Abstract
The C-terminal domains of holins are highly hydrophilic and contain clusters of consecutive basic and acidic residues, with the overall net charge predicted to be positive. The C-terminal domain of λ S was found to be cytoplasmic, as defined by protease accessibility in spheroplasts and inverted membrane vesicles. C-terminal nonsense mutations were constructed in S and found to be lysis proficient, as long as at least one basic residue is retained at the C terminus. In general, the normal intrinsic scheduling of S function is deranged, resulting in early lysis. However, the capacity of each truncated lytic allele for inhibition by the S107 inhibitor product of S is retained. The K97am allele, when incorporated into the phage context, confers a plaque-forming defect because its early lysis significantly reduces the burst size. Finally, a C-terminal frameshift mutation was isolated as a suppressor of the even more severe early lysis defect of the mutant SA52G, which causes lysis at or before the time when the first phage particle is assembled in the cell. This mutation scrambles the C-terminal sequence of S, resulting in a predicted net charge increase of +4, and retards lysis by about 30 min, thus permitting a viable quantity of progeny to accumulate. Thus, the C-terminal domain is not involved in the formation of the lethal membrane lesion nor in the “dual-start” regulation conserved in lambdoid holins. Instead, the C-terminal sequence defines a cytoplasmic regulatory domain which affects the timing of lysis. Comparison of the C-terminal sequences of within holin families suggests that these domains have little or no structure but act as reservoirs of charged residues that interact with the membrane to effect proper lysis timing.
Holins are small integral membrane proteins which are required for host lysis by most double-stranded DNA bacteriophage. The best-studied holin gene is the λ S gene, which spans only 107 codons and is the first gene in the sole λ late gene transcript (6, 41, 42). Endolysins are diverse soluble bacteriolytic enzymes which accumulate in the cytoplasm of the infected cell during the vegetative phase. Examples of endolysins are the λ gene R transglycosylase, the T7 gene 3.5 amidase, and the P22 gene 19 lysozyme, enzymes with unrelated mechanisms and different evolutionary lineages (2, 8, 11). The function of the holin is to cause the formation of membrane lesions (“holes”) which allow the escape of the soluble endolysin and the subsequent destruction of the cell wall. Equally important, the holin has an essential timing function which causes it to form the holes at a genetically programmed time. The holin is a “clock” which thus determines the length and productive burst size of the phage infective cycle (41).
Nothing is known about the structure of the putative hole or the mechanism of hole formation, topics which may be fruitfully approached now that purified holin protein is available (34). However, substantial genetic information has been obtained regarding the components of the timing function. The first three codons of λ S are Met1-Lys2-Met3-, and it has been established that translational initiations occur at both Met codons in vivo, generating two gene products, S107 and S105, that differ only in the first two residues. Remarkably, the two proteins have opposing functions, with S105 being the active holin and S107 acting as a holin inhibitor (6). Thus, the lysis clock is set, to a significant extent, by the proportion of initiation events at the two start codons (5, 10, 31). The operational difference between S107 and S105 is the cationic side chain at position two in S107, and putting a second basic residue between the two start codons makes the inhibitor form even more inhibitory (4, 28). The inhibitory capacity of S107 depends on the energized membrane (4), leading to the simplest model that the N-terminal positive charge of S107 interacts electrostatically with the predominantly anionic inner surface of the cytoplasmic membrane. Interpretation of these facts is hampered by the absence of firm topological information regarding the membrane topology of S, which, from inspection of its primary sequence, has three potential transmembrane domains (Fig. 1).
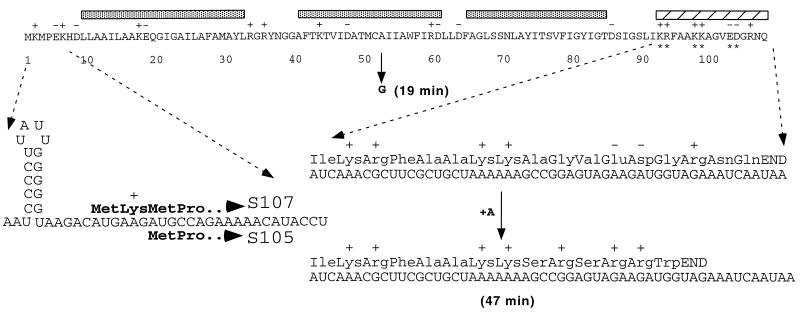
FIG. 1.
Features of the primary sequence of S. The predicted sequence of the λ S107 gene product is shown, with shaded bars over the three potential transmembrane domains and a hatched bar above the hydrophilic C-terminal domain characteristic of holins. Shown on the left is a blow-up of the amino-terminal sequences of the S107 and S105 products, including the corresponding mRNA sequence of the translational initiation region and the sdi stem-loop structure (31). Shown on the right is a blow-up of the C-terminal hydrophilic domain, including the corresponding mRNA sequence and, below that, the +1 frameshift mutation of λaj1, isolated as a suppressor of the plaque-forming defect of SA52G (also shown) (21). The time of lysis onset is shown in parentheses for both the SA52G early-lysis mutant and λaj1.
Many holin sequences have been reported since the holin-endolysin model was first formalized as a fundamental strategy for phage lysis (41, 42). Many holins apparently have the “dual-start” motif described above, although evidence for homologous function for the motif is available only for the holin genes of the lambdoid phage 21 and the B. subtilis phage φ29 (1, 7, 38). An even more universal characteristic is the existence of a C-terminal sequence which is rich in residues charged at physiological pH and usually with a significant net positive charge (41, 42) (Fig. 2). This makes it likely that the C terminus is localized to the cytoplasm, according to the von Heijne “positive inside” rule (39). Moreover, in many cases, there are multiple charged residues in a sequence, including 10 residues in a row in the N15 holin sequence. In addition, in several cases, there is, within an orthologous group of holins, at least one ortholog in which the C-terminal domain is completely dissimilar or in which the sequence similarity ends abruptly in the middle of the domain (Fig. 2; see P2 Y and its ortholog from phage 186, 21 S and its ortholog from φ80, and Hp1 Orf78 and its ortholog from the cryptic prophage of H. somnus). Assuming the C termini of holins have a common function, it is unlikely that such changes are compatible with a highly structured domain but rather that this domain may serve as a reservoir of charged residues with a function analogous to that of the N-terminal motif. If the C-terminal domain is highly charged and largely unstructured, we reasoned that it might be possible to effect truncations of the C terminus which left the lytic capacity of the S protein intact, although the timing function might be deranged. Here we report the results of a molecular and physiological analysis of the C-terminal domain of the λ holin. These results are discussed in terms of a model for holin function and regulation.
FIG. 2.
The C-terminal hydrophilic domain of holin proteins from bacteriophages of gram-negative bacteria. The C-terminal sequence of the first member identified from each of the orthologous groups of holins is shown. Holin sequences are grouped into class I (three potential transmembrane domains) and class II (two potential transmembrane domains). Each holin is given with the total number of residues for the longest form of the predicted protein product. Class I orthologous groups: λ S (four orthologs, including P22 gp13), N15 (no orthologs), P2 Y (one ortholog, 186 Orf24), P1 LydA (no orthologs), and PRD1 OrfM (no orthologs). Class II orthologous groups: 21 S (six orthologs, including φ80 S); Hp1 Orf78 (one ortholog, Haemophilus somnus cryptic holin), T7 gp17.5 1 ortholog, and T3 Lys. If an ortholog has two or fewer residues changed, the changes are shown below the representative sequence. More-divergent termini within the orthologous group are shown in their entirety. Acidic and basic residues are shown in capital letters, and two or more consecutive charged residues are underlined. The H. somnus sequence comes from a cryptic prophage (29). A compilation of holin sequences can be found in Bläsi and Young (42) and in Young (41). The phage N15 holin sequence was obtained from R. Hendrix (19). Also shown are the C-terminal sequences of the predicted products of modified S alleles where a nucleotide sequence coding for an oligohistidine tag was inserted correctly (Sτ94) and incorrectly (Sτ94x, Sτ94z) after codon 94 (35).
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, and growth conditions.
The Escherichia coli strains, bacteriophages, and plasmids used in this study are listed in Table 1. Standard bacterial broth media, plates, and top agar (Luria-Bertani [LB] and tryptone broth [TB]) were used (26). Unless indicated otherwise, bacterial cultures were grown in LB broth medium supplemented with ampicillin (Ap) (100 mg/ml; Sigma) to maintain selection of plasmids. For some plating assays, TB was also used as indicated. Growth and lysis were monitored by measuring the optical density at either 550 or 600 nm, as indicated, by using a Gilford Stasar III sipping spectrophotometer.
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain, plasmid, or bacteriophage | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | araD139 Δ(argF-lac)U169 thiA rpsL150 relA1 deoC1 ptsF25 rbsR flbB5301 | 33 |
| MC4100F′ | MC4100/F′ proAB lacIqlacZΔM15 Tn10 (Tcr) | 36 |
| MS59 | MC4100 supF | M. Syvanen collection |
| JM103 | Δ(lac-proAB) supE hsdR rpsL endA thi sbcBC/F′ traD36 proAB lacIqlacZΔM15 | 25 |
| CJ236 | dut1 ung1 thi recA pCJ105 (Cmr) | 23 |
| MH760λ | MC4100 ompR472 (λ) (thermostable λ lysogen) | 33 |
| Plasmids | ||
| pBH20 | pBR322 with wild-type lacPO (Apr) | 20 |
| pACS107 | P15A ori; S107RRz cloned under lacPO control | 4 |
| pACSam | Same as pACS107 but with Sam7 null allele | 3 |
| pRG1 | pBH20 with SRRz cloned under lacPO control | 30 |
| pBS112 | S107 on high-copy-number vector with the translational initiation signals of phage T7 gene 10 | 4 |
| pPF101 | pRG1 SK92am | This study |
| pPF102 | pRG1 SR93am | This study |
| pPF110 | pRG1 S105 | This study |
| pPF113 | pPF110 SK97am | This study |
| pPF114 | pPF110 SK98am | This study |
| pPF115 | pPF110 SE102am | This study |
| pPF116 | pPF110 SD103am | This study |
| Bacteriophages | ||
| λCE6 | Sam7 cI857 int::(T7 gene 1) | 37 |
| M13SRRz | M13mp18 (carrying the λ lysis cassette SRRz) | 4 |
| M13S105RRz | M13mp18 (carrying S105RRz) | 3 |
| λkan | λ b519 b515 Tn903 cI857 nin5 | 21 |
| λkan Δ(SR) | λ b519 b515 Tn903 cI857 nin5 Δ(SR) | 31 |
| λbj1 | λkanSA52G (early lysis, non-plaque forming) | 21 |
| λbj1 | Plaque-forming pseudorevertant of λbj1 | This study |
| λS105 | λkan SM1L (start codon for S107 inactivated; produces only S105) | 31 |
λ methods.
Plating of phage λ was done by dilution with λdil (5 mM MgSO4, 0.01% gelatin, 10 mM Tris-HCl [pH 7.4]) and mixing 0.1 ml of the dilution with 0.1 ml of a fresh overnight of MC4100 (nonsuppressing lawn) or MS59 (suppressing lawn), grown either in TB or LB as indicated. This was followed by incubation at room temperature for 30 min and mixing with 3 ml of molten top LB or TB agar; the mixtures were then poured onto fresh LB or TB plates. The plates were incubated for at least 7 h at 37°C before they were scored for plaques. Lysogens were selected by infecting saturated mixtures that had been incubated overnight with a lysate or plaque resuspended in λdil, streaking onto a LB-kanamycin (Km) plate, and incubation overnight at 30°C. The passive lysogeny assay was done as previously described (21). Briefly, lysates of non-plaque-forming bacteriophage carrying a Km resistance marker are incubated with the lysogen MH760λ for 1 h and then plated at various dilutions on LB-Km plates. Colonies arising after overnight incubation at 37°C are counted and compared with counts obtained with a known titer of a plaque-forming bacteriophage carrying the same drug resistance marker.
Selection of plaque-forming pseudorevertant of SA52G mutant.
A lysate of λbj1, which carries the SA52G allele and has a severe defect in plaque formation (Table 1) (21), was plated on an MC4100 lawn. Small plaques were picked, purified by replating, and then picked again with a Pasteur pipet into λdil. After vortex mixing with a drop of CHCl3, the suspension was used to lysogenize MC4100 on LB-Km as described previously (21). Individual lysogens were purified and tested for lysis kinetics after thermal induction as described previously (21).
Site-directed mutagenesis and cloning of the S mutant alleles.
Amber (UAG) nonsense codons were individually introduced by replacing C-terminal codons specifying charged amino acids starting at codon position 92 (see Fig. 1). The mutant sequences were generated by oligonucleotide site-directed mutagenesis according to the method of Kunkel et al. (23) by using the single-stranded DNA templates M13SRRz and M13S105RRz prepared from the dut ung strain CJ236. For clarity, S+ alleles are defined as those in which the normal translational start is used (i.e., so that both S107 and S105 proteins are produced), whereas S105 and S107 refer to S alleles in which a CUG codon replaces the start codon at position 1 and position 3, respectively, and thus only the indicated S gene product is produced. All oligonucleotides used as mutagenic primers were complementary to the S coding sequence, except for the underlined mismatches. The mutations generated are given in parentheses: 5′-GAAGCGCTAGATAAGC-3′ (Lys92am), 5′-GCAGCGAACTATTTGATAA-3′ (Arg93am), 5′-CTCCGGCTTTCTAAGCAGCG-3′ (Lys97am), 5′-CTACTCCGGCCTATTTAGCAGC-3′ (Lys98am), 5′-GATTTCTACCATCCTATACTCCGG-3′ (Glu102am), and 5′-GATTTCTACCCTATTCTACTC-3′ (Asp103am).
The λ S105RRz cassette was reisolated on an EcoRI/HindIII fragment from M13S105RRz DNA and cloned into the corresponding sites downstream of the lacPO promoter-operator region in pBH20, which resulted in plasmid pPF110. After the mutations were verified by sequencing, the respective S alleles were isolated together with the accessory lysis genes R and Rz on an EcoRI/HindIII fragment and cloned into the EcoRI/HindIII restriction sites of the cloning ector pBH20 (Table 1). Plasmids pPF101 and pPF102 carry the amber mutations at positions K92 and R93, respectively, in S. Cloning of the mutant S105 alleles into vector pBH20 resulted in plasmids pPF113 (K97am), pPF114 (K98am), pPF115 (E102am), and pPF116 (D103am). Transfer of the K92am, K97am, E102am, and D103am mutant alleles to λ by recombination and the determinations of the burst size were done out as previously described (31).
Determination of membrane topology.
E. coli MC4100(pBS112), carrying the S107 allele (5) was grown in M9 maltose medium for λCE6 infection as described previously (9), except that rifampin (100 μg/ml) was added at 15 min after infection. A 10-ml volume of culture was pulse-labelled with 200 μCi of [35S]methionine (>1,000 Ci/mmol; NEN) for 5 min, beginning at 25 min after infection. Preparation of inverted membrane vesicles (IMV) and spheroplasts and the proteinase K digestions were done as described elsewhere (15, 22, 27), with the following modifications. For spheroplast samples, 4 ml of labelled culture was divided into 1-ml aliquots and placed in four microfuge tubes. After the cells were pelleted by centrifugation for 1 min, the pellet was resuspended in 100 μl of spheroplasting buffer (40% sucrose, 20 mM EDTA, 60 mM Tris-HCl [pH 8]) and treated with lysozyme (10 μg/ml; Sigma) at 37°C. One sample was treated with 2% Triton X-100 to lyse the cells prior to proteinase K treatment. Proteinase K digestion was halted with 2 mM phenylmethylsulfonyl fluoride (Sigma). After proteinase K treatment, spheroplasts were collected by pelleting in a microfuge and then resuspended in 25 μl of 1% sodium dodecyl sulfate (SDS)–1 mM EDTA for 40 s before dilution to a final volume of 500 μl with TSET (150 mM NaCl, 1 mM EDTA, 2% Triton X-100, 50 mM Tris-HCl [pH 8.0]) for immunoprecipitation. For IMV samples, the remaining 6 ml of labelled culture was pelleted as described above and resuspended in 1 ml of French press buffer (20 mM EDTA, 60 mM Tris-HCl [pH 8]). The suspension was disrupted by passage through a French pressure cell (SLM-Aminco) at 16,000 lb/in2, generating the IMV sample, which was freed of unlysed cells by centrifugation in a microfuge tube at 3,000 × g for 1 min. The resulting IMV sample was divided into aliquots and placed in 6 microfuge tubes and then treated with proteinase K as described above for different time intervals. One sample was lysed with detergent prior to digestion, as described above. After the digestion was stopped with 2 mM phenylmethylsulfonyl fluoride, the IMV were harvested by centrifugation at 100,000 × g as described earlier (22). The cytoplasmic membrane protein fraction was obtained by resuspending the membrane pellet in 200 μl of membrane extraction buffer (10 mM Tris-HCl [pH 8], 35 mM MgCl2, 1% Triton X-100), extraction with stirring for 1 h, and removal of the insoluble debris by recentrifugation at 100,000 × g. The extract (ca. 50 μl) was diluted with 450 μl of TSET buffer for immunoprecipitation.
Immunoprecipitation was carried out with antibody raised against a synthetic peptide corresponding to the C-terminal 16 residues of S, as described previously (10). In vitro translation of S mRNA and autoradiography was performed as described previously (9).
RESULTS
C-terminal domain of the λ holin is disposed in the cytoplasm.
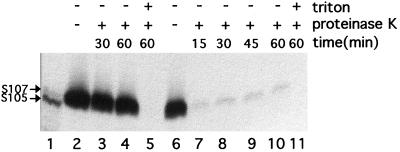
As a first step in determining the membrane topology of the S holin, the localization of the C-terminal domain was investigated by protease accessibility analysis in inverted membrane vesicles and in spheroplasts. To maximize labelling of the S protein and to reduce the background, the labelling was done with cells carrying plasmids in which the S reading frame is transcribed from a T7 RNA polymerase promoter and fused to the efficient ribosome binding site of T7 gene 10. S protein has been shown to accumulate in a fully functional form in the cytoplasmic membrane under these conditions (34). Initial experiments were done with the S107 allele because expression of the S+ allele is incompatible with efficient spheroplasting (32). The results show that the C-terminal domain of the S107 protein is susceptible to exogenous protease in inverted membrane vesicles but not in spheroplasts (Fig. 3). The faint protease-resistant band presumably corresponds to the small fraction of inner membrane vesicles which are not inverted. Although spheroplasts could not be reproducibly prepared, similar results were obtained with protease treatment of inner membrane vesicles using the S+ allele (not shown). The simplest interpretation is that, as expected from sequence analysis of the S gene, the highly charged C-terminal domain is cytoplasmic. Although in any model for S topology at least one loop between transmembrane domains should be accessible from the periplasmic face, the fact that the S protein is completely protected in spheroplasts indicates that the loop is not sufficiently exposed to be accessible to the protease. (The accessibility of the periplasmic face of the spheroplasts prepared by this technique was verified by determining the proteolysis products of labelled Tsr protein [not shown]).
FIG. 3.
The C-terminal domain of the S protein is accessible to proteases in inverted membrane vesicles. E. coli MC4100(pBS112) cells were infected with λCE6 and labelled with [35S]methionine at 25 min after infection. Spheroplasts and IMV, treated with proteinase K, with or without detergent, for various times as indicated. Each sample was subjected to immunoprecipitation with anti-S antibody (9), and the immunoprecipitate was analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. Lanes: 1, in vitro translation products of S+ mRNA, as described previously (9); 2 to 5, spheroplast samples; 6 to 11, IMV samples.
Effect of C-terminal amber mutations on lysis and its timing.
To test the hypothesis that some or all of the C-terminal domain might be dispensable for lysis, nonsense mutations in a number of distal codons of the S gene were created by site-directed mutagenesis. The different S alleles were cloned as SRRz lysis gene “cassettes” under lacPO control on a moderate-copy-number plasmid, resulting in the pPF series of plasmids (Table 1). Remarkably, all of the S amber mutant alleles truncated at codon 93 or beyond were lysis proficient, confirming that most of the C-terminal domain of S is unnecessary for “hole formation” (Table 2). In addition, all of the lysis-proficient alleles shared with the parental allele the property of premature triggering by the addition of an energy poison, indicating that the intrinsic components of lysis timing were retained in these mutants. However, induction of the K92am allele does not result in lysis. Addition of chloroform, which permeabilizes the inner membrane, results in the immediate loss of turbidity, which shows that the lysis defect is not due to a deficiency in endolysin activity (not shown). We conclude that the K92am is an absolute-lysis-defective allele. The K92am allele is recessive, as determined by its lack of an effect on lysis kinetics when expressed in trans to the S+ allele borne on a compatible plasmid (not shown). It is unclear whether the defect is due to the inability of the mutant protein to accumulate in the membrane, because the available anti-S antibody was raised against an oligopeptide corresponding to the C-terminal sequence, which prevents detection of any of the C-terminal amber mutants in immunoblots (13). Nevertheless, the recessive character of the K92am lysis defect is consistent with the idea that at least one positively charged residue at the C-terminal end is required for insertion in the membrane or the attainment of proper topology.
TABLE 2.
Lysis phenotypes of C-terminal S mutants
| S allele | C-terminal sequence | Net charge | Lysis onset (min) for allelea:
|
Lysis rateb | Pre-mature lysisc | |
|---|---|---|---|---|---|---|
| S | S105 | |||||
| ++ ++ −− + | ||||||
| Wild type | ..LIKRFAAKKAGVEDGRNQ | +3 | 35 | 28 | Fast | + |
| 90 107 | ||||||
| ++ ++ − | ||||||
| D103am | ..LIKRFAAKKAGVE | +3 | >45 | Slow | + | |
| ++ ++ | ||||||
| E102am | ..LIKRFAAKKAGV | +4 | 40 | Slow | + | |
| ++ + | ||||||
| K98am | ..LIKRFAAK | +3 | 24 | Fast | + | |
| ++ | ||||||
| K97am | ..LIKRFAA | +2 | 22 | Fast | + | |
| + | ||||||
| R93am | ..LIK | +1 | 21 | Fast | + | |
| K92am | ..LI | 0 | No | No | None | − |
The S allele has both wild-type translational start codons (M1, M3). The S105 allele has the M1L translational start codon (no start codon for the S107 reading frame).
Rate of lysis after onset. Fast, A500 decreases by ≥50% in 5 min; slow, A500 decreases by <50% in 15 min.
Lysis observed after the addition of 10 mM KCN at 5 min before the normal lysis time.
As detailed above, the N terminus of the S107 gene product of S determines its inhibitor function. We wanted to test if the C terminus, although largely dispensable for lysis, was also required for the ability of S107 to modulate lysis time. When the S alleles were expressed from a moderate-copy-number plasmid in trans to an S107 allele (which produces only the S107 product) on a compatible plasmid, the lysis times for all of the C-terminal truncations were normally retarded, relative to the retardation seen for full-length S (Table 3). Thus, the C-terminal domain does not seem to be required for intermolecular interactions, at least not for those involved in the inhibitor function.
TABLE 3.
Effect of endolysin or S107 inhibitor supplied in trans
| C-terminal sequence | Allelea | Lysis onset (min) with pACSamRb | Lysis ratec | Lysis onset (min) with pACS107Rd |
|---|---|---|---|---|
| Wild type | S105 | 30 | Fast | 47 |
| D103am | S105 | 34 | Fast | 41 |
| E102am | S105 | 32 | Fast | 40 |
| K98am | S105 | 25 | Fast | 29 |
| K97am | S105 | 23 | Fast | 36 |
| R93am | Wild type | 21 | Fast | 26 |
S105, M1 start codon was altered to CUG (Leu) codon; wild type, both M1 and M3 start codons were present.
Time of onset of lysis, after induction, in the presence of compatible plasmid pACSamR.
Same as Table 2, footnote b.
Time of onset of lysis, after induction, in the presence of compatible plasmid pACS107R.
For the truncations at codon 93 or beyond, there is a general correlation between the length of the residual C-terminal domain and the timing of lysis (Table 2). The two shortest truncations which were lysis proficient both lysed earlier than the wild-type allele (Table 2). However, in two cases, the D103am and E102am, the onset of lysis was followed by a very gradual loss of turbidity. Moreover, even the addition of CHCl3 did not accelerate lysis dramatically, indicating a reduced level of R expression (not shown). It seemed likely that the placement of the nonsense codons in these alleles were disrupting translation of the R gene and thus disturbing the stability of the polycistronic mRNA produced from the cloned lysis gene cassette in the lacPO plasmid vector. Inspection of the sequence shows that the D103am mutation eliminates the normal start codon of R, and thus the residual bacteriolytic activity must come from downstream translational start sites (Fig. 4). The E102am mutation does not directly affect the Shine-Dalgarno sequence or the start codon, but the intervening nucleotide sequence is altered. Thus, the lack of endolysin activity here, too, is likely to be due to a translational defect. In both cases, supplying the R gene product in trans from a compatible plasmid resulted in more-saltatory lysis profiles, although the timing of lysis onset was not altered (Table 3).
FIG. 4.
Sequence of S-R intergenic region. The sequence of the S-R intergenic region is shown, with the corresponding C-terminal sequences of S and N-terminal sequences of R. Also shown are the sequences of the D103am and E102am mutants. In each case, the presumptive Shine-Dalgarno sequence for R is underlined and both the wild-type start codon and the first downstream alternative start codon for R are shown in boldface. Asterisks denote the sequence alterations resulting from the creation of the corresponding amber codons in the S reading frame.
Characterization of λSam recombinants.
To achieve a more robust expression system, the nonsense alleles were recombined into the normal phage context, where S is under its cognate transcriptional controls and where the Q-mediated transcription is much less susceptible to polarity effects (12). The standard λ vector used for recombining the plasmid S alleles is λkan, which carries deletions of three nonessential regions of the phage, a Tn903 insertion to provide Kmr for selecting lysogens and a deletion of the S and R genes (31). Recombination with the plasmids results in transfer of the S and R alleles to the phage context. Thermal induction of the lysogen by brief aeration at 42°C allows synchronous initiation of the phage vegetative cycle throughout the culture and thus allows rather precise assessment of the lysis kinetics associated with each S allele by monitoring the A550 during growth under standard conditions (21, 31). Moreover, plating efficiency and plaque size could be assessed as measures of the biological function of the S allele. Figure 5 shows a representative induction experiment which shows that the K92am allele is lysis defective in the phage context. In addition, lysis can be accomplished in the induced K92am lysogen by adding CHCl3 but not cyanide, indicating that the defect is due to an absence of functional S protein and not to the production of endolysin (not shown). Three other truncation alleles are lysis proficient, although the lysis profile of D103am is significantly more gradual, distinctly different from the sharply defined, saltatory lysis profiles exhibited by the parental and K97am alleles. The E102 allele exhibited variable lysis kinetics, although it was always triggered somewhat earlier than the parental allele (Fig. 5). In addition, the K97am allele, which had the shortest lysis-proficient C-terminal domain of the alleles crossed into the phage context, exhibited onset of lysis much earlier than the parental allele. The plating characteristics of these S mutants were also different. As expected, PFU were detected in the lysate from the nonlysing alleles at a level consistent with spontaneous reversion (Table 4). PFU were produced at about equal efficiency for the parental and E102am alleles, and at a slightly reduced level, with significantly smaller plaque size, for the D103am allele. In contrast, the early lysis allele K97am exhibited a pronounced plating defect, with reduced numbers of pinpoint plaques under optimal plating conditions and with no plaques under more-stringent conditions (Table 4). On a suppressor lawn, all of the mutant lysates generated normal numbers of plaques. To determine plating efficiency, the number of Kmr transducing particles was assessed by a passive recombination assay, where cells with a resident prophage are infected with each lysate and plated on antibiotic medium. This assay revealed that the burst size in the K97am allele was reduced to about 20% that of the parental allele, presumably because the early onset of lysis aborts the vegetative phase before its most productive period.
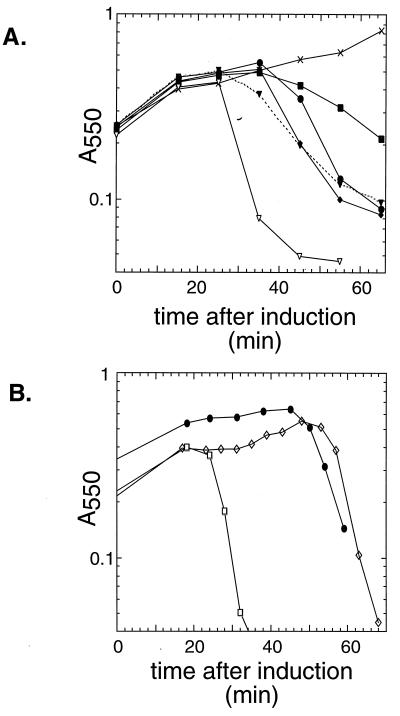
FIG. 5.
Lysis profiles for induced lysogens with C-terminal alterations in S. (A) Lysogens of λb515 b519 Tn903 cI857 nin5 S105 carrying S alleles with different C-terminal amber mutations were thermally induced in logarithmic phase and monitored for lysis by measuring the A550. Representative lysis profiles are shown for all alleles, including two profiles showing the variability of the K97am lysis kinetics. Symbols: ●, parental (no truncation); ■, D103am; ⧫, E102am; ▿ and ▾, K97am; ×, K92am. (B) Lysogens of λkan (●) and the isogenic phages λbj1 (SA52G [□]) and λaj1 (plaque-forming revertant of λbj1 [◊]) were thermally induced and monitored as in panel A.
TABLE 4.
Plating phenotypes of λSam recombinants
| S allele | Plating efficiencya | Plaque morphologyb | Relative burst sizec |
|---|---|---|---|
| S105 | 1 | Large | 1.0 |
| S105 D103am | 1 | Tiny | 1.0 |
| S105 E102am | 1 | Large | 1.0 |
| S105 K97am | 10−4 | Tiny | 0.2 |
| S105 K92am | 10−7 |
PFU per milliliter, as determined by titering on MC4100, divided by particles per milliliter, as determined by passive lysogeny assay. The result was normalized to the value for the S105 parental allele as 1.0.
Large, ≥2 mm; tiny, ≤0.2 mm.
Number of particles produced per cell after thermal induction of MC4100 lysogen; the value was normalized to the value for the S105 parental as 1.0.
C-terminal frameshift mutation suppresses a severe early lysis defect.
A similar but more extreme plating defect was found for the allele A52G due to a more exacerbated early lysis time, 19 min after induction, before the first progeny virion has been assembled intracellularly in the average induced cell (21). This mutant has an absolute plating defect under stringent plating conditions. Pseudorevertants were isolated as small plaque-formers under these conditions and were shown to have regained an extended vegetative period (Fig. 5B). Sequence analysis of a pseudorevertant, λaj1, revealed that the suppressing mutation was a frameshift within the 97th codon of S. The new reading frame specifies a C-terminal sequence which, although shorter and completely unrelated to the original S sequence, has a predicted net increase of four positive charges (Fig. 1). Thus, a C-terminal change resulting in a change in the electrostatic charge of the C terminus, without conservation of sequence, can compensate for an alteration in the intrinsic timing of the holin. Consistent with this finding, other S distal frameshifts, created by errors in the process of inserting oligohistidine tags into sites in the C-terminal domain (35), were found to retain normal lysis timing. In these two alleles, the nonhomologous sequence begins after position 94 and generates C-terminal sequences unrelated to the parental allele but with net predicted charges similar to that of the parental allele (Fig. 2). In both cases, the S alleles are fully functional, with similar or even accelerated lysis kinetics (35). We conclude that the sequence of the C-terminal domain does not serve a specific functional role in the ability of S to mediate hole formation but instead serves as a reservoir of charge which serves to regulate the timing of lysis.
DISCUSSION
Implications for holin topology.
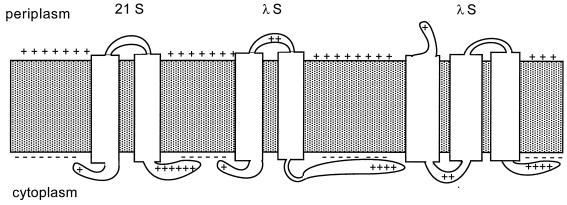
We have demonstrated that the C-terminal domain of S is located in the cytoplasm. While this result is not unexpected since its sequence is so rich in acidic and basic residues, it does reduce the number of reasonable models for the transmembrane topology of S. As noted above, the S sequence has three putative transmembrane domains, although the putative TM3 domain is rather rich in hydroxylated residues and is not recognized by available computer algorithms as likely to be membrane embedded (40). Requiring the C-terminal domain to be cytoplasmically disposed results in opposite locations for the N terminus, depending on whether two or three transmembrane domains are present (Fig. 6, center and right models). The most successful primary sequence tool for predicting membrane topology is defined by the von Heijne rules, which state that the cytoplasmic disposition of positively charged residues immediately flanking each transmembrane domain will be maximized (14, 39). By this criterion, a model with three transmembrane domains is favored (Fig. 6, right), not only for S but also for the other nonorthologous class I holins identified in phages of gram-negative hosts (P1 LydA, P2 Y, N15 holin, and PRD1 OrfM). However, there are numerous class II holins which have shorter primary sequences which, by inspection, clearly can have only have two transmembrane domains (42). These holins, at least one of which has already been shown to complement λ S efficiently (7), also have highly charged C-terminal domains, which presumably are also located in the cytoplasm (Fig. 2 and 6). The membrane topology of class I holins has been elusive (6). If all three potential transmembrane domains are in the bilayer and since the C terminus was found to be in the cytoplasm, then the N terminus should be on the periplasmic side of the membrane. Chemical modification studies of S in isolated membrane vesicles support the existence of the third transmembrane domain (18). Moreover, recent experiments have shown that a secretory signal sequence fused to the N terminus of S facilitates S hole formation in a signal peptidase-dependent manner, strongly supporting the notion that S function requires its N terminus of S to cross the membrane at some point (17). Thus, according to this view, the positive charge on the N terminus, which confers inhibitor function on S107, does so by retarding this translocation (6). Interestingly, the dual-start motif also functions as a regulatory feature in the class II holins, where translocation of the N terminus to the periplasm is, by inspection, highly unlikely (1).
FIG. 6.
Possible membrane topologies of S. Depicted are putative membrane topologies for prototype class II (S from phage 21; left) and class I (S from phage λ; middle and right) holin proteins. The inner membrane is shown as shaded area with positively charged periplasmic and negatively charged cytoplasmic surfaces. Transmembrane helical domains are represented as white rectangles spanning the membrane. Basic residues in putative solvent-exposed domains are shown for both 21 and λ holins.
Regulatory, but not functional role, for the C terminus.
In addition, we have shown that most of the C-terminal charged domain of S is not required for the ability to form the inner membrane lesions (“holes”) necessary for the R endolysin to gain access to the cell wall and effect lysis. Only one positively charged residue, at codon 92, is required for lytic function. The positively charged residue may be required for the ability of S to integrate in the membrane or, as demonstrated for derivatives of the Lep protein, to define the orientation of the adjacent transmembrane domain (14). Moreover, the timing of lysis is loosely correlated with the number of positively charged residues or the total net positive charge in the C terminus. This conclusion stems from the lysis kinetics exhibited by the C-terminal amber mutants. Additional evidence is provided by the fact that the frameshift mutation isolated as an intragenic suppressor of an early lysis mutation has a scrambled sequence beyond codon 97. In the frameshift mutant which suppresses A52G, the C-terminal nine residues, containing one basic and two acidic residues, are replaced with a different six-residue sequence containing three basic residues. Apparently, this change of +4 positive charges acts to retard the intrinsic tendency of the A52G mutation to trigger hole formation early. Similar frameshifts were fortuitously found during oligohistidine tag mutagenesis of the S sequence (35). The insertion frameshifts also scramble the C-terminal sequence but result in predicted charges of either +3 or +4, and the lysis onset is either as fast or even faster than with the parental S. Moreover, in a selection for S genes with reduced lethality, Raab et al. (30, 31) isolated an S missense mutant, E102K, a change which increases the net positive charge of the C-terminal domain by two and results in a 19-min delay in lysis onset. Interestingly, the E102K allele is recessive, indicating that the timing delay associated with the C-terminal charged domain is a cis effect. Thus, the primary function of the C-terminal domain appears not to be as a structural element in the lethal “hole” but instead in the proper scheduling of the hole-forming event, with an increasing positive charge being directly correlated to a slowing of the onset of lysis. This does not imply that the C-terminal domain is unnecessary for the biological function of the S protein. It should be noted that the K97am allele is also formally a Sam defective allele, like the Sam7 null allele (16), despite the fact that it supports complete lysis of the host. This illustrates that the timing and hole-forming functions of the S gene are both essential to λ.
Lytic function defined by the hydrophobic core.
A similar regulatory role has been suggested for the N-terminal sequence that precedes the first transmembrane domain (6, 36). To a significant extent, the lysis “clock” is set by the relative proportion of S105 and S107 products. In effect, this proportion specifies the average charge on the N-terminal side chains of the population of S molecules in the membrane, and the delay in lysis needed for accumulation of progeny virions appears to be a direct function of the positive charge on the N terminus (4, 28). Moreover, as an adventitious result of PCR-mediated site-directed mutagenesis, mutants have been isolated with additional basic residues in the first 10 positions, and these alleles exhibit negative-dominant lysis delay phenotypes (35). This suggests that the N-terminal domain primarily acts to regulate lysis timing, again with an increasing positive charge being correlated with the retardation of lysis onset. Thus, both N- and C-terminal domains appear to function primarily as inhibitory domains which primarily regulate by electrostatic means. In other words, the intrinsic hole-forming capacity of the S protein appears to be specified by its hydrophobic core (6). This in turn suggests that the protein-protein interactions which determine the specificity of the oligomeric hole structure are probably within the membrane-embedded sequences. This notion is consistent with the fact that the S107 inhibitor can still exert its inhibitory function on the products of the C-terminal truncations (see Table 3). Moreover, the great preponderance of missense mutations which inactivate the lethal capacity of S cluster in the first two transmembrane domains (30, 31). The fact that many of these mutations are relatively subtle and do not reduce the hydrophobic character (i.e., A48V, M50I, A52V, and L25F) suggests that an intimate interaction between transmembrane domains is essential for hole formation. Similar conclusions have been given a biochemical and structural basis by Engelman and coworkers, who have shown that altering the bulk of residues in the transmembrane domain of glycophorin can abolish the ability of that transmembrane domain to form dimers (24). Biochemical analysis of S may be accelerated by the finding that both N- and C-terminal extrema are unnecessary for hole formation.
ACKNOWLEDGMENTS
We thank other members of the two laboratories for their help and constructive criticism. Amanda Jochen is specifically acknowledged for isolating the aj1 pseudorevertant. We also appreciate, as always, the patient clerical services of Sharyll Pressley.
Work in the Young laboratory is supported by funding from project Public Health Service grant GM27099 and by funds from the College of Agriculture, Texas A&M University, and the Texas Agricultural Experiment Station. Work in the Bläsi laboratory is supported by grant P12220MOB from the Austrian Science Foundation and grant 6170 from the Austrian National Bank.
REFERENCES
- 1.Barenboim, M., C.-Y. Chang, F. dib Hajj, and R. Young. A holin and its inhibitor are both encoded by the phage 21 S gene. Mol. Microbiol., in press.
- 2.Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage λ is the murein transglycosylase. Mol Gen Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- 3.Bläsi, U., C.-Y. Chang, and R. Young. Unpublished data.
- 4.Bläsi U, Chang C-Y, Zagotta M T, Nam K, Young R. The lethal λ S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bläsi U, Nam K, Hartz D, Gold L, Young R. Dual translational initiation sites control function of the λ S gene. EMBO J. 1989;8:3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonovich M T, Young R. Dual start motif in two lambdoid S genes unrelated to λ S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens S R, Eppler K, Parr R, Poteete A R. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology. 1989;171:588–598. doi: 10.1016/0042-6822(89)90628-4. [DOI] [PubMed] [Google Scholar]
- 9.Chang C-Y, Nam K, Bläsi U, Young R. Synthesis of two bacteriophage λ S proteins in an in vivo system. Gene. 1993;133:9–16. doi: 10.1016/0378-1119(93)90218-r. [DOI] [PubMed] [Google Scholar]
- 10.Chang C-Y, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng X, Zhang X, Pflugrath J W, Studier F W. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc Natl Acad Sci USA. 1994;91:4034–4038. doi: 10.1073/pnas.91.9.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forbes D, Herskowitz I. Polarity suppression by the Q gene product of phage lambda. J Mol Biol. 1982;160:549–569. doi: 10.1016/0022-2836(82)90314-x. [DOI] [PubMed] [Google Scholar]
- 13.Fraisl, P., and R. Young. Unpublished data.
- 14.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 15.Geller B L. Electrochemical potential releases a membrane-bound secretion intermediate of maltose-binding protein in Escherichia coli. J Bacteriol. 1990;172:4870–4876. doi: 10.1128/jb.172.9.4870-4876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldberg A R, Howe M. New mutations in the S cistron of bacteriophage lambda affecting host cell lysis. Virology. 1969;38:200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- 17.Graschopf, A., and U. Bläsi. Molecular function of the dual-start motif in the lambda S holin. Submitted for publication. [DOI] [PubMed]
- 18.Gründling, A., and R. Young. Unpublished data.
- 19.Hendrix, R. 1992. Personal communication.
- 20.Itakura K, Hirose T, Crea R, Riggs A, Heyneker H L, Boliver F, Boyer H W. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1979;198:1056–1063. doi: 10.1126/science.412251. [DOI] [PubMed] [Google Scholar]
- 21.Johnson-Boaz R, Chang C-Y, Young R. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol Microbiol. 1994;13:495–504. doi: 10.1111/j.1365-2958.1994.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn A, Zhu H-Y, Dalbey R E. Efficient translocation of positively charged residues of M13 procoat protein across the membrane excludes electrophoresis as the primary force for membrane insertion. EMBO J. 1990;9:2385–2389. doi: 10.1002/j.1460-2075.1990.tb07413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Lemmon M A, Flanagan J M, Treutlein H R, Zhang J, Engelman D M. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 25.Messing J, Crea R, Seeburg P H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 27.Müller M, Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nam K, Bläsi U, Zagotta M T, Young R. Conservation of a dual-start motif in P22 lysis gene regulation. J Bacteriol. 1990;72:204–211. doi: 10.1128/jb.172.1.204-211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontarollo R A, Rioux C R, Potter A A. Cloning and characterization of bacteriophage-like DNA from Haemophilus somnus homologous to phages P2 and HP1. J Bacteriol. 1997;179:1872–1879. doi: 10.1128/jb.179.6.1872-1879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raab R, Neal G, Garrett J, Grimaila R, Fusselman R, Young R. Mutational analysis of bacteriophage lambda lysis gene S. J Bacteriol. 1986;167:1035–1042. doi: 10.1128/jb.167.3.1035-1042.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 32.Reader R W, Siminovitch L. Lysis defective mutants of bacteriophage lambda: on the role of the S function in lysis. Virology. 1971;43:623–637. doi: 10.1016/0042-6822(71)90287-x. [DOI] [PubMed] [Google Scholar]
- 33.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 34.Smith D L, Struck D K, Scholtz J M, Young R. Purification and biochemical characterization of the lambda holin. J Bacteriol. 1998;180:2531–2540. doi: 10.1128/jb.180.9.2531-2540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith D L, Young R. Oligohistidine tag mutagenesis of the λ holin gene. J Bacteriol. 1998;180:4199–4211. doi: 10.1128/jb.180.16.4199-4211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 37.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 38.Tedin K, Resch A, Steiner M, Bläsi U. Dual translational start motif evolutionarily conserved in the holin gene of Bacillus subtilis phage φ29. Virology. 1995;206:479–484. doi: 10.1016/s0042-6822(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 40.Young, R. Unpublished data.
- 41.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]