our previous finding, CMPD101 treatment (Figure 2C) and APH1AS103D/S105D expression (Figures 4C-4E), which increase βarr2 recruitment to APH1A, result in increased γ-secretase activity and Aβ generation. Interestingly, our data in this study also reveal that phosphorylation of APH1A at distinct sites can differentially regulate βarr2 recruitment and γ-secretase activity (Figures 4C-4E). Accordingly, APH1A phosphorylation patterns that reduce βarr2 recruitment do not necessarily reduce γ-secretase activity, as observed with the APH1AS103A, APH1AS105D, and APH1AS251A mutants. APH1A contains two histidine residues, H171 and H197, within TM5 and TM6, respectively, that are critical for γ-secretase complex assembly and stability, APP-C99 substrate recognition, and aspartyl proteolytic cleavage (Chen et al., 2010; Pardossi-Piquard et al., 2009). We postulate that APH1A ICL2 and C-terminal phosphorylation allows differential βarr2 binding modes to ICL2/3 of the APH1A TM core, which in turn induce conformational shifts in TM5 and TM6 of APH1A to regulate APP-C99 binding and γ-secretase activity. Additionally, the binding conformation of βarr2 may also sterically prevent the entry of γ-secretase substrates. Collectively, we argue that γ-secretase activity is regulated by both the extent of βarr2 recruitment and the distinct APH1A-βarr2 conformation following binding (Figure 7).
Structural analysis of β-arrestin interaction with GPCRs indicates that there is a biphasic interaction between β-arrestin and the receptor (Ranjan et al., 2017). Initial β-arrestin binding to a GPCR is mediated via GPCR C-terminal phosphorylation. However, β-arrestin can also transition into a fully engaged conformation with the receptor where a small structural region in β-arrestins known as the finger loop engages with the cytoplasmic TM core of the receptor (Chen et al., 2017; Latorraca et al., 2018; Shukla et al., 2014; Yin et al., 2019). Notably, our computational modeling data suggest a similar, critical role of the APH1A TM cytoplasmic core (i.e., ICL2 and ICL3) in binding to βarr2 finger loop residues, which resembles a fully engaged GPCR-β-arrestin complex. Our MD simulation studies are supported by APH1A MS and mutagenesis studies, which suggest that ICL2 phosphorylation, in particular, is a critical determinant of βarr2 binding and functional effect on γ-secretase activity and APP-C99 cleavage. A recent study investigated how GPCR phosphorylation patterns direct β-arrestin-mediated signaling. Latorraca et al. (2020) determined that phosphorylation at different positions, rather than the total number of phosphorylated residues, determines the β-arrestin binding conformations and the scaffolding of different signaling proteins. Importantly, the authors conclude that phosphorylation patterns that favor β-arrestin binding are not necessarily the same as those that favor β-arrestin signaling (Latorraca et al., 2020). Furthermore, additional recent studies have highlighted the importance of the spatial positioning of phosphorylation sites to regulate both β-arrestin recruitment and downstream functional responses (Baidya et al., 2020; Dwivedi-Agnihotri et al., 2020). We observe a similar mechanism with APH1A phosphorylation, βarr2 binding, and the downstream functional effect on APP-C99 cleavage by the γ-secretase complex where APH1A phosphorylation mutants that decrease βarr2 recruitment do not always reduce γ-secretase activity (Figures 4C-4E).
We predict that changes to the APH1A phosphorylation barcode may occur via direct or indirect changes in GRK activity in the brain. Our lab recently demonstrated that GRKs are differentially expressed across AD-affected regions and in different cell types in the brain. Significantly, we observed decreases in GRK2, GRK5, and GRK6 expression in AD brain tissue relative to age-matched control samples (Guimarães et al., 2021). As such, changes in GRK levels across brain regions and during disease progression may directly affect constitutive GRK phosphorylation of APH1A and alter γ-secretase activity. Furthermore, we cannot rule out the possibility of unidentified, endogenous interacting partners of APH1A that can induce conformational changes in APH1A to directly recruit GRKs. Alternatively, changes in GPCR signaling in different brain regions with age and by other AD etiologies may indirectly alter the activity of GRKs, thus changing the APH1A phosphorylation barcode and affecting γ-secretase activity. Multiple GPCRs have been demonstrated to contribute to the pathophysiology of AD, including regulating APP proteolytic processing (Huang et al., 2017b; Thathiah and De Strooper, 2011). As both GPCR signaling complexes and the γ-secretase complex preferentially partition into DRMs (Hur et al., 2008; Teng et al., 2010; Villar et al., 2016), it is conceivable that GPCR dysregulation (e.g., changes in expression level or ligand stimulation) will alter GRK activity in cells and in the local DRM environment, which may lead to changes in APH1A phosphorylation and γ-secretase activity. Notably, we observe an increased distribution of the γ-secretase subunits in DRM domains in GRK2 KO cells. Intriguingly, the constitutively active, orphan GPCR GPR3 is increased in the AD brain, and in vitro GPR3 expression increases DRM localization of the γ-secretase complex (Thathiah et al., 2009). Furthermore, genetic deletion of Gpr3 in mice reduces βarr2 interaction with APH1A in cortical brain tissue (Thathiah et al., 2013), reduces γ-secretase activity and Aβ generation, and alleviates cognitive deficits in four AD transgenic mouse models (Huang et al., 2015). Notwithstanding, βarr1 has also been shown to interact with APH1A to regulate formation of an NCT/APH1A γ-secretase subcomplex (Liu et al., 2013). However, whether GPR3 modulates the interaction between βarr1 and APH1A has not been investigated. Thus, constitutive GPR3 activity may modulate the APH1A phosphorylation barcode by recruiting specific GRKs to the local DRM environment of the γ-secretase complex to alter the APH1A phosphorylation barcode.
Distinct binding modes and conformations of β-arrestin isoforms have emerged as primary mechanisms driving distinct functional outcomes, including desensitization, endocytosis, and signaling. Interestingly, βarr1 and βarr2 both contain a nuclear localization signal (NLS) (Hoeppner et al., 2012; Wang et al., 2003). However, βarr2 also contains a nuclear export signal (NES) (Wang et al., 2003). Studies suggest that βarr2 is rapidly exported to the cytoplasm, whereas βarr1 may be retained in the nucleus (Wang et al., 2003), and is involved in transcriptional regulation, which indicates that differences in subcellular localization contribute to the functional divergence of βarr1 and βarr2. Functional specialization may also occur as a result of the cell type-specific expression pattern of β-arrestin isoforms (Tobin et al., 2008). Accordingly, different cell types may express different levels and/or β-arrestin isoforms along with different GRKs, which would lead to differential phosphorylation patterns of a GPCR (or APH1A) and result in preferential recruitment of βarr1 or βarr2 and distinct outcomes in different cellular contexts.
Overall, our analysis opens the door for computational drug screening to identify and design small molecules or peptides to modulate βarr2 interaction with APH1A. As advances in amyloid and tau brain imaging along with AD biomarker discovery continue to be made, the opportunity to detect and intervene early in AD progression becomes more possible. The work here suggests an avenue for therapeutic drug discovery to disrupt critical APH1A-βarr2 interactions and reduce Aβ generation early in disease progression in an effort to negate the Aβ concentration-dependent pathogenic cascade cumulating in neurodegeneration and dementia.
Limitations of the study
A limitation in the scope of this study is the absence of the direct link between specific phosphorylated residues on APH1A and the responsible GRK(s). Although the use of GRK KO cell lines and the GRK2/3 inhibitor allows us to establish a putative role for the GRK family of kinases in phosphorylation of APH1A, further in vitro GRK kinase assays with purified proteins would permit identification of the specific residues phosphorylated by each GRK. The caveat is that in vitro kinase assays lack the physiological relevance of cell-based assays. Furthermore, the MD simulation studies of βarr2 in complex with APH1A, which predict the role of individual sites of interaction, will need to be addressed in conjunction with the contribution of each individual APH1A phosphorylation site. Our current models investigate only the APH1A S103A/S110A and S105D/110D double-phosphorylation mutants, which suggest that different phosphorylation patterns may lead to distinct βarr2 conformations. Nevertheless, the contribution of each individual phosphorylation site will need to be investigated with unphosphorylated APH1A or a fully phosphorylated APH1A, using both MD simulations and validated in in vitro assays. Finally, our study does not address the specific APH1A-βarr2 conformations that are formed by genetic deletion of each Grk or by APH1A serine mutagenesis. Currently, we can only speculate on the movement of TM5 and TM6 of APH1A and the positioning of H171 and H197 relative to the APP-C99 binding pocket of presenilin. Additional computational and structural analyses are necessary to gain a deeper understanding of the conformationally distinct APH1A-βarr2 complexes induced by the APH1A phosphorylation barcodes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Amantha Thathiah (amantha@pitt.edu).
Materials availability
This study generated HEK293 GRK knockout monoclonal cell lines by CRISPR/Cas9, APH1A-PK wild-type and mutant plasmids, and 3xHA-βarr2 mutant plasmids. All cell lines and plasmids will be shared by the lead contact upon request.
Data code and availability
All data reported in this paper will be shared by the Lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the Lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
The HEK293 cell line was purchased from American Type Culture Collection. The CHO-KI β-arrestin 2 cell line (CHO-βarr2) was purchased from DiscoveRx. The ReNCell GA2 familial Alzheimer’s disease (fAD) human neural progenitor cell (hNPC) line was a kind gift from Dr. Rudolph E. Tanzi and Dr. Doo Yeon Kim (Harvard University, Cambridge, MA) and generated as previously described (Choi et al., 2014).
Cell culture
HEK293 cells were maintained in complete media containing DMEM/F12 with GlutaMAX (Fisher Scientific) and supplemented with 10% fetal bovine serum (ThermoFisher Scientific). Cells were maintained at 37°C and 5% CO2. CHO-K1 βarr2 cells were maintained in F12 media supplemented with 10% heat-inactivated fetal bovine serum, 1x Pen-Strep-Glutamine, and 3μg/mL hygromycin. Cells were maintained at 37°C and 5% CO2. Human fAD NPCs (ReNCell) were plated and maintained on Matrigel GFR matrix (Corning) – coated 6-well plates and T75 cell culture flasks, respectively. Human fAD NPCs were expanded in ReNCell proliferation media containing DMEM/F12 with GlutaMAX (Fisher Scientific) supplemented with 1x B-27 supplement (Gibco), 2 μg/mL heparin (StemCell Technologies), 20 ng/mL hEGF (Millipore Sigma), 20 ng/mL bFGF (Millipore Sigma) and filtered through a 0.2 μm Fisherbrand disposable PES filter (Fisher Scientific). Human fAD NPCs were maintained at 37°C, 5% O2, and 5% CO2.
METHOD DETAILS
Antibodies and compounds
Rabbit polyclonal antibodies to human PS1-NTF (B19.3, 1:20,000), APH1AL (B82.3, 1:1,000), PEN-2 (B126.2, 1:1,000) and the APP C terminus (B63.3, 1:10,000) were the gift of Dr. Bart De Strooper (VIB and KU Leuven, Leuven, Belgium; UK Dementia Research Institute and University College London, London, United Kingdom) (Annaert et al., 2001; Esselens et al., 2004). Antibodies to the following were purchased: GRK2 (mouse, C-9, SantaCruz Biotechnology, 1:1000), GRK3 (rabbit, D8G6V Cell Signaling Technologies, 1:1000), GRK5 (goat, AF4539, R&D Systems, 1:1000), GRK5 (mouse, D-9, SantaCruz Biotechnology, 1:1000), GRK6 (rabbit, D1A4, Cell Signaling Technologies, 1:1000), NCT (mouse, 9C3, Millipore Sigma, 1:3,000), FLAG (mouse, M2, Millipore Sigma, 1:1,000), GAPDH (rabbit, Millipore Sigma, 1:1,000), βarr2 (mouse, H-9, SantaCruz Biotechnology, 1:1000), hemagglutinin (HA) (mouse, HA.11, BioLegend, 1:1000), hemagglutinin (HA) (rabbit, C29F4, Cell Signaling Technologies, 1:1000), ProLink (PK) (Mouse, DiscoverX, 1:500). Takeda Compound101 was purchased from Hello Bio, and octyl-β-D-glucopyranoside was purchased from EMD Millipore. The compound 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate (CHAPS) was purchased from Sigma-Aldrich.
Plasmid construction
All mutations in APH1A and βarr2 were generated with the XL II Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions.
Generation of HEK293 GRK knockout monoclonal cell lines by CRISPR/Cas9
Genetic deletion of Adrbk1, Adrkb2, Grk5, and Grk6 was performed using the vector pSpCas9(BB)-2A-Puro (Px459v2.0; Addgene plasmid no. 62988; deposited by F. Zhang) (Ran et al., 2013). Two sets of forward and reverse, small guide RNAs (sgRNAs) were designed targeting exon 1 of the Adrbk1 gene, exon 1 of the Adrbk2 gene, exon 3 of the Grk5 gene, and exon 2 of the Grk6 gene. The DNA sequences for each sgRNA are listed in Table S1. Underlined letters indicate additional nucleotides added to the sgRNA sequence corresponding to the Bbs1 restriction site. Each set of target sgRNAs was annealed at 37°C and cloned into the Px459v2.0 plasmid via the Bbs1 restriction site. Low passage HEK293 cells plated at a density of 1.4 × 106 cells /well in 6-well plates were transfected with 1μg of each target plasmid (2μg DNA total/well) using X-tremeGENE HP DNA reagent (Sigma-Aldrich) according to manufacturer’s instructions. After 48 h, medium supplemented with puromycin (3μg/mL) was added for 24 h. Cells were washed with PBS and allowed to recover from puromycin for 24 h in complete media. Cells were then serially diluted and plated in 96-well plates to generate individual clones. The CRISPR control cell line was generated by transfection with the empty Px459v2.0 plasmid. Successful deletion of Adrbk1, Adrbk2, Grk5, and Grk6 was verified by Western blot analysis for GRK2, GRK3, GRK5, GRK6 and polymerase chain reaction genotyping using primers flanking exon 1, 1, 3, and 2 of Adrbk1, Adrbk2, Grk5, and Grk6 listed in Table S2. gDNA was extracted using KAPA genotyping kit (Kapa Biosystems) according to manufacturer’s instructions. Monoclonal Adrbk1-, Adrbk2-, Grk5-, and Grk6 –deleted cell lines were then expanded and used in subsequent assays.
Mass spectrometry
Pelleted cells were solubilized in 10% SDS; 100mM TEAB, probe sonicated, vortexed, and centrifuged to remove insoluble material. Total protein was quantified by Micro BCA (Pierce). 1.5 mg protein was digested with trypsin on Straps (Protifi), and desalted on Peptide Desalted Columns (ThermoFisher Scientific). Phosphopeptides were enriched with Fe cartridges on an AssayMAP Bravo (Agilent). Phosphopeptide enrichments were loaded onto an EASY C18, 1.7μm 2.1 × 50cm column at 300 nL/min with an UltiMate™ 3000 RSLCnano HPLC system, eluted over a 120-min gradient, and analyzed on Orbitrap Eclipse™. The instrument was operated in MS2. MS1 spectra were acquired at a resolving power of 120,000. MS2 spectra acquired in the Orbitrap with CID normalized collision energy = 38. Dynamic exclusion was enabled to minimize the redundant selection of peptides previously selected for MS/MS. Phosphopeptides were identified (FDR 0.05) and quantified in Proteome Discoverer (2.5).
Aβ enzyme-linked immunosorbent assay (ELISA)
Aβ40 and Aβ42 levels were determined by standard sandwich ELISA using end-specific antibodies provided by Janssen Pharmaceutica as previously described (Huang et al., 2015). Briefly, 96-well plates were coated and incubated overnight with monoclonal antibodies JRFcAb40/28 and JRFcAb42/26, which recognize the C terminus of Aβ species terminating at amino acid 40 or 42, respectively. Horseradish peroxidase (HRP)–conjugated JRFAbN/25 was used as the detection antibodies for human Aβ. Culture media from HEK293 cells with C99-FLAG overexpression and ReNCell fAD neuronal culture experiments were subjected to Aβ40 and Aβ42 ELISA. Aβ levels in each condition were normalized to C99-FLAG expression.
β-arrestin PathHunter assay
On day 1, the CHO-K1 β-arrestin cell line was seeded on black, clear-bottom 96-well plates at a cell density of 30,000 cells per well. Two plates were plated per experiment – one for PathHunter assays and one in parallel Aβ ELISAs. On day 2, cells were transfected with wild-type or mutant pCMV-APH1A-PK1 and pSG5,-C99-FLAG using X-tremeGENE HP DNA transfection reagent (Millipore Sigma) according to the manufacturer’s instructions. On day 3, the media was changed to F12 media (serum-free) for 16h. On day 4, one plate of cells was rinsed with PBS and analyzed with the PathHunter β-arrestin assay from DiscoveRx according to the manufacturer’s protocol. Media and cells were collected from the second plate for Aβ ELISA and Western blot analysis of APH1A-PK and C99. RLU values were normalized to APH1A-PK expression levels in each condition.
Co-immunoprecipitation assays
On day 1, HEK293 cells were plated in 10cm dishes at a density of 6 × 106 cells/dish. On day 2, cells were transfected with 3xHA-βarr2 WT, S75R, or L72E mutant and APH1A WT, APH1A-PK WT, or APH1A-PK mutant cDNA using X-tremeGENE HP DNA transfection reagent (Millipore Sigma) according to manufacturer’s instructions. On day 3, the culture media was refreshed. On day 4, cells were collected and pelleted via centrifugation at 4°C. Cell pellets were lysed on ice for 1 h in 1mL of ice-cold lysis buffer (1% octyl-β-D-glucopyranoside or CHAPS, 1x complete protease inhibitor cocktail (Roche), 1x phosphatase inhibitor cocktails 2 and 3 (Millipore Sigma) in 25mM HEPES/150mM NaCl/1mM EDTA buffer. 20μL of pre-washed Protein G Dynabeads (Invitrogen) were incubated in 3% BSA at 1hr at room temperature on a rotator with 2μg of HA.11 or negative control antibody. Following incubation, 500μg of sample lysate was added to the 20μL of pre-coupled Dynabeads and incubated with rotation at 4°C for 16 h (overnight). On day 5 following overnight incubation, the unbound fraction was collected, and beads were washed 3 times with lysis buffer. Following the final wash, bound fractions were either collected in 20μL of 0.1M Glycine pH2.8 or boiled with 2x Tris-Glycine Sample Buffer (Invitrogen). For the samples eluted with 0.1M Glycine, 10μL of 1M Tris pH8.0 was added to each sample to neutralize, and 10μL 4x Laemmeli Sample Buffer (Bio-Rad) + 4% β-mercaptoethanol (Bio-Rad) was added to bring the final volume to 40 μL. Samples were then heated for 10 min at 70°C and separated by SDS-PAGE and transferred to 0.45μm nitrocellulose membranes (Bio-Rad) using the Power Blotter Station (Invitrogen). Membranes were blocked for 1 h at room temperature with agitation in 5% milk and incubated overnight on a shaker in appropriate antibody dilutions. The following day, membranes were washed 3 times for 10 min in 1XTBS-Tween (0.1% v/v) and incubated in a goat anti-Rabbit IgG (H + L) HRP-conjugated secondary antibody (1:10,000, Bio-Rad) or a goat anti-mouse IgG (H + L) HRP-conjugated secondary antibody (1:10,000, Bio-Rad) for 1 h at room temperature. Membranes were washed for 10 min, 3 times in 1XTBS-Tween (0.1% v/v) and 5 min, 3 times in 1XTBS. Chemiluminescence was measured upon addition of Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate (Perkin Elmer).
Knock-down of GRKs in HEK293 by siRNA
HEK293 cells were transfected with siRNA directed against human GRK2, GRK3, GRK5, GRK6, or control siRNA (Qiagen) using Lipofectamine RNAiMAX reagent (Invitrogen). The siRNA sequences targeting GRKs were: GRK2: 5′-AAGAAGUACGAGAAGCUGGAG-3′ (NM_001619, position 268–288); GRK3: 5′-AAGCAAGCUGUAGAACACGUA-3′ (X69117, position 376–396); GRK 5: 5′-AAGCCGUGCAAAGAACUCUUU-3′ (NM_005308, position 406–426); GRK6: 5′-AACAGUAGGUUUGUAGUGAGC-3′ (AF040751, position 724–744) as previously published (Kim et al., 2005). A nonsilencing RNA duplex (5′-AAUUCUCCGAACGUGUCACGU-3′), as the manufacturer indicated, was used as a control. Briefly, HEK293 cells were seeded at a cell density of 1 × 106 cells /well in 6-well plates in DMEM supplemented with 10% FBS. One days after seeding, cells were transfected with the 120pmol/siRNA. Two days after seeding, cells were transfected with 1μg of pSG5-C99-FLAG (1μg DNA total/well) using X-tremeGENE HP DNA reagent (Sigma Aldrich) according to manufacturer’s instructions. After 24 h, the medium was refreshed with 600 μL of serum free medium, and the cells were allowed to accumulate Aβ in the conditioned medium overnight prior to analysis by ELISA for the detection of Aβ40. Cells were collected in STE buffer (250 mM sucrose, 5 mM Tris-HCl, and 1 mM EGTA, pH 7.4) supplemented with 1% Triton X-100, 1x complete protease inhibitor cocktail (Roche), 1x phosphatase inhibitor cocktails 2 and 3 (Millipore Sigma) on ice for 20 min. Lysates were centrifuged at 14,000 rpm for 15 min at 4°C. Equivalent amounts of protein samples were mixed with sample buffer (4x LDS and 4% 2-mercaptoethanol) followed by incubation in a heater for 10 min at 70°C. Samples were then subjected to Western blot analysis for the detection of GRK2, GRK3, GRK5, GRK6, C99, and GAPDH.
Subcellular fractionation
The subcellular fractionation follows our previously published protocol (Thathiah et al., 2009). One T175 flask of HEK CRISPR control cells and GRK2 KO cells was rinsed twice with ice-cold 1X PBS, and solubilized with 2 mL of MES buffer (25 mM MES, pH 6.5, 150 mM NaCl) containing 1% CHAPS and 1X complete protease inhibitor cocktail. Cells were lysed by sequential passage through 18-gauge (5 times) and 26-gauge (10 times) needles and then placed on ice for 1 h. Following the removal of insoluble material by centrifugation at 15, 000 x g for 15 min, the protein concentrations of the supernatant samples were determined. The same concentration of total protein extracted from each cell line was mixed with an 80% sucrose solution dissolved in MES buffer, adjusted to a 45% sucrose concentration, and transferred to the bottom of ultracentrifugation tubes. A discontinuous sucrose gradient was prepared by layering 35% and 5% sucrose in MES buffer. Samples were centrifuged at 200, 000 × g for 18 h. Thirteen 960 μL fractions were collected from the top of the gradient and used for Western blot analysis.
Generation of βarr2-APH1A complex based on structural alignment
As APH1A bears structural similarities to the GPCRs, our original model for βarr2-APH1A complex was based on the X-ray structure of rhodopsin-arrestin complex (PDB id 4zwj) (Kang et al., 2015). We aligned the APH1A subunit from the cryo-EM resolved γ-secretase complex (PDB id 5fn5) (Bai et al., 2015) against rhodopsin and the βarr2 molecules (PDB id 3p2d) (Zhan et al., 2011) against the arrestin chain, then built the ‘alignment’ model for the βarr2-APH1A complex with the aligned chains and further refinement by energy minimization and MD simulations (see Figure S8).
Generation of structural models for βarr2-APH1A complex using docking simulations
We used the HADDOCK 2.2 Server (Dominguez et al., 2003; van Zundert et al., 2016) for docking βarr2 onto APH1A. APH1A subunit was taken from the relaxed cryo-EM structure of γ-secretase complex (PDB id 5fn2, chain C) (Bai et al., 2015). Two independent dockings were performed based on the active and inactive states of arrestin, named as DOCK1 and DOCK2. The active state for Dock1 comes from the crystal structure of visual arrestin chain (PDB id: 5w0p, chain A) (Zhou et al., 2017) bound to rhodopsin. Dock2 is performed with the inactive conformer of βarr2 (PDB id: 3p2d) (Zhan et al., 2011) in the apo state. The finger loop adopts a helical conformation in the active state bound to rhodopsin; whereas it is unstructured in the apo βarr2. HADDOCK performs flexible protein-protein docking when the active residues, i.e. those involved in the protein-protein interaction, are provided. In our case, finger loop residues of arrestin and their counterparts in the APH1A were defined as active residues to allow for their conformational flexibility. As a result of each docking, 200 poses were produced and clustered. We use the best-scoring cluster/pose from DOCK1 and DOCK2 for further refinements by full atomic simulations (see below). The first cluster from DOCK1 contained 186 out of 200 generated poses (HADDOCK score: −89.2). Modeling was performed on this complex to convert the visual arrestin chain into βarr2. The first cluster from DOCK2 included 59 out of 200 generated poses with a HADDOCK score of −99.0.
MD simulations of the βarr2-APH1A complex (WT and mutated)
All atom MD systems with explicit membrane were set-up using GHARMM-GUI membrane builder (Jo et al., 2009), and simulations were performed using NAMD (Phillips et al., 2005) with the CHARMM36m force field (Huang et al., 2017a) for proteins and the CHARMM36 lipids (Klauda et al., 2010), and the TIP3P water model. We performed two independent runs on each model of the complex explained above, namely Alignment, DOCK1 and DOCK2. Typically, a given system comprised approximately 675 POPC lipid molecules, 78,080 water molecules, 215 Na+ ions, and 228 Cl− ions, i.e. a total of 340,560 atoms in a 160 Å × 160 Å × 142 Å box. We relaxed the systems using the equilibration steps in CHARMM-GUI and performed NPT dynamics for 40 ns with 2 fs time step. Nosé-Hoover constant pressure (1 bar) and temperature (310 K) were used. To study the effect of mutations on the complex, we generated three additional runs each containing one mutation (L72E and S75R in βarr2, and R184D in APH1A), for DOCK1 model and the same protocol as that described above. For trajectory analysis and visualization we used VMD (Humphrey et al., 1996) and Pymol (version 1.8).
QUANTIFICATION AND STATISTICAL ANALYSIS
Total abundance of identified phospho-peptides for each APH1A residue was quantified across independent experiments and in HEK293 control and each GRK KO cell line. Total abundance of phosphorylated peptides were then normalized to the abundance of the respective peptide in the control cells. Samples from each independent experiment were processed and run at the same time under the same conditions. Sample number n represent the number of independent experiments or transfection. Two-way analysis of variance (ANOVA) were used to make comparisons between multiple samples across multiple groups. Data in figures are presented as means ± SEM *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-GRK2 (C-9) | SantaCruz Biotechnology | Cat# sc-13143, RRID:AB_626751 |
| Rabbit anti-GRK3 (D8G6V) | Cell Signaling Technologies | Cat# 80362, RRID:AB_2799951 |
| Goat anti-GRK5 (AF4539) | R&D Systems | Cat# AF4539, RRID:AB_2248068 |
| Mouse anti-GRK5 (D-9) | SantaCruz Biotechnology | Cat# sc-518005 |
| Rabbit anti-GRK6 (D1A4) | Cell Signaling Technologies | Cat# 5878, RRID:AB_11179210 |
| Rabbit anti-GAPDH | Millipore Sigma | Cat# G9545, RRID:AB_796208 |
| Rabbit anti-Caveiolin-1 | Abcam | Cat# ab2910, RRID:AB_303405 |
| Mouse anti-HA (HA.11) | BioLegend | Cat# 901522, RRID:AB_2716059 |
| Rabbit anti-HA (C29F4) | Cell Signaling Technologies | Cat# 3724, RRID:AB_1549585 |
| Mouse anti-FLAG (M2) | Millipore Sigma | Cat# P2983, RRID:AB_439685 |
| Mouse anti-ProLink | DiscoveRx | Cat# 922-0010 |
| Mouse anti-βarr2 (H-9) | SantaCruz Biotechnology | Cat# sc-13140, RRID:AB_626701 |
| Rabbit anti-APP (B63.3) | Annaert et al. (2001) | N/A |
| Rabbit anti-APH1AL (B82.3) | Esselens et al. (2004) | N/A |
| Rabbit anti-PEN2 (B126.2) | Esselens et al. (2004) | N/A |
| Mouse anti-NCT (9C3) | Millipore Sigma | Cat# MAB5556, RRID:AB_2235791 |
| Rabbit anti-PS1-NTF (B19.3) | Annaert et al. (2001) | N/A |
| Mouse anti-PS1-CTF (PS1-loop) | Millipore Sigma | Cat# MAB5232, RRID:AB_95175 |
| Goat anti-mouse IgG (H + L) HRP-conjugated secondary antibody | Bio-Rad | Cat# 1706516, RRID:AB_11125547 |
| Goat anti-rabbit IgG (H + L) HRP-conjugated secondary antibody | Bio-Rad | Cat# 1706515, RRID:AB_11125142 |
| Chemicals, peptides, and recombinant proteins | ||
| Takeda Compound 101 (CMPD101) | Hello Bio | Cat# HB2840 |
| B-27 Supplement | Fisher Scientific | Cat# 17504044 |
| Matrigel GFR matrix | Fisher Scientific | Cat# 354230 |
| hEGF | Millipore Sigma | Cat# GF144 |
| bFGF | Millipore Sigma | Cat# GF003AF |
| CHAPS | Millipore Sigma | Cat# 10810118001 |
| Critical commercial assays | ||
| PathHunter β-arrestin Assay | DiscoveRx | Cat# 93-0001 |
| XL II Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200521 |
| Pierce Micro BCA Assay Kit | Fisher Scientific | Cat# 23227 |
| S-trap Midi | Protifi | Cat# C02-midi-20 |
| Peptide Desalting Spin Columns | Thermo Scientific | Cat# 89851 |
| Deposited data | ||
| rhodopsin-arrestin complex | Kang et al. (2015) | PDB: 4ZWJ |
| γ-secretase complex | Bai et al. (2015) | PDB: 5FN5 |
| Beta-arrestin-2 | Zhan et al. (2011) | PDB: 3P2D |
| APH1A subunit of γ-secretase complex | Bai et al. (2015) | PDB: 5FN2 |
| Visual arrestin | Zhou et al. (2017) | PDB: 5W0P |
| Experimental models: Cell lines | ||
| HEK293T | American Type Culture Collection (ATCC) | Cat# ATCC CRL-3216 |
| HEK293T ΔAdrbk1 (GRK2 KO) | This manuscript | N/A |
| HEK293T ΔAdrbk2 (GRK3 KO) | This manuscript | N/A |
| HEK293T ΔGrk5 (GRK5 KO) | This manuscript | N/A |
| HEK293T ΔGrk6 (GRK6 KO) | This manuscript | N/A |
| CHO-K1 β-arrestin 2 cells (CHO-βarr2) | DiscoveRx | Cat# 93-0309C2 |
| ReNCell GA2 human neural progenitor cells | Choi et al. (2014) | N/A |
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro | Ran et al. (2013) | Addgene #62988 |
| pSG-C99-FLAG | Thathiah et al. (2009) | N/A |
| pCMV-PK1 | DiscoveRx | Cat# 93-0167 |
| pCMV-APH1A-PK1 (and mutants) | This manuscript | N/A |
| pEZ-3xHA-βarr2 (and mutants) | This manuscript | N/A |
| Software and algorithms | ||
| Prism 8.0 | GraphPad | |
| Proteome Discoverer 2.5 | ThermoFisher | |
| HADDOCK 2.2 | van Zundert et al. (2016) | https://milou.science.uu.nl/services/HADDOCK2.2/ |
| GHARMM-GUI membrane builder v2.0 | Jo et al. (2009) | http://www.charmm-gui.org/input/membrane |
| NAMD version 2.13 | Phillips et al., 2005 | https://www.ks.uiuc.edu/Research/namd/ |
| VMD version 1.9.3 | Humphrey et al. (1996) | https://www.ks.uiuc.edu/Research/vmd/ |
| Pymol version 1.8 | Schrodinger, LLC | https://www.schrodinger.com/products/pymol |
| Other | ||
| GRK2 siRNA | Kim et al. (2005) | N/A |
| GRK3 siRNA | Kim et al. (2005) | N/A |
| GRK5 siRNA | Kim et al. (2005) | N/A |
| GRK6 siRNA | Kim et al. (2005) | N/A |
Highlights.
GRKs differentially regulate phosphorylation of the γ-secretase complex subunit APH1A
APH1A phosphorylation barcodes differentially regulate βarr2 recruitment to APH1A
The finger loop domain of βarr2 interacts with the cytoplasmic TM core of APH1A
Differential APH1A and βarr2 conformations regulate Aβ generation
ACKNOWLEDGMENTS
We would like to thank Dr. Cheng Zhang (University of Pittsburgh) for his kind help in revising the final manuscript and providing feedback. We greatly appreciate Dr. Bart De Strooper (VIB and KU Leuven, Leuven, Belgium; UK Dementia Research Institute and University College London, London, United Kingdom) for gifts of the APH1A, PS1-NTF, and PEN2 antibodies. We thank Dr. Marc Mercken (Johnson & Johnson Pharmaceuticals R&D, Beerse, Belgium) for the Aβ antibodies. We also would like to thank Dr. Rudolph Tanzi and Dr. Doo Yeon Kim (Harvard University, Cambridge, MA) for the ReNCell human fAD neural progenitor cells. Finally, we would also like to thank Thais Rafael Guimarães and Bryan Hurtle for their valuable and constructive insights toward the completion of this study. This study was supported by the National Institute on Aging (grant R01 AG058851 to A.T.), the National Institute of General Medical Sciences (grants R01 GM139297 and P41 GM103712 to I.B.), the National Institute of Mental Health (grants R01 MH125235 and R01 MH118497 to M.M.), and the Clear Thoughts Foundation (A.T.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111110.
REFERENCES
- Annaert WG, Esselens C, Baert V, Boeve C, Snellings G, Cupers P, Craessaerts K, and De Strooper B (2001). Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32, 579–589. 10.1016/s0896-6273(01)00512-8. [DOI] [PubMed] [Google Scholar]
- Bai XC, Rajendra E, Yang G, Shi Y, and Scheres SHW (2015). Sampling the conformational space of the catalytic subunit of human γ-secretase. Elife 4, e11182. 10.7554/elife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidya M, Kumari P, Dwivedi-Agnihotri H, Pandey S, Chaturvedi M, Stepniewski TM, Kawakami K, Cao Y, Laporte SA, Selent J, et al. (2020). Key phosphorylation sites in GPCRs orchestrate the contribution of β-Arrestin 1 in ERK1/2 activation. EMBO Rep. 21, e49886. 10.15252/embr.201949886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, and Brunak S (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol 294, 1351–1362. 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Chen AC, Guo LY, Ostaszewski BL, Selkoe DJ, and LaVoie MJ (2010). Aph-1 associates directly with full-length and C-terminal fragments of gamma-secretase substrates. J. Biol. Chem 285, 11378–11391. 10.1074/jbc.m109.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Perry NA, Vishnivetskiy SA, Berndt S, Gilbert NC, Zhuo Y, Singh PK, Tholen J, Ohi MD, Gurevich EV, et al. (2017). Structural basis of arrestin-3 activation and signaling. Nat. Commun 8, 1427. 10.1038/s41467-017-01218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, et al. (2014). A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278. 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Staus DP, Wingler LM, Ahn S, Pani B, Capel WD, and Lefkowitz RJ (2018). G protein-coupled receptor kinases (GRKs) orchestrate biased agonism at the β2-adrenergic receptor. Sci. Signal 11, eaar7084. 10.1126/scisignal.aar7084. [DOI] [PubMed] [Google Scholar]
- De Strooper B (2003). Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-secretase complex. Neuron 38, 9–12. 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, and Shenoy SK (2007). β-Arrestins and cell signaling. Annu. Rev. Physiol 69, 483–510. 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, and Bonvin AMJJ (2003). HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc 125, 1731–1737. 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Drube J, Haider RS, Matthees ESF, Reichel M, Zeiner J, Fritzwanker S, Ziegler C, Barz S, Klement L, Filor J, et al. (2022). GPCR kinase knockout cells reveal the impact of individual GRKs on arrestin binding and GPCR regulation. Nat. Commun 13, 540. 10.1038/s41467-022-28152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi-Agnihotri H, Chaturvedi M, Baidya M, Stepniewski TM, Pandey S, Maharana J, Srivastava A, Caengprasath N, Hanyaloglu AC, Selent J, and Shukla AK (2020). Distinct phosphorylation sites in a prototypical GPCR differently orchestrate β-arrestin interaction, trafficking, and signaling. Sci. Adv 6, eabb8368. 10.1126/sciadv.abb8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselens C, Oorschot V, Baert V, Raemaekers T, Spittaels K, Serneels L, Zheng H, Saftig P, De Strooper B, Klumperman J, and Annaert W (2004). Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J. Cell Biol 166, 1041–1054. 10.1083/jcb.200406060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães TR, Swanson E, Kofler J, and Thathiah A (2021). G protein-coupled receptor kinases are associated with Alzheimer’s disease pathology. Neuropathol. Appl. Neurobiol 47, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJ, Mushegian A, and Gurevich VV (2012). G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol. Ther 133, 40–69. 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, and Gurevich EV (2019). GPCR signaling regulation: the role of GRKs and arrestins. Front. Pharmacol 10, 125. 10.3389/fphar.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, and Gurevich VV (2020). Grks as modulators of neurotransmitter receptors. Cells 10, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q-T, Xiao P, Huang S-M, Jia Y-L, Zhu Z-L, Lin J-Y, Yang F, Tao X-N, Zhao R-J, Gao F-Y, et al. (2021). Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2. Nat. Commun 12, 2396. 10.1038/s41467-021-22731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner CZ, Cheng N, and Ye RD (2012). Identification of a nuclear localization sequence in β-arrestin-1 and its functional implications. J. Biol. Chem 287, 8932–8943. 10.1074/jbc.m111.294058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Skwarek-Maruszewska A, Horré K, Vandewyer E, Wolfs L, Snellinx A, Saito T, Radaelli E, Corthout N, Colombelli J, et al. (2015). Loss of GPR3 reduces the amyloid plaque burden and improves memory in Alzheimer’s disease mouse models. Sci. Transl. Med 7, 309ra164. 10.1126/scitranslmed.aab3492. [DOI] [PubMed] [Google Scholar]
- Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, and MacKerell AD (2017a). CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Todd N, and Thathiah A (2017b). The role of GPCRs in neurodegenerative diseases: avenues for therapeutic intervention. Curr. Opin. Pharmacol 32, 96–110. 10.1016/j.coph.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dy-namics. J. Mol. Graph 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hur J-Y, Welander H, Behbahani H, Aoki M, Frånberg J, Winblad B, Frykman S, and Tjernberg LO (2008). Active gamma-secretase is localized to detergent-resistant membranes in human brain. FEBS J. 275, 1174–1187. 10.1111/j.1742-4658.2008.06278.x. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Carrillo MC, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, et al. (2012). National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 8, 1–13. 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, and Im W (2009). CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J 97, 50–58. 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. (2015). Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567. 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O-J, Gardner BR, Williams DB, Marinec PS, Cabrera DM, Peters JD, Mak CC, Kim K-M, and Sibley DR (2004). The role of phosphorylation in D1 dopamine receptor desensitization: evidence for a novel mechanism of arrestin association. J. Biol. Chem 279, 7999–8010. 10.1074/jbc.m308281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Ahn S, Ren X-R, Whalen EJ, Reiter E, Wei H, and Lefkowitz RJ (2005). Functional antagonism of different G protein-coupled receptor kinases for beta-arrestin-mediated angiotensin II receptor signaling. Proc. Natl. Acad. Sci. USA 102, 1442–1447. 10.1073/pnas.0409532102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Choi SH, D’Avanzo C, Hebisch M, Sliwinski C, Bylykbashi E, Washicosky KJ, Klee JB, Brüstle O, Tanzi RE, and Kim DY (2015). A 3D human neural cell culture system for modeling Alzheimer’s disease. Nat. Protoc 10, 985–1006. 10.1038/nprot.2015.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, and Pastor RW (2010). Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843. 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komolov KE, and Benovic JL (2018). G protein-coupled receptor kinases: past, present and future. Cell. Signal 41, 17–24. 10.1016/j.cellsig.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhen OME, Wang G, Solberg J, Erickson LJ, Law PY, and Loh HH (2000). Hierarchical phosphorylation of delta-opioid receptor regulates agonist-induced receptor desensitization and internalization. J. Biol. Chem 275, 36659–36664. 10.1074/jbc.m006788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Srivastava A, Banerjee R, Ghosh E, Gupta P, Ranjan R, Chen X, Gupta B, Gupta C, Jaiman D, and Shukla AK (2016). Functional competence of a partially engaged GPCR-β-arrestin complex. Nat. Commun 7, 13416. 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorraca NR, Wang JK, Bauer B, Townshend RJL, Hollingsworth SA, Olivieri JE, Xu HE, Sommer ME, and Dror RO (2018). Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456. 10.1038/s41586-018-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorraca NR, Masureel M, Hollingsworth SA, Heydenreich FM, Suomivuori C-M, Brinton C, Townshend RJ, Bouvier M, Kobilka BK, and Dror RO (2020). How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell 183, 1813–1825.e18. 10.1016/j.cell.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, and Shenoy SK (2005). Transduction of receptor signals by beta-arrestins. Science 308, 512–517. 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhao X, Zeng X, Bossers K, Swaab DF, Zhao J, and Pei G (2013). β-arrestin1 regulates γ-secretase complex assembly and modulates amyloid-β pathology. Cell Res. 23, 351–365. 10.1038/cr.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang T-Y, Bressler EA, Hara MR, et al. (2011). Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal 4, ra51. 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorato JJ, Palczewski K, Regan JW, Caron MG, Lefkowitz RJ, and Benovic JL (1991). Role of acidic amino acids in peptide substrates of the .beta.-adrenergic receptor kinase and rhodopsin kinase. Biochemistry 30, 5118–5125. 10.1021/bi00235a002. [DOI] [PubMed] [Google Scholar]
- Palmer TM, and Stiles GL (2000). Identification of threonine residues controlling the agonist-dependent phosphorylation and desensitization of the rat A(3) adenosine receptor. Mol. Pharmacol 57, 539–545. 10.1124/mol.57.3.539. [DOI] [PubMed] [Google Scholar]
- Pardossi-Piquard R, Yang S-P, Kanemoto S, Gu Y, Chen F, Böhm C, Sevalle J, Li T, Wong PC, Checler F, et al. (2009). APH1 polar transmembrane residues regulate the assembly and activity of presenilin complexes. J. Biol. Chem 284, 16298–16307. 10.1074/jbc.m109.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, and Schulten K (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J,Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Dwivedi H, Baidya M, Kumar M, and Shukla AK (2017). Novel structural insights into GPCR-β-arrestin interaction and signaling. Trends Cell Biol. 27, 851–862. 10.1016/j.tcb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Ren X-R, Reiter E, Ahn S, Kim J, Chen W, and Lefkowitz RJ (2005). Different G protein-coupled receptor kinases govern G protein and beta-arrestin-mediated signaling of V2 vasopressin receptor. Proc. Natl. Acad. Sci. USA 102, 1448–1453. 10.1073/pnas.0409534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas C, Penela P, Murga C, Salcedo A, García-Hoz C, Jurado-Pueyo M, Aymerich I, and Mayor F (2007). The G protein-coupled receptor kinase (GRK) interactome: role of GRKs in GPCR regulation and signaling. Biochim. Biophys. Acta 1768, 913–922. 10.1016/j.bbamem.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Serneels L, Dejaegere T, Craessaerts K, Horré K, Jorissen E, Tousseyn T, Hébert S, Coolen M, Martens G, Zwijsen A, et al. (2005). Differential contribution of the three Aph1 genes to gamma-secretase activity in vivo. Proc. Natl. Acad. Sci. USA 102, 1719–1724. 10.1073/pnas.0408901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Pozo A, Frosch MP, Masliah E, and Hyman BT (2011). Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med 1, a006189. 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla AK,Westfield GH, Xiao K, Reis RI, Huang L-Y, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. (2014). Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222. 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Zhao J, Wang F, Ma L, and Pei G (2010). A GPCR/secretase complex regulates β- and γ-secretase specificity for Aβ production and contributes to AD pathogenesis. Cell Res. 20, 138–153. 10.1038/cr.2010.3. [DOI] [PubMed] [Google Scholar]
- Thal DM, Yeow RY, Schoenau C, Huber J, and Tesmer JJG (2011). Molecular mechanism of selectivity among G protein-coupled receptor kinase 2 inhibitors. Mol. Pharmacol 80, 294–303. 10.1124/mol.111.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thathiah A, and De Strooper B (2011). The role of G protein-coupled receptors in the pathology of Alzheimer’s disease. Nat. Rev. Neurosci 12, 73–87. 10.1038/nrn2977. [DOI] [PubMed] [Google Scholar]
- Thathiah A, Spittaels K, Hoffmann M, Staes M, Cohen A, Horré K, Vanbrabant M, Coun F, Baekelandt V, Delacourte A, et al. (2009). The orphan G protein-coupled receptor 3 modulates amyloid-beta peptide generation in neurons. Science 323, 946–951. 10.1126/science.1160649. [DOI] [PubMed] [Google Scholar]
- Thathiah A, Horré K, Snellinx A, Vandewyer E, Huang Y, Ciesielska M, De Kloe G, Munck S, and De Strooper B (2013). β-arrestin 2 regulates Aβ generation and γ-secretase activity in Alzheimer’s disease. Nat. Med 19, 43–49. 10.1038/nm.3023. [DOI] [PubMed] [Google Scholar]
- Tobin AB, Butcher AJ, and Kong KC (2008). Location, location, location…site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol. Sci 29, 413–420. 10.1016/j.tips.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, and Bonvin AMJJ (2016). The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol 428, 720–725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Cheng H, Lin W, Sakurai T, Li T, Nukina N, Wong PC, Xu H, and Thinakaran G (2004). Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes. J. Biol. Chem 279, 44945–44954. 10.1074/jbc.m407986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar VAM, Cuevas S, Zheng X, and Jose PA (2016). Localization and signaling of GPCRs in lipid rafts. Methods Cell Biol. 132, 3–23. 10.1016/bs.mcb.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Wang P, Wu Y, Ge X, Ma L, and Pei G (2003). Subcellular localization of beta-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J. Biol. Chem 278, 11648–11653. 10.1074/jbc.m208109200. [DOI] [PubMed] [Google Scholar]
- Watari K, Nakaya M, and Kurose H (2014). Multiple functions of G protein-coupled receptor kinases. J. Mol. Signal 9, 1. 10.1186/1750-2187-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yang F, Zhang D, Liu Z, Lin A, Liu C, Xiao P, Yu X, and Sun J-P (2017). Phosphorylation of G Protein-Coupled receptors: from the barcode hypothesis to the flute model. Mol. Pharmacol 92, 201–210. 10.1124/mol.116.107839. [DOI] [PubMed] [Google Scholar]
- Yin W, Li Z, Jin M, Yin Y-L, de Waal PW, Pal K, Yin Y, Gao X, He Y, Gao J, et al. (2019). A complex structure of arrestin-2 bound to a G protein-coupled receptor. Cell Res. 29, 971–983. 10.1038/s41422-019-0256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Gimenez LE, Gurevich VV, and Spiller BW (2011). Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J. Mol. Biol 406, 467–478. 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Thompson R, Zhang H, and Xu H (2011). APP processing in Alzheimer’s disease. Mol. Brain 4, 3. 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. (2017). Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell 170, 457–469.e13. 10.1016/j.cell.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.