Abstract
Bacterial lipoproteins are hydrophilic proteins that are anchored to a cell membrane by N-terminally linked fatty acids. It is widely believed that nearly all lipoproteins produced by Gram-negative bacteria are either retained in the inner membrane (IM) or transferred to the inner leaflet of the outer membrane (OM). Lipoproteins that are exposed on the cell surface have also been reported, but are generally considered to be rare. Results from a variety of recent studies, however, now suggest that the prevalence of surface exposed lipoproteins has been underestimated. In this review we describe the evidence that the surface exposure of lipoproteins in Gram-negative bacteria is a widespread phenomenon and discuss possible mechanisms by which these proteins might be transported across the OM.
Keywords: Gram-negative bacteria, lipoproteins, outer membrane, protein secretion, protein translocation
Lipoproteins Are a Diverse Family of Membrane-associated Proteins
Bacterial lipoproteins (see Glossary) are peripheral membrane proteins that are attached to a cell membrane by N-terminally linked fatty acids. Following their synthesis in the cytoplasm, lipoproteins are transported across the cytoplasmic membrane [also known as the inner membrane (IM) in Gram-negative bacteria] via the Sec or Tat pathway. Like all proteins that are exported through these pathways lipoproteins contain a cleavable N-terminal signal peptide, but the C-terminus of lipoprotein signal peptides is unique in that it contains a four-residue motif known as a lipobox. Although the lipobox consensus sequence was originally defined as L(A/S)(G/A)|C [1], the only truly conserved residue is the cysteine at the +1 position that is the target of acylation and the N-terminal residue of the protein following signal peptide cleavage. After lipoproteins cross the cytoplasmic membrane/IM the thiol group of the cysteine residue is acylated by a preprolipoprotein diacylglyceryl transferase (Lgt) (Figure 1). The presence of the lipid moieties facilitates the attachment of the protein to the membrane. Subsequently the signal peptide is cleaved by a specialized lipoprotein signal peptidase (Lsp or signal peptidase II) and the new N-terminal amide group is acylated by an N-acyl transferase (Lnt). Recent results indicate that the last step is dispensable in some organisms including Francisella tularensis and Neisseria gonorrhoeae [2]. Lipoproteins are heterogeneous in sequence and have been shown to be involved in a wide variety of cellular processes including molecular transport reactions, signal transduction, maintenance of envelope integrity, and pathogenesis. Based on algorithms that search for lipobox motifs in genome sequences and other criteria, most bacteria are predicted to encode ~100–200 lipoproteins ([3]; see also http://www.mrc-lmb.cam.ac.uk/genomes/dolop/).
Figure 1.
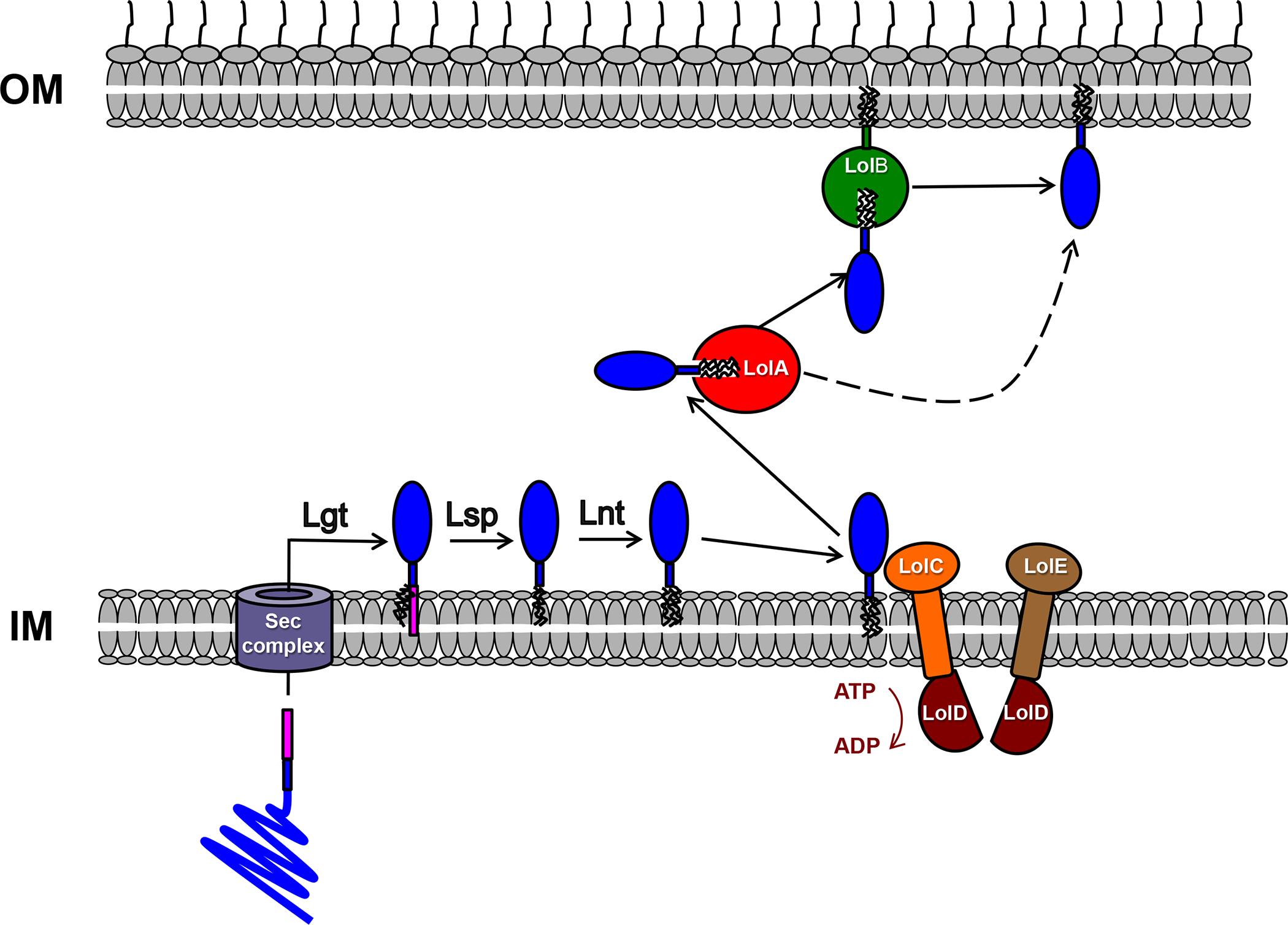
Post-translational Modification of Lipoproteins and Trafficking to the OM by the Lol Pathway. Following their translocation across the IM by the Sec pathway (or, in a few cases, by the Tat pathway) proteins that contain a lipobox motif are acylated by a prelipoprotein diacylglyceryl transferase (Lgt). The signal peptide (pink) is then removed by a lipoprotein signal peptidase (Lsp) and, in many organisms, the new N-terminal amide group is acylated by an N-acyl transferase (Lnt). Lipoproteins that do not contain a ‘Lol avoidance’ signal are then released from the IM by the ABC transporter LolCDE and shuttled to the OM by LolA. In some organisms the lipoprotein is transferred to LolB, which facilitates integration into the inner leaflet of the OM, but in organisms that lack LolB membrane insertion is facilitated by LolA.
Early Evidence For the Surface Localization of Lipoproteins in Gram-negative Bacteria
While lipoproteins produced by Gram-positive bacteria are automatically exposed on the cell surface, the ultimate fate of lipoproteins produced by Gram-negative bacteria is not apparent a priori. Experiments conducted as early as the 1970s showed that while some Escherichia coli lipoproteins are retained in the IM, many others are localized to the outer membrane (OM) [4, 5]. It is now known that lipoproteins are transferred to the inner leaflet of the OM by the Lol pathway [6]. This pathway uses an ABC transporter (LolCDE) to release lipoproteins from the IM, a carrier protein to shuttle cargo from the IM to the OM (LolA), and an OM lipoprotein receptor (LolB) (Figure 1). While LolA and LolCDE are highly conserved in Gram-negative taxa, LolB has been found only in β- and γ-proteobacteria. The only lipoproteins that remain associated with the IM are those that contain a so-called ‘Lol avoidance’ signal near the N-terminus. In E. coli and other Enterobacteriaciae the presence of an aspartate at +2 leads to the retention of a lipoprotein in the IM, but in other organisms the identity of residues at +3 and +4 dictates lipoprotein localization [7, 8].
A few lipoproteins that are either partially or completely exposed on the cell surface or that are secreted were also reported in studies conducted as early as the 1980s. An interest in conjugation led to the discovery that TraT, an E. coli protein that inhibits the transfer of DNA into F+ cells, is a surface exposed lipoprotein [9, 10]. Studies on membrane proteins that are immunogenic—and therefore presumably surface exposed—led to the identification of multiple surface exposed lipoproteins in the spirochetes Treponema pallidum and Borrelia burgdorferi, the etiologic agent of Lyme disease [11, 12]. VacJ, an exposed lipoprotein produced by Shigella flexneri, emerged in a genetic screen for factors that promote intercellular spreading [13]. In the same time period an enzyme that is secreted into the growth medium by Klebsiella pneumoniae called pullulanase (PulA) was also shown to be a lipoprotein [14].
Recent Studies Indicate That Surface Exposed Lipoproteins Are Widespread
The number of confirmed or putative surface exposed lipoproteins has been growing over the years, but these proteins have been perceived to be rare for several reasons. First, reports of extracellular lipoproteins have been sporadic, and have often emerged fortuitously from studies that focused on bacterial pathogenesis or vaccine targets rather than on protein localization per se. In general, these studies describe protein topology only in passing and do not place the localization results in a broader context. Second, because extracellular lipoproteins have often been observed in a few organisms that have been the subject of intense vaccine research—including organisms such as B. burgdorferi that have unusual biological properties—the commonality of surface localization in the larger class of Gram-negative bacteria has been unclear. Finally, while the Lol machinery that transfers lipoproteins to the inner leaflet of the OM has been well characterized, the mechanism by which most surface exposed lipoproteins are localized has remained obscure. The lack of evidence for a general transport process may contribute to the perception that surface localized lipoproteins are outliers.
The picture that now emerges from combining the results of studies conducted over three decades (including some very recent studies) on an increasing number of organisms is that surface exposed lipoproteins are widely produced by Gram-negative bacteria. Furthermore, while only a few of the lipoproteins produced by some species appear to be surface exposed, a much higher percentage of the lipoproteins produced by others reach the cell surface. Due to an interest in identifying vaccine targets and in understanding the functional properties of surface localized virulence factors, cell surface lipoproteins have recently been reported in a variety of pathogens including Helicobacter pylori, Porphyromonas gingivalis, Campylobacter jejuni, Legionella pneumophila, Vibrio cholerae, Francisella tularensis and Moraxella catarrhalis (Table 1). B. burgdorferi has been shown to produce multiple cell surface lipoproteins including OspA/B, OspC, Vsp1, VlsE, Erps, BBE31, BBA52, and BBA33 that function in immune system evasion and adhesion both in ticks and mammalian hosts [12, 15–20]. Interestingly, there is evidence that about two-thirds of the lipoproteins produced by this organism localize to the cell surface [21]. Continuing interest in potential vaccine candidates has also resulted in the discovery of multiple cell surface localized lipoproteins in Neisseria meningitidis and N. gonorrhoeae [22–31]. Functionally characterized proteins include LP2086 (or GNA1870), which binds Factor H and thereby plays a role in complement evasion [32], and the heparin binding protein GNA2132 [33]. Both proteins are components of a recently approved meningococcal vaccine, 4CMenB (Bexsero®) [34]. The surface lipoproteins TbpB and LbpB promote iron acquisition by serving as cofactors for transferrin and lactoferrin binding, respectively [35, 36]. Finally, NalP functions as a serine protease that catalyzes the proteolytic processing of other cell surface proteins [24, 37].
Table 1.
Surface Exposed Lipoproteins in Pathogenic Gram-negative Bacteria (Other Than Borrelia and Neisseria)
| Organism | Protein | Function | Evidence for surface exposure | Refs |
|---|---|---|---|---|
| Klebsiella pneumoniae | PulA | Pullulanase | Immunofluorescence microscopy and protease treatment | [14, 76] |
| Shigella flexneri | VacJ | Involved in intercellular spreading | Biotinylation | [13] |
| Helicobacter pylori | AlpA | Autotransporter | Adhesion assay | [61] |
| Lpp20 | Unknown | Immunogold labeling/electron microscopy (EM) | [77] | |
| Porphyromonas gingivalis | IhtB | Hemin binding | Immunogold labeling/EM | [78] |
| Bordetella pertussis | SphBI | Autotransporter | Protease treatment | [79] |
| Campylobacter jejuni | JlpA | Adhesin | Protease treatment | [80] |
| CapA | Autotransporter | Immunogold labeling/EM | [63] | |
| Legionella pneumophila | LdtA | Unknown | Immunofluorescence microscopy and protease treatment | [81] |
| Vibrio cholerae | VolA | Lysophopholipase A | Biotinylation and immunogold labeling/EM | [82] |
| Haemophilus influenzae | P6 | Pal homolog | Immunofluorescence microscopy and flow cytometry | [83] |
| PH | Factor H binding | Flow cytometry | [84] | |
| Francisella tularensis | FipB | Oxidoreductase and protein disulfide isomerase | Immunofluorescence microscopy, biotinylation, and protease treatment | [85] |
| Moraxella catarrhalis | ORF13 | Survival in nasopharynx | Biotinylation | [86] |
Perhaps unexpectedly, recent studies that focus on basic research questions rather than pathogenesis have also yielded evidence for the surface exposure of multiple lipoproteins in E. coli. The E.coli K-12 genome encodes 86 putative lipoproteins [3], but for many years TraT was the only lipoprotein that was known to be localized on the cell surface. Lpp, which is probably the best-studied bacterial lipoprotein, was shown to exist more than 40 years ago in a ‘bound’ form that is tethered to the peptidoglycan layer and a more abundant ‘free’ form [38–41], but evidence that the free form is surface exposed emerged only in 2011 [42]. A subpopulation of another lipoprotein that binds to peptidoglycan (Pal) also appears to be exposed on the cell surface [43]. This finding is consistent with the observation that the Pal homolog produced by Haemophilus influenzae is a target for human bacteriocidal antiboides [44]. Recent studies showed that RcsF, a lipoprotein that detects envelope stress, is exposed on the cell surface through interactions with integral OM proteins [45, 46]. Similar to most proteins that are embedded in the OM, these proteins form a closed cylindrical structure known as a β barrel. There is also evidence that BamC, a component of the heteooligomer that catalyzes the membrane integration of β barrel proteins (the Bam complex), is localized on the outside of the cell [47]. Finally, x-ray crystallographic analysis has shown that small segments of two lipoproteins that form multimeric pore-forming structures in the OM (Wza and CsgG) are exposed on the cell surface [48, 49].
Although the vast majority of studies on Gram-negative bacteria have focused on Proteobacteria because of their experimental tractability, their long experimental history, or their role in infectious disease, there have been reports of cell surface lipoproteins in an evolutionarily distant genus—the Bacteroides–-for many years. These organisms have become the subject of intense interest recently because of their abundance in the gut microbiome and their connection to multiple aspects of human health. The well-characterized Sus system for starch utilization produced by Bacteroides thetaiotaomicron relies on several lipoproteins, SusDEF, to enhance substrate binding and uptake by a TonB-dependent transporter (TBDT) called SusC. These lipoproteins were found to be surface exposed using a whole cell proteolysis assay [50]. SusD is protease sensitive only when SusC is absent and therefore presumably forms a complex with SusC that renders it protease resistant. SusD has many paralogs in the Bacteroides that likely promote nutrient binding that may also be surface exposed. Indeed a Bacteroides fragilis paralog (NanU) was found to be surface localized by immunofluorescence microscopy [51]. Furthermore, a Sus-like locus in Bacteroides ovatus encodes a surface exposed lipoprotein that has endo-xyloglucanase activity [52]. Additional lipoproteins for which there is evidence of surface exposure include the plasminogen binding protein Pbp [53], the adhesins AapA, and AapF [54], the glycoproteins BF0522 and BF3567 [55], and the collagen binding protein Cpb1 [56].
A comprehensive analysis of OM protein topology recently confirmed that the surface exposure of lipoproteins in B. fragilis is very common [57]. Treatment of whole cells with a protease or a membrane impermeable biotinylation reagent followed by quantitative LC-MS/MS revealed that a significant proportion of lipoproteins produced under laboratory growth conditions (35% in the protease sensitivity experiment) are surface exposed. As expected, many of the exposed lipoproteins are SusD-like proteins or proteins encoded adjacent to TBDT genes [57], but some of the exposed lipoproteins have no known function. Interestingly, several lipoproteins of unknown function were also found to be localized partially or exclusively in the culture medium. A similar proteomic analysis revealed that Capnocytophaga canimorsus, a component of the canine oral microbiome that belongs to the same phylum as B. fragilis (the Bacteroidetes) likewise produces a large number of surface exposed lipoproteins [58].
Caveats and Pitfalls in Assessing Lipoprotein Localization
Lipoprotein topology is generally assessed in whole cell assays that examine sensitivity to exogenous proteases or membrane-impermeable chemical reagents, release into the extracellular environment, or antibody binding, but all of these methods have their limitations. Some lipoproteins that are exposed on the cell surface may fold tightly or, in certain organisms, be buried under a thick capsule and therefore remain resistant to protease treatment. Antibodies may also fail to bind to buried proteins, and some antibodies that are useful to detect a protein in cell lysates (e.g., by Western blot) are not useful to detect the same protein on the cell surface (e.g., by immunofluorescence). Similar to SusD, some surface lipoproteins may be protected as the result of interactions with binding partners. Most of the available chemical modifying reagents react with the sides chains of specific amino acids (typically lysine and cysteine residues), so the modifiability of a given cell surface protein depends on both its sequence composition and the accessibility of its reactive groups. Indeed the observation that some of the candidate surface exposed lipoproteins detected in the aforementioned global analysis of the OM proteome in B. fragilis [57] were protease sensitive but only weakly biotinylated whereas others were protease resistant but highly biotinylated illustrates that single methods can be insufficient to determine the localization of a lipoprotein.
Standard methods may not only underestimate the number of surface localized lipoproteins, but can also generate potentially false positive results. Non-quantitative methods are especially prone to generating ambiguous results. In principle, a lipoprotein might be immunogenic or detectable by immunofluorescence even if a small fraction of the molecules are surface localized. The presence of a few molecules of a lipoprotein on the cell surface might reflect dual localization or transient exposure, but might also be due to sorting errors or the aberrant behavior of a small population of the protein. Likewise, a chemically modified form of a lipoprotein might be detected if a small fraction of the cells lyse or become permeabilized. The potential pitfall of chemical modification analysis is illustrated by the fact that multiple residues of several abundant intracellular factors including molecular chaperones, ribosomal proteins, and IM proteins were biotinylated by a cell-impermeable reagent in a recent proteomic analysis [57]. Furthermore, experiments in which a gene encoding a lipoprotein is cloned into a plasmid and overexpressed or expressed from a non-native promoter might generate localization artifacts.
Because the methods that are used to determine the localization of lipoproteins each has its strengths and weaknesses, ideally a combination of methods should be used. At a minimum, careful controls for cell integrity and a quantitative analysis should be performed. In addition, functional and structural information can also be used to bolster evidence for the surface localization of a lipoprotein. The finding that the Neisseria lipoprotein TbpB greatly increases the efficiency of iron uptake by the TBDT TbpA taken together with crystallographic data that provide a compelling model for the interaction of TbpA and TbpB with its ligand leave little doubt that TbpB is surface localized [29, 59]. Historically this sort of rigor has not always been applied, and as a consequence the quality of evidence for the localization of lipoproteins is variable. Nevertheless, while the conclusions that have been drawn about the localization of specific lipoproteins might be faulty, given the accumulated evidence it would be safe to say that the cell surface localization of lipoproteins in Gram-negative bacteria is more common than currently believed.
How Do Lipoproteins Reach the Cell Surface?
The surface exposed lipoproteins that have been identified to date are highly heterogeneous in sequence and structure and lack common features that might explain how they are secreted. Furthermore, in some cases the entire protein and the lipid anchor are transported across the OM, but in other cases only part of the protein is exposed on the cell surface while the lipid anchor remains in the inner leaflet of the OM. This structural and topological diversity may help to explain why different mechanisms have been implicated in the localization of the small number of surface exposed lipoproteins that have been examined. It has been known for many years that the Klebsiella PulA protein is secreted in its entirety by a type II pathway, one of about a dozen numbered pathways used by Proteobacteria and some other Gram-negative organisms to secrete or inject many different types of proteins into the environment or target cells [14, 60]. Unlike several other pathways that transport their substrates across both cell membranes through a single continuous channel (e.g., the type III, IV, and VI pathways), the type II pathway recruits substrates following their translocation across the IM and can therefore access lipoproteins. A few other lipoproteins including the N. meningitidis NalP protein, the H. pylori AlpA protein, the C. jejuni CapA protein, and the Bordetella pertussis SphB1 protein are exposed on the cell surface by the autotransporter (type Va) pathway [24, 61–63]. Autotransporters are proteins that contain an N-terminal extracellular (‘passenger’) domain and a C-terminal β barrel domain that resides in the OM. In addition to catalyzing the membrane insertion of the β barrel domain, the Bam complex also appears to play an important role in facilitating the secretion of the passenger domain [64]. Although most passenger domains move progressively across the OM in a C-to-N terminal direction until they are fully secreted [64, 65], the lipid moiety of NalP remains tethered to the inner leaflet of the OM while the bulk of the ~760 residue passenger domain is exposed on the cell surface [24].
Entirely different mechanisms have been implicated in the surface exposure of small segments of lipoproteins. The E. coli RcsF protein has a transmembrane topology in which the acylated N terminus and a short unstructured region are exposed while the C terminus remains in the periplasm. Remarkably, the exposure results from the threading of RcsF through the lumen of β barrel proteins [45, 46]. The formation of these unusual protein complexes occurs during OM protein assembly and requires the Bam complex. When OM defects compromise the activity of the Bam complex RcsF remains in the periplasm, where an interaction between its C-terminal domain and an IM protein activate a stress response [46]. In another scenario, the pore-forming proteins Wza and CsgG are first targeted to the OM by the Lol system as monomers. C-terminal or internal segments of the monomers subsequently interact to form homooligomeric α-helical or β-stranded barrels that have a hydrophobic exterior and that insert into the OM in a potentially spontaneous, Bam-complex independent fashion [48, 49, 66]. The short loops that connect the structural elements of the barrels become exposed upon membrane integration. The C-terminus of the 58 residue E. coli lipoprotein Lpp may also be exposed through a similar oligomerization reaction [42]. It should be noted, however, that lipoproteins that are threaded through other proteins or that form large oligomeric complexes probably represent special cases that may reach the cell surface by unusual processes.
To date, the most systematic effort to identify a general pathway for the transfer of lipoproteins across the OM has involved an analysis of proteins produced by B. burgdorferi, an organism that lacks all of the numbered secretion pathways found in other Gram-negative bacteria. The best characterized proteins, OspA and OspC/Vsp, have been shown to be secreted in their entirety and anchored to the outer leaflet of the OM [67]. The secretion of OspA requires that the protein remain in at least a partially unfolded translocation-competent conformation [67–69]. Translocation is mediated by an unstructured N-terminal ‘tether’ peptide that appears to prevent premature folding in the periplasm. The observation that C-terminal epitope tags attached to OspA derivatives that are trapped in the periplasm are selectively exposed on the cell surface provides evidence that translocation is initiated at the C-terminus but does not depend on a specific targeting peptide [68]. Surprisingly, a fluorescent reporter protein is exposed on the cell surface when it is fused to either the OspA tether or an N-terminal peptide derived from a periplasmic lipoprotein [70]. This finding suggests that lipoproteins are localized to the cell surface by default but can be retained in the periplasm by sequence-specific signals. Based on the available evidence, a model has been proposed in which many lipoproteins are transiently anchored to the inner surface of the OM and kept unfolded by periplasmic ‘holding’ chaperones [67]. The proteins would then subsequently be transferred to an unidentified ‘flippase’ that mediates cell surface exposure. In this regard it is noteworthy that LolB is only found in β- and γ-proteobacteria and not in organisms such as B. burgdorferi and B. fragilis that secrete a large number of lipoproteins. It is conceivable that in the absence of LolB lipoproteins are often transferred from LolA to a periplasmic factor that directs its cargo to a surface localization pathway.
While it is not yet clear if the results obtained from studies on Borrelia can be generalized or if there is a common pathway that is used for the extracellular localization of the majority of lipoproteins in Gram-negative bacteria, at least in theory there are many different ways that lipoproteins might reach the cell surface. It is tempting to imagine that surface exposed lipoproteins are first targeted to the inner leaflet of the OM and then secreted by a dedicated transport system analogous to the hypothetical Borrelia flippase that recognizes substrates based on sequence or structure (Figure 2, Key Figure, pathway 1). Because the periplasm lacks ATP, the transport reaction would presumably be driven by a different energy source (e.g., folding of the protein in the extracellular space). Although no obvious candidates have emerged in any organism, it is conceivable that the Bam complex plays a role in this transport process (Figure 2, pathway 2). Indeed the depletion of BamA reduces the levels of the surface lipoproteins OspA and CspA in B. burgdorferi [71]. Given that the Bam complex is required for the assembly of β barrel proteins (including potential lipoprotein transporters), however, it is not clear if the decrease is a direct or indirect effect of Bam complex depletion. While the Bam complex might not function as a lipoprotein transporter per se, in cases where a lipoprotein acts in conjunction with a β barrel protein (e.g., a TBDT) it might facilitate interactions between the lipoprotein and its binding partner. The lipoprotein might then reach the cell surface by piggybacking onto the β barrel protein during its assembly (Figure 2, pathway 3). Such a mechanism does not appear to account for the localization of the B. thetaiotaomicron SusD and N. gonorrhoeae TbpB proteins, however, which are exposed on the cell surface in the absence of their β barrel protein binding partners [50].
Figure 2.
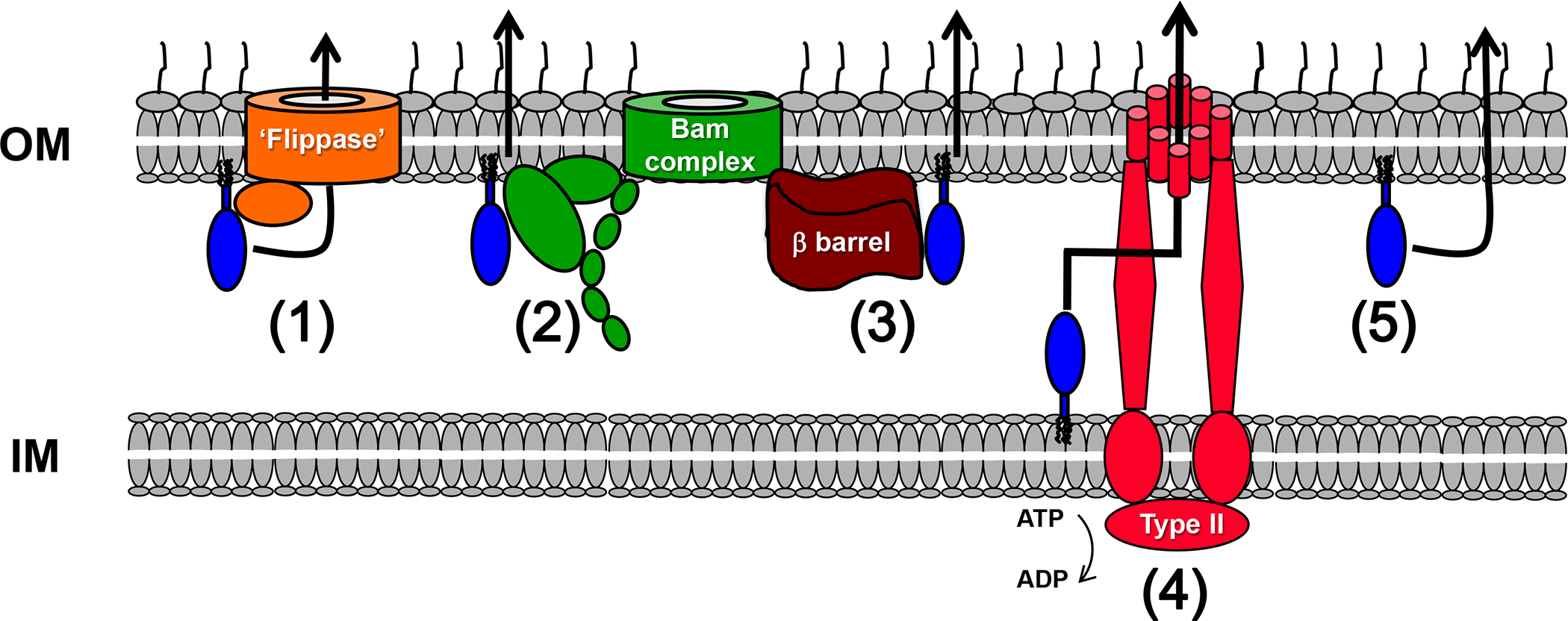
Possible Mechanisms for the Surface Exposure of Lipoproteins. In principle, a subset of lipoproteins that are localized to the inner leaflet of the OM by the Lol pathway might be recognized by a dedicated ‘flippase’ based on sequence or structural features and transported to the outer leaflet of the OM (pathway 1). The Bam complex, a heterooligomer that catalyzes the insertion of β barrel proteins into the OM, might directly mediate the surface exposure of lipoproteins (pathway 2) or indirectly mediate the exposure of lipoproteins that piggyback onto β barrel proteins during their assembly (pathway 3). Some lipoproteins might reach the extracellular space by one of the known secretion pathways that recruit substrates from the periplasmic space (e.g., type I or II pathway) (pathway 4). Cytoplasmic ATPases associated with these secretion pathways might facilitate release of the lipoproteins from the IM. Finally, lipoproteins might be transported across the OM by a spontaneous mechanism, possibly during periods of OM remodeling (pathway 5).
Despite their limited distribution in Gram-negative taxa, a few of the numbered secretion pathways might also play important roles in lipoprotein surface exposure (Figure 2, pathway 4). In addition to the type II pathway, the type I pathway can also recruit proteins from the periplasmic space [72]. Indeed it is conceivable that some of the surface localized lipoproteins synthesized by B. fragilis are secreted by several type I systems that are produced simultaneously but that have no known substrates [57]. Similar to soluble proteins that are secreted by the type II pathway, PulA appears to contain a complex targeting signal that is present in its folded structure [73]. Interestingly, the secretion of PulA does not require its lipidation [74, 75]. This finding suggests that the protein is not ordinarily localized to the OM, but rather released from the IM by a cytoplasmic ATPase activity associated with the type II pathway. Other lipoproteins that are secreted by numbered pathways are probably also detected on the basis of specific targeting signals. Although there is no evidence that TbpB is secreted by a numbered pathway, its localization does not require lipidation and therefore must be based on the recognition of an internal targeting signal [74, 75]. Presumably lipoproteins that are engaged by the numbered pathways are first secreted into the extracellular space. While some of these proteins may remain free in the environment, others may become attached to the cell surface through the partitioning of their lipid moieties into the outer leaflet of the OM before they have a chance to diffuse away.
One intriguing possibility is that lipoproteins other than those that form barrel-like structures are exposed on the surface by a spontaneous mechanism that does not rely on a proteinaceous transporter (Figure 2, pathway 5). Segments of lipoproteins that remain unfolded or that adopt specific conformations might have a unique ability to insert into or pass through the lipid bilayer, possibly under circumstances in which the OM undergoes remodeling (e.g., during cell division).
Concluding Remarks
The evidence accumulated over many years now indicates that the surface exposure of lipoproteins in Gram-negative bacteria is a widespread phenomenon and not just an oddity, but the analysis of this class of proteins is in its infancy and many questions remain unanswered (see Outstanding Questions). The proportion of lipoproteins that reach the cell surface in most organisms is not yet clear largely because there have been very few attempts to analyze lipoprotein localization globally. In the future such studies should be useful not only to generate important statistical information, but also to identify sequence or structural patterns that might provide clues about the mechanism of surface localization. Because proteomic studies can now be conducted in a wide variety of bacteria, it should ultimately be possible to perform a systematic analysis of lipoprotein localization to search for evolutionary trends and to determine, for example, whether organisms that secrete a large number of lipoproteins produce common factors that might serve as components of a conserved transport pathway. Of course an in depth analysis of the localization of specific model lipoproteins using genetic and biochemical methods will be necessary to fully elucidate the mechanisms by which these proteins are transported across the OM. In conjunction with further functional and structural studies on surface exposed lipoproteins, mechanistic studies may also help to explain why these proteins are first acylated and are not simply transported into the extracellular space as non-acylated proteins by one of the many known secretion and surface localization pathways.
Outstanding Questions.
What proportion of the lipoproteins produced by different Gram-negative bacteria are surface exposed? Are there phylogenetic or biological links between organisms that secrete many different lipoproteins?
Do surface exposed lipoproteins tend to have specific structural features or functions?
What sequence or structural features earmark lipoproteins for surface exposure?
Are there dedicated transport systems that expose lipoproteins on the cell surface?
Are there many lipoproteins that have a dual localization or that are conditionally exposed on the cell surface? Can surface exposed lipoproteins ever be reimported into the periplasm?
Trends.
For many years it was believed that almost all lipoproteins produced by Gram-negative bacteria are confined to either the inner membrane or the inner leaflet of the outer membrane, but recent results indicate that the surface exposure of lipoproteins is a widespread phenomenon.
A recent proteomic study provided evidence that a large fraction of the lipoproteins produced by at least a subset of Gram-negative are surface exposed.
Although the localization of surface exposed lipoproteins has not been extensively investigated, available evidence indicates that lipoproteins can reach the cell surface by variety of distinct mechanisms.
ACKNOWLEDGEMENTS
We thank Jessica Pierce for helpful comments on the manuscript. Research conducted in the authors’ laboratory is supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
GLOSSARY
- Barrel assembly machine (Bam) complex
a heterooligomeric complex that catalyzes the integration of β barrel proteins (proteins that fold into a closed cylindrical structure) into the bacterial outer membrane. In E. coli the Bam complex consists of BamA [an integral membrane protein that contains a β barrel domain and five periplasmic polypeptide transport-associated (POTRA) domains] and four lipoproteins designated BamB-E. BamA and BamD are conserved in all Gram-negative bacteria. The mechanism by which the Bam complex catalyzes the membrane integration of β barrel proteins is currently unknown.
- Gram-negative bacteria
a large group of bacteria (sometimes also referred to as diderms) that contain two cell membranes, a cytoplasmic or inner membrane and an outer membrane. These organisms have a thin peptidoglycan layer between the two membranes that does not retain the crystal violet stain used in the identification of bacteria by Gram staining (hence the name Gram-negative). The best known Gram-negative bacteria are the Proteobacteria, which include human pathogens such as E. coli, Salmonella enteric serovar Typhimurium, Helicobacter pylori and Legionella pneumophila. Other important Gram-negative phyla are the Bacteroidetes (major components of the human microbiome), Spirochetes (corkscrew-shaped bacteria), and Cyanobacteria (photosynthetic bacteria).
- Inner membrane (IM)
the membrane of Gram-negative bacteria that surrounds the cytoplasm.
- Lipoprotein
a protein that is modified in the periplasm by the attachment of lipids to an N-terminal cysteine residue and the N-terminal amino group. The lipid groups anchor the protein to a membrane.
- Numbered secretion pathways
a collection of specialized, structurally distinct secretion pathways numbered I-IX that deliver proteins into the extracellular space (or directly into other bacteria or eukaryotic cells). While these pathways are widely distributed in Protebacteria, some of them are also found in other Gram-negative phyla. In some pathways proteins are secreted across both membranes in one step (e.g., type III and IV pathways), while in other pathways proteins are secreted sequentially across the two cell membranes (e.g., type II and V pathways). The structure and function of the proteins secreted by each pathway vary considerably, but in general the proteins play important roles in nutrient acquisition, survival in specific environments, or virulence.
- Outer membrane (OM)
the outermost layer of the Gram-negative bacterial cell envelope. While phospholipids are the predominant lipid component of the IM and other biological membranes, in many organisms the outer leaflet of the OM contains a large amount of a complex glycolipid called lipopolysaccharide. Integral OM proteins lack the highly hydrophobic membrane-spanning segments that are hallmarks of IM proteins presumably because they must avoid retention in the IM and pass through the water-filled periplasm. Instead they contain amphipathic β strands that insert into the OM only when they fold into a barrel-like structure that exposes the hydrophobic residues on the outer surface.
- Periplasm
the space between the two membranes in Gram-negative bacteria. The periplasm contains a wide variety of nutrient binding proteins, enzymes, and molecular chaperones, but lacks ATP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hayashi S and Wu HC (1990) Lipoproteins in bacteria. J Bioenerg Biomembr 22, 451–471 [DOI] [PubMed] [Google Scholar]
- 2.LoVullo ED, et al. (2015) Revisiting the Gram-negative lipoprotein paradigm. J Bacteriol 197, 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu MM, et al. (2006) A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J Bacteriol 188, 2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun V (1975) Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 415, 335–377 [DOI] [PubMed] [Google Scholar]
- 5.Ichihara S, et al. (1981) Characterization of new membrane lipoproteins and their precursors of Escherichia coli. J Biol Chem 256, 3125–3129 [PubMed] [Google Scholar]
- 6.Okuda S and Tokuda H (2011) Lipoprotein sorting in bacteria. Annu Rev Microbiol 65, 239–259 [DOI] [PubMed] [Google Scholar]
- 7.Narita S and Tokuda H (2007) Amino acids at positions 3 and 4 determine the membrane specificity of Pseudomonas aeruginosa lipoproteins. J Biol Chem 282, 13372–13378 [DOI] [PubMed] [Google Scholar]
- 8.Masuda K, et al. (2002) Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc Natl Acad Sci U S A 99, 7390–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning PA, et al. (1980) Outer membrane of Escherichia coli: properties of the F sex factor traT protein which is involved in surface exclusion. J Bacteriol 142, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perumal NB and Minkley EG (1984) The product of the F sex factor traT surface exclusion gene is a lipoprotein. J Biol Chem 259, 5357–5360 [PubMed] [Google Scholar]
- 11.Chamberlain NR, et al. (1989) Major integral membrane protein immunogens of Treponema pallidum are proteolipids. Infect Immun 57, 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt ME, et al. (1990) Immunogenic integral membrane proteins of Borrelia burgdorferi are lipoproteins. Infect Immun 58, 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T, et al. (1994) Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11, 31–41 [DOI] [PubMed] [Google Scholar]
- 14.Pugsley AP, et al. (1986) Extracellular pullulanase of Klebsiella pneumoniae is a lipoprotein. J Bacteriol 166, 1083–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunikis J and Barbour AG (1999) Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect Immun 67, 2874–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zückert WR, et al. (2004) Cross-species surface display of functional spirochetal lipoproteins by recombinant Borrelia burgdorferi. Infect Immun 72, 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Hage N, et al. (2001) Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147, 821–830 [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. (2011) Molecular interactions that enable movement of the Lyme disease agent from the tick gut into the hemolymph. PLoS Pathog 7, e1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar M, et al. (2011) Borrelia burgdorferi BBA52 is a potential target for transmission blocking Lyme disease vaccine. Vaccine 29, 9012–9019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhi H, et al. (2015) The BBA33 lipoprotein binds collagen and impacts Borrelia burgdorferi pathogenesis. Mol Microbiol 96, 68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zückert WR (2014) Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond. Biochim Biophys Acta 1843, 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pizza M, et al. (2000) Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 [DOI] [PubMed] [Google Scholar]
- 23.Masignani V, et al. (2003) Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Ulsen P, et al. (2003) A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol Microbiol 50, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 25.Fletcher LD, et al. (2004) Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun 72, 2088–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leuzzi R, et al. (2005) Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol Microbiol 58, 669–681 [DOI] [PubMed] [Google Scholar]
- 27.Arenas J, et al. (2006) Locus NMB0035 codes for a 47-kDa surface-accessible conserved antigen in Neisseria. Int Microbiol 9, 273–280 [PubMed] [Google Scholar]
- 28.Pettersson A, et al. (2006) Vaccine potential of the Neisseria meningitidis lactoferrin-binding proteins LbpA and LbpB. Vaccine 24, 3545–3557 [DOI] [PubMed] [Google Scholar]
- 29.DeRocco AJ and Cornelissen CN (2007) Identification of transferrin-binding domains in TbpB expressed by Neisseria gonorrhoeae. Infect Immun 75, 3220–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu CA, et al. (2008) Immunoproteomic identification of the hypothetical protein NMB1468 as a novel lipoprotein ubiquitous in Neisseria meningitidis with vaccine potential. Proteomics 8, 2115–2125 [DOI] [PubMed] [Google Scholar]
- 31.Sardiñas G, et al. (2009) Assessment of vaccine potential of the Neisseria-specific protein NMB0938. Vaccine 27, 6910–6917 [DOI] [PubMed] [Google Scholar]
- 32.Madico G, et al. (2006) The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol 177, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serruto D, et al. (2010) Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci U S A 107, 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serruto D, et al. (2012) The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30 Suppl 2, B87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson JE, et al. (1994) Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J Bacteriol 176, 3162–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersson A, et al. (1998) Molecular characterization of LbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol Microbiol 27, 599–610 [DOI] [PubMed] [Google Scholar]
- 37.Roussel-Jazédé V, et al. (2013) Lipidation of the autotransporter NalP of Neisseria meningitidis is required for its function in the release of cell-surface-exposed proteins. Microbiology 159, 286–295 [DOI] [PubMed] [Google Scholar]
- 38.Braun V and Rehn K (1969) Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10, 426–438 [DOI] [PubMed] [Google Scholar]
- 39.Braun V and Bosch V (1972) Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur J Biochem 28, 51–69 [DOI] [PubMed] [Google Scholar]
- 40.Braun V and Wolff H (1970) The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur J Biochem 14, 387–391 [DOI] [PubMed] [Google Scholar]
- 41.Braun V and Sieglin U (1970) The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur J Biochem 13, 336–346 [DOI] [PubMed] [Google Scholar]
- 42.Cowles CE, et al. (2011) The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol Microbiol 79, 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel LV, et al. (2015) Dual Orientation of the Outer Membrane Lipoprotein Pal in Escherichia coli. Microbiology 161, 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy TF, et al. (1986) Identification of a 16,600-dalton outer membrane protein on nontypeable Haemophilus influenzae as a target for human serum bactericidal antibody. J Clin Invest 78, 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konovalova A, et al. (2014) Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc Natl Acad Sci U S A 111, E4350–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho SH, et al. (2014) Detecting envelope stress by monitoring β-barrel assembly. Cell 159, 1652–1664 [DOI] [PubMed] [Google Scholar]
- 47.Webb CT, et al. (2012) Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J Mol Biol 422, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong C, et al. (2006) Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature 444, 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goyal P, et al. (2014) Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shipman JA, et al. (2000) Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol 182, 5365–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phansopa C, et al. (2014) Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J 458, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsbrink J, et al. (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sijbrandi R, et al. (2008) Pbp, a cell-surface exposed plasminogen binding protein of Bacteroides fragilis. Microbes Infect 10, 514–521 [DOI] [PubMed] [Google Scholar]
- 54.Weinacht KG, et al. (2004) Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol Microbiol 53, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 55.Fletcher CM, et al. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galvão BP, et al. (2014) Identification of a collagen type I adhesin of Bacteroides fragilis. PLoS One 9, e91141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson MM, et al. (2015) Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One 10, e0117732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manfredi P, et al. (2011) The genome and surface proteome of Capnocytophaga canimorsus reveal a key role of glycan foraging systems in host glycoproteins deglycosylation. Mol Microbiol 81, 1050–1060 [DOI] [PubMed] [Google Scholar]
- 59.Noinaj N, et al. (2012) Structural basis for iron piracy by pathogenic Neisseria. Nature 483, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.d'Enfert C, et al. (1987) Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J 6, 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Odenbreit S, et al. (1999) Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol 31, 1537–1548 [DOI] [PubMed] [Google Scholar]
- 62.Coutte L, et al. (2003) Surface anchoring of bacterial subtilisin important for maturation function. Mol Microbiol 49, 529–539 [DOI] [PubMed] [Google Scholar]
- 63.Ashgar SS, et al. (2007) CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut. J Bacteriol 189, 1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ieva R and Bernstein HD (2009) Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A 106, 19120–19125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Junker M, et al. (2009) Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol 71, 1323–1332 [DOI] [PubMed] [Google Scholar]
- 66.Dunstan RA, et al. (2015) Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol Microbiol 97, 616–629 [DOI] [PubMed] [Google Scholar]
- 67.Chen S and Zückert WR (2011) Probing the Borrelia burgdorferi surface lipoprotein secretion pathway using a conditionally folding protein domain. J Bacteriol 193, 6724–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulze RJ, et al. (2010) Translocation of Borrelia burgdorferi surface lipoprotein OspA through the outer membrane requires an unfolded conformation and can initiate at the C-terminus. Mol Microbiol 76, 1266–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumru OS, et al. (2011) Surface localization determinants of Borrelia OspC/Vsp family lipoproteins. J Bacteriol 193, 2814–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schulze RJ and Zückert WR (2006) Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol 59, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 71.Lenhart TR and Akins DR (2010) Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol Microbiol 75, 692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamanaka H, et al. (2008) MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J Bacteriol 190, 7693–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Francetić O and Pugsley AP (2005) Towards the identification of type II secretion signals in a nonacylated variant of pullulanase from Klebsiella oxytoca. J Bacteriol 187, 7045–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ostberg KL, et al. (2013) Conserved regions of gonococcal TbpB are critical for surface exposure and transferrin iron utilization. Infect Immun 81, 3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poquet I, et al. (1993) Stable periplasmic secretion intermediate in the general secretory pathway of Escherichia coli. EMBO J 12, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.d'Enfert C, et al. (1987) Export and secretion of the lipoprotein pullulanase by Klebsiella pneumoniae. Mol Microbiol 1, 107–116 [DOI] [PubMed] [Google Scholar]
- 77.Keenan J, et al. (2000) Immune response to an 18-kilodalton outer membrane antigen identifies lipoprotein 20 as a Helicobacter pylori vaccine candidate. Infect Immun 68, 3337–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dashper SG, et al. (2000) Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis. J Bacteriol 182, 6456–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coutte L, et al. (2001) Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J 20, 5040–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin S, et al. (2001) JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol Microbiol 39, 1225–1236 [DOI] [PubMed] [Google Scholar]
- 81.Arambula D, et al. (2013) Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc Natl Acad Sci U S A 110, 8212–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pride AC, et al. (2013) The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. MBio 4, e00305–00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michel LV, et al. (2013) Dual orientation of the outer membrane lipoprotein P6 of nontypeable haemophilus influenzae. J Bacteriol 195, 3252–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fleury C, et al. (2014) Identification of a Haemophilus influenzae factor H-Binding lipoprotein involved in serum resistance. J Immunol 192, 5913–5923 [DOI] [PubMed] [Google Scholar]
- 85.Qin A, et al. (2014) FipB, an essential virulence factor of Francisella tularensis subsp. tularensis, has dual roles in disulfide bond formation. J Bacteriol 196, 3571–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W, et al. (2014) Identification of an outer membrane lipoprotein involved in nasopharyngeal colonization by Moraxella catarrhalis in an animal model. Infect Immun 82, 2287–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]


