Abstract
RNA N6-methyladenosine (m6A) modification and its regulators fine tune gene expression and contribute to tumorigenesis. This study aims to uncover the essential role and the underlying molecular mechanism(s) of the m6A reader YTHDC1 in promoting triple negative breast cancer (TNBC) metastasis.
Methods: In vitro and in vivo models were employed to determine the pathological function of YTHDC1 in TNBC metastasis. To identify bona fide YTHDC1 target RNAs, we conducted RNA-seq, m6A-seq, and RIP-seq, followed by integrative data analysis and validation assays.
Results: By analyzing The Cancer Genome Atlas (TCGA) dataset, we found that elevated expression of YTHDC1 is positively correlated with poor prognosis in breast cancer patients. Using a mammary fat pad mouse model of TNBC, YTHDC1 significantly promoted lung metastasis of TNBC cells. Through multiple transcriptome-wide sequencing and integrative data analysis, we revealed dysregulation of metastasis-related pathways following YTHDC1 depletion and identified SMAD3 as a bona fide YTHDC1 target RNA. Depletion of YTHDC1 caused nuclear retention of SMAD3 mRNA, leading to lower SMAD3 protein levels. Loss of YTHDC1 led to impaired TGF-β-induced gene expression, leading to inhibition of epithelial-mesenchymal transition (EMT) and suppressed TNBC cell migration and invasion. SMAD3 overexpression was able to restore the response to TGF-β in YTHDC1 depleted TNBC cells. Furthermore, we demonstrated that the oncogenic role of YTHDC1 is mediated through its recognition of m6A as m6A-binding defective mutants of YTHDC1 were unable to rescue the impaired cell migration and invasion of YTHDC1 knockout TNBC cells.
Conclusions: We show that YTHDC1 plays a critical oncogenic role in TNBC metastasis through promoting the nuclear export and expression of SMAD3 to augment the TGF-β signaling cascade. Overall, our study demonstrates that YTHDC1 is vital for TNBC progression by enhancing TNBC cell survival and TGF-β-mediated EMT via SMAD3 to enable the formation of distant metastasis and highlights the therapeutic potential of targeting the YTHDC1/m6A/SMAD3 axis for TNBC treatment.
Keywords: N6-methyladenosine, YTHDC1, TGF-β, SMAD3, metastasis
Introduction
Breast cancer is the most frequently diagnosed cancer in the United States, making up 15% of all cancer diagnoses in 2021 1. Triple negative breast cancer (TNBC) patients with distant metastases only have a 5-year survival rate of 12%, due to its heterogenous and highly invasive nature coupled with a dearth of treatment options compared to other breast cancer subtypes 1, 2. Therefore, it is critical to better understand the molecular pathways and mechanisms that contribute to tumorigenesis and metastasis of TNBC and thereby identify novel therapeutic targets.
In normal cells, TGF-β signaling pathway plays a major role in driving organismal development, immune tolerance, and stem cell maintenance 3. Breast cancer cells exploit the TGF-β pathway to promote epithelial-mesenchymal transition (EMT) leading to elevated migratory and invasive characteristics to enhance metastasis to distant sites like the lungs and bones 4, 5. Serum levels of TGF-β are often elevated in breast cancer patients and is correlated to worse survival, underscoring the effects of the tumor microenvironment on metastasis 6. The canonical TGF-β pathway is activated by binding of extracellular TGF-β to the TGF-β receptor which then phosphorylates SMAD2 and SMAD3. Following this, SMAD2 and SMAD3 form a complex with a co-activator, SMAD4, and other transcription factors, culminating in the activation of TGF-β responsive genes which promote EMT 7. Thus, it is essential to better understand the mechanisms that augment the sensitivity of cells to TGF-β, which could lead to additional therapeutic avenues to treat metastatic TNBC.
N6-methyladenosine (m6A) is the most abundant internal chemical modification in eukaryotic messenger RNA (mRNA) which is able to fine-tune gene expression 8, 9. It is found within the RRACH motif and is associated with regulation of transcription, splicing, nuclear export, RNA stability, and translation 9-15. Importantly, m6A binding proteins (i.e., “readers”) bind to methylated transcripts and direct these transcripts to other complexes in the cell, thus governing the fate of methylated transcripts 8, 9, 16. The YT521 homology (YTH) domain family of proteins are the most well-studied m6A readers, and among them, YTHDC1 has been linked to nuclear export, transcription, splicing, and RNA stability of methylated transcripts 12, 15, 17-19.
Dysregulated m6A methylation is a known contributor to tumorigenesis and metastasis of numerous cancers in part due to the upregulation or downregulation of various m6A “writers”, “erasers”, or “readers” 20-22. To date, METTL3, ALKBH5, FTO, and YTHDF2 have been implicated in promoting breast cancer tumorigenesis by affecting cancer cell survival/proliferation and cancer stem cell pluripotency 23-26. YTHDC1 has been shown to promote endometrial cancer, glioblastoma, and acute myeloid leukemia (AML) progression 27-31; however, its role in breast cancer remains unknown.
In the present study, we identify YTHDC1 as a novel facilitator of TNBC metastasis, with our in vivo animal model studies demonstrating a pivotal role for YTHDC1 in promoting lung metastasis. Mechanistically, YTHDC1 enhances nuclear export of SMAD3 mRNA in an m6A-dependent manner to sensitize TNBC cells to the effects of TGF-β treatment.
Results
Elevated expression of YTHDC1 predicts poorer survival in breast cancer patients
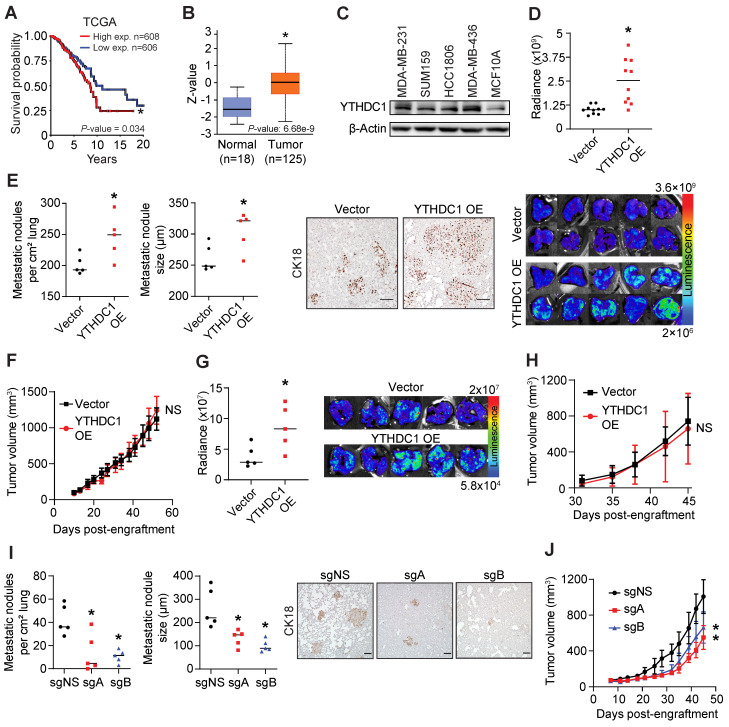
We analyzed patient survival and YTHDC1 gene expression data from The Cancer Genome Atlas (TCGA) cohorts and found that higher levels of YTHDC1 in breast tumors resulted in a significantly poorer prognosis (Figure 1A). Mass spectrometry data from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) showed that primary tumors express YTHDC1 protein at significantly higher levels compared to normal cells (Figure 1B). Notably, YTHDC1 protein expression is significantly higher in tumors having a proteomic profile representing upregulation of collagen VI (k7) and endoplasmic reticulum-related proteins (k10), which are consistent with cells undergoing metastasis/EMT, compared to either normal cells or tumors with a basal-like breast cancer proteomic profile (k4) 32-34 (Figure S1A). Higher YTHDC1 protein levels were observed in the TNBC cell lines MDA-MB-231, SUM159, HCC1806, and MDA-MB-436 compared to non-tumorigenic MCF10A mammary cells (Figure 1C). Taken together, both clinical and laboratory evidence imply an oncogenic role for YTHDC1 in TNBC.
Figure 1.
YTHDC1 promotes TNBC metastasis. A) Breast cancer patient survival in patients expressing high or low levels of YTHDC1 from TCGA data. Log-rank test. B) Box-whisker plot of YTHDC1 protein levels in tumor and normal patient samples from CPTAC data. Z-values represent standard deviations from the median across samples. t-test. C) Western Blot of YTHDC1 levels across different TNBC cells and non-tumorigenic MCF10A cells. D-F) Cells were engrafted into the 4th mammary fat pad of 6-9 week-old NSG mice. 1×106 MDA-MB-231 cells overexpressing YTHDC1 (OE) or vector were engrafted into each mouse. n = 10/group. D) Quantification of lung metastases by chemiluminescence (top) and associated images (bottom). t-test. E) Number and size of lung metastases (left) and corresponding images of lung samples (right) stained for human CK18. Each point represents the average number or size of 10 metastatic nodules in each mouse. Scale bar: 100 μm. n = 5/group. t-test. F) Volume of primary tumor measured twice weekly. Two-way ANOVA. G-H) 2×105 SUM159 cells overexpressing YTHDC1 (OE) or vector were engrafted into each mouse. n = 5/group. G) Quantification of lung metastases by chemiluminescence and associated images. t-test. H) Volume of primary tumor measured twice weekly. Two-way ANOVA. I-J) 1×106 MDA-MB-231 cas9 cells transduced with either control sgRNA (sgNS) or sgRNAs targeting YTHDC1 (sgA and sgB) were engrafted into each mouse. n = 5/group. I) Number and size of lung metastases and images of lung samples stained for human CK18. Each point represents the average number or size of 10 metastatic nodules in each mouse. n = 5/group. Scale bar: 100 μm. One-way ANOVA compared to sgNS. J) Volume of primary tumor measured twice weekly. Two-way ANOVA. The data are presented as the mean ± SD for tumor volume plots; NS = not significant, *P < 0.05.
YTHDC1 promotes lung metastasis in a xenograft model
To determine the pathological role of YTHDC1 in TNBC, we utilized an NSG mouse model and engrafted MDA-MB-231 cells overexpressing YTHDC1 in the 4th mammary fat pad. Strikingly, mice engrafted with TNBC cells overexpressing YTHDC1 developed significantly more lung metastases compared to control cells (Figure 1D). Immunohistochemistry staining of the lungs with anti-human CK18 antibody revealed greater numbers and larger sizes of metastatic nodules in the YTHDC1 overexpressing group (Figure 1E). Despite an increase in lung metastases, YTHDC1 overexpressing MDA-MB-231 cells formed similar sized primary tumors to those in the control mice (Figure 1F and Figure S1B). We repeated this model using another TNBC cell line overexpressing YTHDC1, SUM159, and likewise found significantly higher levels of lung metastases without an increase in primary tumor size compared to control mice (Figure 1G, 1H, and Figure S1C). Conversely, YTHDC1 KO in TNBC cells (MDA-MB-231) resulted in significantly decreased number and size of lung metastases in xenograft recipient mice (Figure 1I) and resulted in a moderate decrease in primary tumor size compared to control mice (Figure 1J and Figure S1D). Collectively, our in vivo animal model data support a tumor-promoting role for YTHDC1, which is consistent with clinical evidence. Notably, we found that YTHDC1 plays a greater role in promoting in vivo lung metastasis with a lesser effect on the growth of the primary tumor.
YTHDC1 is a key regulator of pathways important for TNBC metastasis
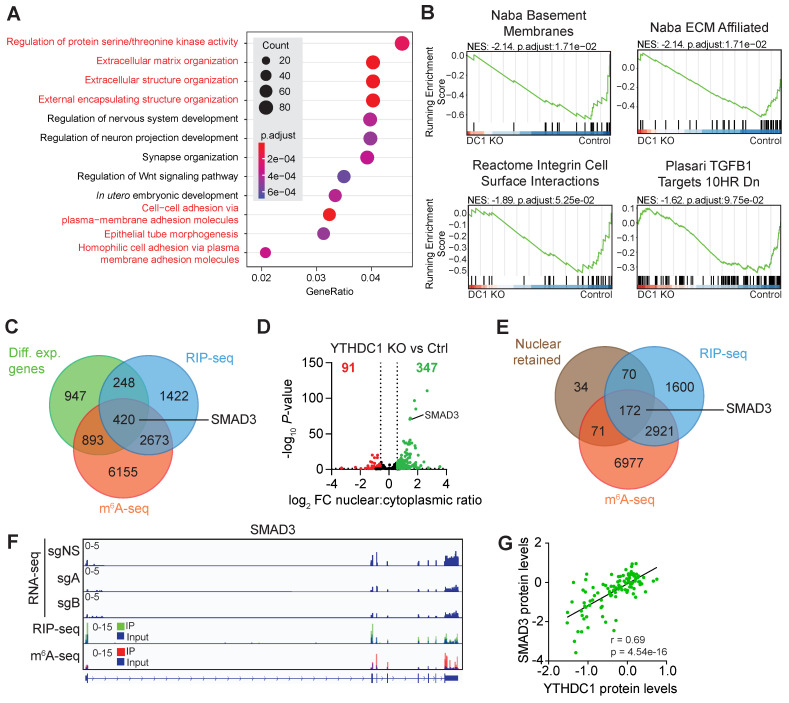
To decipher the molecular mechanism by which YTHDC1 plays a critical role in TNBC metastasis, we conducted RNA-seq with poly(A) RNA from MDA-MB-231 YTHDC1 KO (sgA and sgB) and control cells (Figure S2A). Differentially expressed genes enriched for pathways related to the extracellular matrix, cell-cell adhesion, and epithelial to mesenchymal transition (EMT) were found by Gene Ontology (GO) analysis (Figure 2A). Similarly, Gene Set Enrichment Analysis (GSEA) identified significantly enriched gene sets involved in metastasis such as basement membranes, the extracellular matrix, cell surface interactions, and transforming growth factor beta (TGF-β) (Figure 2B). These enriched pathways reflect YTHDC1's major role in promoting metastasis of TNBC cells as seen in our in vivo studies.
Figure 2.
Identification of YTHDC1 target mRNAs in MDA-MB-231 cells by integrated analysis of sequencing data. A) Gene Ontology (GO) analysis of significantly differentially expressed genes in MDA-MB-231 YTHDC1 KO cells compared to control. Pathways related to metastasis are highlighted in red. B) Pathways related to metastasis enriched in Gene Set Enrichment Analysis (GSEA) of significantly differentially expressed genes in MDA-MB-231 YTHDC1 KO cells compared to control. C) Venn diagram of differentially expressed genes overlapped with RIP-seq and m6A-seq data. D) Volcano plot showing changes in mRNA nuclear export following YTHDC1 KO compared to control. E) Venn diagram of nuclear retained transcripts overlapped with RIP-seq and m6A-seq data. F) RNA-seq, RIP-seq and m6A-seq tracks for SMAD3. G) Correlation between SMAD3 protein and YTHDC1 protein expression levels from CPTAC breast cancer tumor samples. Pearson's correlation test.
SMAD3 is a bona fide YTHDC1 target, whose nuclear export is regulated by YTHDC1 in TNBC cells
Next, we elected to identify high confidence targets of YTHDC1 by combining four sequencing strategies: m6A-seq, RNA-immunoprecipitation (RIP)-seq, whole cell RNA-seq, and nuclear vs cytoplasmic RNA-seq (Figure S2A-S2E). We utilized m6A-seq to identify m6A-methylated transcripts and RIP-seq to identify YTHDC1 bound transcripts in MDA-MB-231 cells. Since YTHDC1 has been reported to play a role in RNA stability and transcription 12, 27, 28, we focused on identifying YTHDC1 targets that were differentially expressed by overlapping RIP-seq, m6A-seq, and differentially expressed transcripts from RNA-seq. Among the 420 genes found was SMAD3 (Figure 2C), a vital TGF-β-activated transcription factor that transactivates many genes involved in epithelial to mesenchymal transition (EMT), cell motility, and metastasis 3. Since YTHDC1 has also been shown to promote nuclear export of mRNA to the cytoplasm 19, we performed RNA-seq on nuclear and cytoplasmic fractions of MDA-MB-231 YTHDC1 KO cells to identify differentially exported mRNA (Figure S2D and S2E). As expected, YTHDC1 KO resulted in an increase in RNA nuclear retention, with 347 genes retained in the nucleus (Figure 2D). After cross-referencing this list with m6A-seq and RIP-seq data, we identified 172 potential YTHDC1 targets and strikingly, SMAD3 was also among the most significant transcripts retained in the nucleus (Figure 2D and 2E). Tracks of RNA-seq data visualize the downregulation of SMAD3 following YTHDC1 KO in MDA-MB-231 cells (Figure 2F). RIP-seq and m6A-seq tracks also show enrichment of YTHDC1 binding and the presence of m6A, respectively, on exons 1, 2, and 8 of SMAD3 RNA (Figure 2F). In addition, CPTAC data showed a significantly positive correlation between the protein expression levels of YTHDC1 and SMAD3 in breast cancer patient samples, which supports regulation of SMAD3 by YTHDC1 (Figure 2G). Together, our data suggest SMAD3 as a high confidence YTHDC1 target in TNBC.
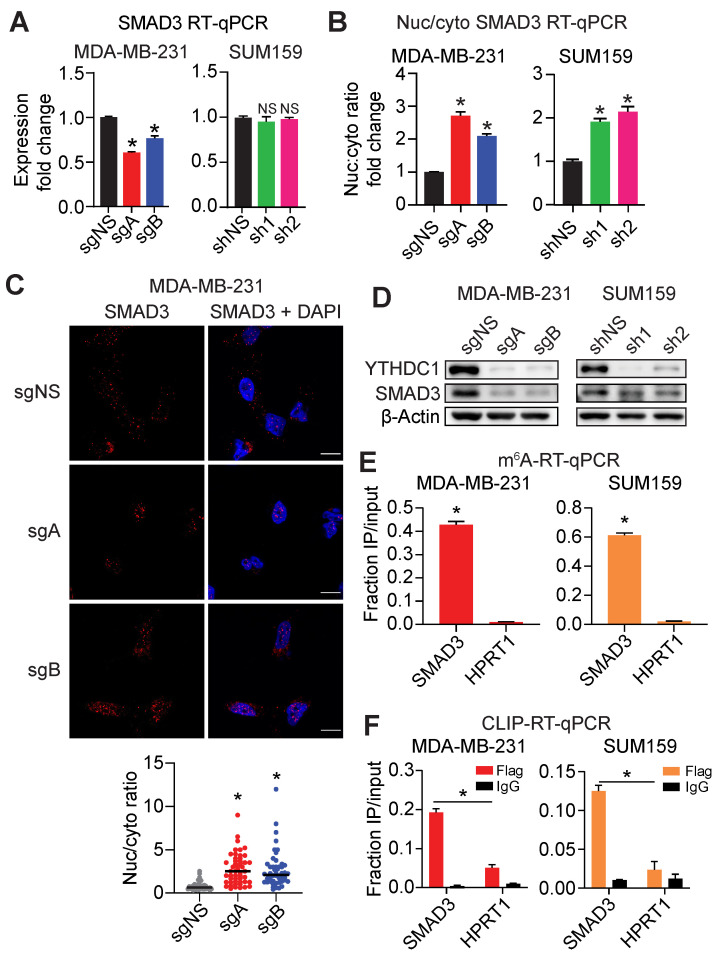
Following YTHDC1 depletion in MDA-MB-231 and SUM159 cells, we observed only either a modest reduction (in MDA-MB-231 cells) or no significant change (in SUM159 cells) in SMAD3 mRNA expression (Figure 3A). We then verified nuclear retention of SMAD3 mRNA by nuclear/cytoplasmic fractionation followed by RT-qPCR and found a 2-3-fold elevation of SMAD3 mRNA in the nucleus following depletion of YTHDC1 in MDA-MB-231 and SUM159 cells (Figure 3B and Figure S3A). In addition, we also observed increased nuclear retention of SMAD3 mRNA by fluorescence in situ hybridization (FISH) upon YTHDC1 depletion in MDA-MB-231 and SUM159 cells (Figure 3C and Figure S3B). At the protein level, the loss of YTHDC1 resulted in decreased SMAD3 protein in both MDA-MB-231 and SUM159 cells (Figure 3D). Additionally, forced expression of YTHDC1 caused a noticeable increased expression of SMAD3 protein in mouse tumors, and the opposite is true when YTHDC1 was knocked out (Figure S1B, S1C, and S1D). To validate m6A-seq data, we conducted m6A-RT-qPCR in both MDA-MB-231 and SUM159 cells and found significantly enriched levels of m6A on SMAD3 mRNA compared to HPRT1 which is an unmethylated control (Figure 3E). Crosslinking and immunoprecipitation (CLIP)-RT-qPCR in both MDA-MB-231 and SUM159 cells demonstrated significantly higher levels of SMAD3 transcripts, compared to HPRT1, bound by flag-tagged-YTHDC1, thus validating RIP-seq data (Figure 3F and Figure S3C). In summary, direct binding of YTHDC1 to SMAD3 mRNA results in its efficient nuclear export, thus augmenting SMAD3 protein expression in TNBC cells.
Figure 3.
YTHDC1 promotes SMAD3 mRNA nuclear export in TNBC cells. A) SMAD3 transcript levels quantified by RT-qPCR in MDA-MB-231 YTHDC1 KO or SUM159 YTHDC1 KD cells. One-way ANOVA compared to control. B) Changes in the nuclear to cytoplasmic ratio of SMAD3 mRNA in MDA-MB-231 YTHDC1 KO or SUM159 YTHDC1 KD cells quantified by RT-qPCR. One-way ANOVA compared to control. C) Representative images and quantification of SMAD3 mRNA localization by FISH following YTHDC1 KO in MDA-MB-231 cells. Scale bar: 10 μm. Each point on the graph represents a single cell. One-way ANOVA compared to sgNS. n = 50/group. D) Western Blots of SMAD3 in MDA-MB-231 YTHDC1 KO or SUM159 YTHDC1 KD cells. E) m6A-RT-qPCR for SMAD3 in MDA-MB-231 and SUM159 cells. HPRT1 was used as a non-target control. One-way ANOVA compared to HPRT1. F) CLIP-RT-qPCR for SMAD3 in MDA-MB-231 and SUM159 cells. HPRT1 was used as a non-target control. One-way ANOVA compared to flag-immunoprecipitated HPRT1. RT-qPCR data are presented as the mean ± SD; n = 3/group, NS = not significant, *P < 0.05.
YTHDC1 is essential for TGF-β-induced EMT of TNBC cells
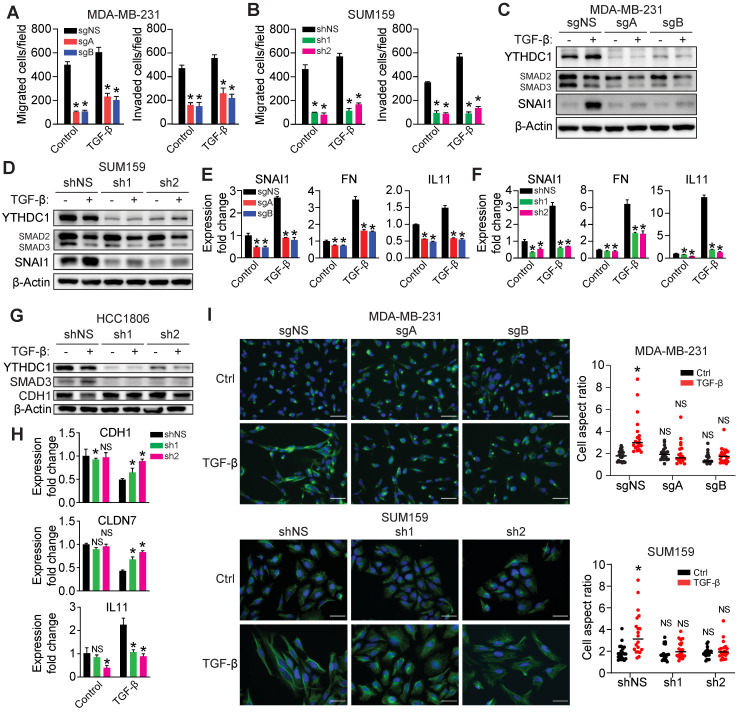
To test whether YTHDC1 is involved in fine-tuning TGF-β signaling, we treated MDA-MB-231 and SUM159 cells (with or without YTHDC1 depletion) with 5 ng/ml TGF-β and measured cell migration and invasion, the expression of TGF-β-induced genes, and cell morphology. The loss of YTHDC1 abrogates TGF-β-induced Transwell migration and invasion of both cell lines (Figure 4A and 4B and Figure S3D and S3E). We observed a prominent lack of SNAI1 protein expression following 24 hours of TGF-β treatment in both YTHDC1 KO MDA-MB-231 and YTHDC1 KD SUM159 cells, indicating that this critical TGF-β-responsive gene is suppressed in YTHDC1 depleted cells (Figure 4C and 4D). Furthermore, we also saw decreased SNAI1, FN, and IL11 RNA levels in YTHDC1 depleted MDA-MB-231 and SUM159 cells following TGF-β treatment (Figure 4E and 4F), indicating a perturbation in TGF-β signaling. In HCC1806 cells, loss of YTHDC1 increased levels of epithelial markers such as CDH1 and CLDN7 and suppressed the mesenchymal marker IL11 following TGF-β treatment (Figure 4G and 4H). Additionally, the induction of an elongated, mesenchymal morphology associated with TGF-β treatment was suppressed in YTHDC1 depleted TNBC cells (Figure 4I). Taken together, our data highlight an essential role for YTHDC1 in promoting TGF-β-signaling and enhancing EMT and cell migration/invasion.
Figure 4.
YTHDC1 is essential for TGF-β-mediated EMT. A-B) Transwell migration and invasion assays for (A) MDA-MB-231 YTHDC1 KO cells (5×104 cells/well incubated for 5 hours for migration and 24 hours for invasion) or (B) SUM159 YTHDC1 KD cells (5×104 cells/well incubated for 24 hours for migration and invasion) treated with 5 ng/ml TGF-β. One-way ANOVA compared to sgNS (MDA-MB-231) or shNS (SUM159) group. *P < 0.05. C-D) Western Blot for protein quantification from (C) MDA-MB-231 YTHDC1 KO cells or (D) SUM159 YTHDC1 KD cells treated with 5 ng/ml TGF-β for 24 hours. E-F) RT-qPCR of TGF-β responsive genes in (E) MDA-MB-231 YTHDC1 KO cells or (F) SUM159 YTHDC1 KD cells treated with 5 ng/ml TGF-β for 24 hours. One-way ANOVA compared to sgNS (MDA-MB-231) or shNS (SUM159) group. *P < 0.05. G) Western Blot for protein quantification from HCC1806 YTHDC1 KD cells treated with 5 ng/ml TGF-β for 72 hours. H) RT-qPCR of TGF-β responsive genes in HCC1806 YTHDC1 KD cells treated with 5 ng/ml TGF-β for 72 hours. One-way ANOVA compared to shNS group. NS = not significant, *P < 0.05. I) Images and quantification of the shapes of control, YTHDC1 MDA-MB-231 KO and SUM159 KD cells treated with TGF-β or vehicle control for 4 days and stained with vimentin. Scale bar: 50 μm. A larger aspect ratio represents a more elongated cell shape. One-way ANOVA compared to control shNS group. NS = not significant, *P < 0.05.
Forced expression of SMAD3 restores migration and invasion of YTHDC1 depleted cells
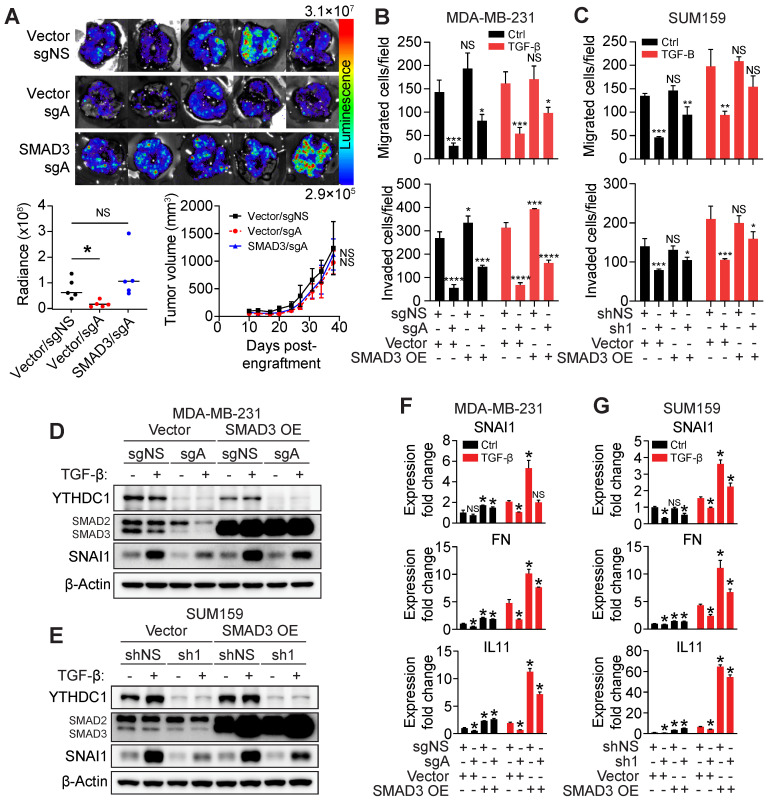
Given that SMAD3 is a critical downstream effector of TGF-β signaling, we hypothesized whether SMAD3 overexpression could rescue the effects of YTHDC1 depletion in TNBC cells. An in vivo rescue experiment was performed and we observed that overexpressing SMAD3 in MDA-MB-231 YTHDC1 KO cells restored metastasis in the lungs, with little effect on primary tumor size (Figure 5A and Figure S4A). In vitro, the impaired migration and invasion phenotypes seen in YTHDC1 depleted cells were partially rescued by SMAD3 overexpression in TNBC cells (Figure 5B and 5C and Figure S4B and S4C). SNAI1 protein level following TGF-β treatment was largely restored by forced expression of SMAD3 in YTHDC1 depleted TNBC cells both in cell culture and in mouse primary tumors (Figure 5D, 5E and Figure S4A). In addition, SMAD3 overexpression also sufficiently restored the expression of other TGF-β-responsive genes such as FN and IL11 in TGF-β treated YTHDC1 depleted cells (Figure 5F and 5G). Thus, overexpressing SMAD3 in YTHDC1 depleted cells is able to restore TGF-β signaling, indicating that YTHDC1 promotes TNBC metastasis at least in part by regulating SMAD3 expression.
Figure 5.
YTHDC1 promotes TGF-β-mediated metastasis by augmenting SMAD3 levels. (A) MDA-MB-231 cas9 cells were engrafted into the 4th mammary fat pad of 6-9 week-old NSG mice. 2×106 cells expressing either vector + sgNS, vector + sgA, or SMAD3 overexpression + sgA were engrafted into each mouse. n = 5/group. Associated lung images (top) and quantification of chemiluminescence signal (bottom left). One-way ANOVA compared to vector + sgNS group. NS = not significant, *P < 0.05. Tumor volumes are also shown (bottom right). Two-way ANOVA compared to vector + sgNS group. NS = not significant, *P < 0.05. B-C) Transwell migration and invasion assays for (B) MDA-MB-231 YTHDC1 KO cells (4×104 cells/well incubated for 24 hours for migration and invasion) or (C) SUM159 YTHDC1 KD (4×104 cells/well incubated for 24 hours for migration and invasion) cells overexpressing SMAD3 treated with 5 ng/ml TGF-β. One-way ANOVA compared to vector + sgNS/shNS group. NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. D-E) Western Blot for protein quantification from (D) MDA-MB-231 YTHDC1 KO cells or (E) SUM159 YTHDC1 KD cells overexpressing SMAD3 treated with 5 ng/ml TGF-β for 24 hours. F-G) RT-qPCR of TGF-β responsive genes in (F) MDA-MB-231 YTHDC1 KO cells or (G) SUM159 YTHDC1 KD cells overexpressing SMAD3 treated with 5 ng/ml TGF-β for 24 hours. One-way ANOVA compared to vector + sgNS/shNS group. *P < 0.05. The data are presented as the mean ± SD; n = 3/group.
YTHDC1 promotes TNBC metastasis as an m6A reader
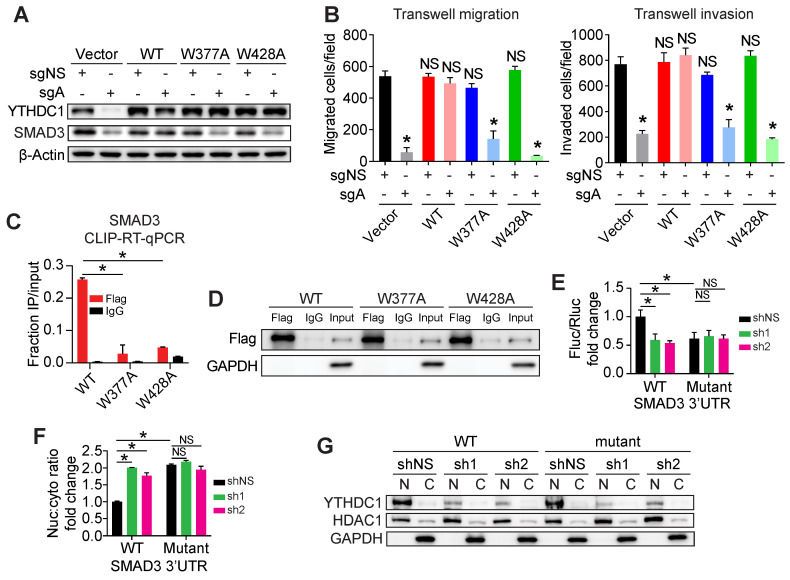
To determine whether YTHDC1's ability to bind to m6A is required for its oncogenic functions, we constructed two YTHDC1 point mutants, W377A and W428A, that have been shown to inhibit its binding to m6A 35. We then overexpressed wild type (WT) YTHDC1 or the W377A and W428A mutants in MDA-MB-231 YTHDC1 KO cells to determine if these mutants were able to rescue YTHDC1 target gene expression and tumorigenic phenotypes (Figure 6A). As expected, overexpression of WT but not the W377A and W428A YTHDC1 mutants was able to restore SMAD3 protein levels in MDA-MB-231 YTHDC1 KO cells (Figure 6A). Furthermore, unlike WT YTHDC1, the W377A and W428A mutants could not rescue the impaired migration and invasion of MDA-MB-231 YTHDC1 KO cells, demonstrating that the metastatic phenotype is dependent on m6A-binding by YTHDC1 (Figure 6B and Figure S4D and S4E). To demonstrate that the phenotypes observed were indeed due to direct binding of YTHDC1 to target transcripts, we performed CLIP-RT-qPCR in MDA-MB-231 cells overexpressing either Flag-tagged WT, W377A or W428A mutants of YTHDC1. We observed significantly decreased binding of SMAD3 RNA to both YTHDC1 mutants compared to WT YTHDC1 (Figure 6C and 6D).
Figure 6.
A functional m6A-binding domain is necessary for the tumorigenic functions of YTHDC1. A) Western Blot analysis of YTHDC1 and SMAD3 in MDA-MB-231 control or YTHDC1 KO cells overexpressing either empty vector, wild type (WT), or m6A-binding defective mutants (W377A and W428A) of YTHDC1. B) Transwell migration and invasion assays of MDA-MB-231 control (sgNS) or YTHDC1 KO cells overexpressing either empty vector, wild type (WT), or mutant (W377A or W428A) YTHDC1 (5×104 cells/well incubated for 24 hours for migration and invasion). One-way ANOVA compared to Vector/sgNS group. C) CLIP-RT-qPCR of SMAD3 bound to wild type (WT) or mutant (W377A or W428A) YTHDC1 in MDA-MB-231 cells. One-way ANOVA compared to WT group. D) Western Blot for immunoprecipitation of wild type or mutant YTHDC1 using anti-flag or control IgG antibodies in CLIP-RT-qPCR assay. (E) Dual luciferase assay in HEK293T cells transfected with WT or mutant SMAD3 3'UTR fused to a firefly luciferase (Fluc) gene. Renilla luciferase (Rluc) used as a loading control. One-way ANOVA compared to shNS+WT SMAD3 3'UTR. (F) Changes in the nuclear to cytoplasmic ratio of firefly luciferase mRNA in HEK293T cells quantified by RT-qPCR. One-way ANOVA compared to shNS+WT SMAD3 3'UTR. The data are presented as the mean ± SD; n = 3/group. NS = not significant, *P < 0.05. (G) Western Blot assay showing nuclear and cytoplasmic distribution of YTHDC1 protein in HEK293T cells and YTHDC1 KD efficiency. HDAC1 and GAPDH were used as nuclear and cytoplasmic marker, respectively.
Our m6A-seq data showed that the first 800 bp of the SMAD3 3'UTR contains the majority of m6A modifications on SMAD3 mRNA (Figure 2F). All the 18 RRACH motif sites in this region were mutated to RRTCH and either the mutant or WT SMAD3 3'UTR was fused to a firefly luciferase gene. Using a dual luciferase assay, we observed that firefly luciferase activity of the mutant SMAD3 3'UTR was significantly decreased compared to that of the WT group in cells without YTHDC1 KD, and the mutations in SMAD3 3'UTR abolished YTHDC1 KD-mediated further down-regulation of the firefly luciferase activity (Figure 6E), indicating the importance of these m6A sites in SMAD3 3'UTR in YTHDC1-meidated promotion of SMAD3 expression. Furthermore, mutant SMAD3 3'UTR caused nuclear retention of firefly luciferase mRNA compared to WT SMAD3 3'UTR (Figure 6F and 6G). This demonstrates that m6A on the SMAD3 3'UTR is recognized by YTHDC1 and results in efficient nuclear export of SMAD3 mRNA. Taken together, these studies provide compelling evidence that YTHDC1's role in promoting TNBC cell migration and invasion is dependent on its ability to read m6A on SMAD3 mRNA.
Discussion
Research into the tumorigenic role of m6A in breast cancer has linked m6A-regulators such as METTL3, ALKBH5, FTO, and YTHDF2 to many facets of breast cancer tumorigenesis such as cell proliferation, survival, and cancer stem cell pluripotency 23-26, 36. In this study, we unravel YTHDC1's role as a significant contributor to TNBC metastasis which reflects the poor prognosis of patients expressing high levels of YTHDC1. In vivo, YTHDC1 promotes lung metastasis from the mammary fat pad despite having a limited effect on primary tumor size. Similar to our observation, previous studies reported that genes involved in migration and invasion such as TGFBR1 and EPHA1 also had a greater effect on tumor dissemination rather than on primary tumor size 5, 37. Further supporting this observation, our RNA-seq data identified significantly dysregulated pathways affecting cell migration, invasion, and metastasis in YTHDC1 KO TNBC cells. Therefore, we uncover a previously unappreciated role of YTHDC1 in promoting TNBC metastasis.
We presumed that YTHDC1 has a wide repertoire of target transcripts as YTHDC1 has been implicated in diverse mechanisms, among them promoting RNA nuclear export by interacting with SRSF3 and enhancing RNA stability by sequestering RNA away from the polyA tail exosome targeting complex (PAXT) and exosome-associated RNA degradation 19, 27. Therefore, we searched for differentially expressed and differentially exported RNAs upon YTHDC1 KO in TNBC cells by RNA-seq, RIP-seq, and m6A-seq to identify high confidence YTHDC1 targets such as SMAD3. Consistent with our discovery, previous studies have shown that m6A writer (METTL14) and eraser (ALKBH5) also modulate SMAD3 expression 38, supporting our conclusion that SMAD3 is a bona fide target of the m6A machinery. Specifically, we showed that YTHDC1 promotes nuclear export of methylated SMAD3 mRNA and thereby affects its protein production. Strikingly, unlike MDA-MB-231 cells where YTHDC1 depletion caused a decrease in both nuclear export and levels of SMAD3 mRNA, loss of YTHDC1 in SUM159 cells caused a decrease in nuclear export of SMAD3 mRNA but did not affect the SMAD3 mRNA stability. Therefore, the main mechanism utilized by YTHDC1 to regulate SMAD3 expression in TNBC cells is nuclear export. Since previous studies demonstrated that elevated nuclear RNA export is a hallmark of breast cancer cells, in part due to upregulation of SRSF3 in HER2+ and triple negative breast cancers 39, our study suggests that YTHDC1 may act in concert with this mechanism to promote the expression of pro-metastatic genes such as SMAD3.
While YTHDC1 plays a role in elevating SMAD3 levels to initiate response of TNBC cells to TGF-β treatment, YTHDC1 may also promote the RNA stability, transcription, or nuclear export of critical downstream TGF-β-responsive genes thus strengthening the TGF-β response. In human pluripotent stem cells (hPSCs), SMAD2/3 was shown to interact with the m6A methyltransferase complex resulting in hypermethylation of downstream SMAD2/3 target mRNAs such as NANOG and SMAD7 40. Another study showed that the loss of METTL14 caused reduced expression of downstream TGF-β genes in breast cancer cells 38. Therefore, this could explain why overexpression of SMAD3 alone does not fully rescue cell migration and invasion of YTHDC1 KO cells in vitro as YTHDC1 could also promote the expression of a set of TGF-β-responsive genes.
In conclusion, our findings contribute to the complexity of TGF-β signaling by linking SMAD3 mRNA nuclear export to the m6A reader YTHDC1, thus promoting TGF-β-responsiveness, EMT, and metastasis of TNBC cells. Given the fundamental role TGF-β plays in metastasis, our findings could potentially be extended to additional invasive cancers such as other breast cancer subtypes, gliomas, and melanomas as intact SMAD signaling machinery is present in most of these cancer types 3. Furthermore, our work provides a rationale for the development of small molecule inhibitors targeting YTHDC1 as a therapeutic strategy to treat breast cancer and other cancers reliant on YTHDC1 such as AML, endometrial cancer, and glioblastoma.
Methods
Database analysis
YTHDC1 RNA-seq expression was correlated to patient survival using data from The Cancer Genome Atlas (TCGA) plotted in XenaBrowser 41. Clinical Proteomic Tumor Analysis Consortium (CPTAC) mass spectrometry data from normal and breast tumor samples and the different proteomic profiles in breast tumors were obtained from UALCAN 42.
Cell culture
MDA-MB-231 and SUM159 cells were kind gifts from Dr. Jun-Lin Guan and Dr. Susan Waltz, respectively. Cells were authenticated using short tandem repeat analysis (Laragen, Inc.). MDA-MB-231 and HEK293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. SUM159 and HCC1806 cells were cultured in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. MCF10A cells (ATCC) were cultured in MEGM Mammary Epithelial Cell Growth Medium (CC-3150, Lonza). HEK293T cells used for lentivirus production were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were kept in a 37°C, 5% CO2 humidified incubator and were routinely tested for mycoplasma contamination using Mycoplasma PCR Detection Kit (Applied Biological Materials) and only mycoplasma free cells were used in experiments. An MDA-MB-231 CRISPR-cas9 single cell clone was isolated by plating lentiCas9-Blast (52962, Addgene) transduced cells into single cells in 96-well plates and selecting with 5 μg/ml of blasticidin.
Plasmids, sgRNAs, and shRNAs
Overexpression plasmids were constructed using the pCDH puromycin resistant or PLX304 blasticidin resistant vectors. Overexpression sequences of YTHDC1, SPRY1, TUG1, and SMAD3 were cloned by PCR from cDNA of MDA-MB-231 cells. The W377A and W428A mutants of YTHDC1 were constructed using the Q5® Site-Directed Mutagenesis Kit (New England Biolabs). For rescue experiments using WT or mutant YTHDC1, the coding sequence was altered to carry synonymous mutations within the sgA site to prevent targeting by sgA. A full list of overexpression vectors is available in Table S1. sgRNA sequences were cloned into the lentiGuide-Puro vector. The sgRNA sequences used were YTHDC1 sgA: 5'-AGATGAGCCGACTTAGTGAA-3' and YTHDC1 sgB: 5'-ACGGAGGATCTCCTATACAC-3'. shRNA targeting YTHDC1 were obtained from Sigma-Aldrich (sh1: TRCN0000243989, sh2: TRCN0000243987). Plasmids used for dual luciferase assay were obtained from Promega. The first 800bp of WT SMAD3 3'UTR was cloned from DNA isolated from MDA-MB-231 cells and fused to a firefly luciferase gene (E1751, Promega). All the 18 RRACH motif sites within the first 800 bp of SMAD3 3'UTR were mutated to RRTCH by site-directed PCR mutagenesis and fused to a firefly luciferase gene to generate mutant SMAD3 3'UTR. As a control, pRL-TK (E2241, Promega) expressing renilla luciferase was used.
Lentiviral production
HEK293T cells were transfected with psPAX2, PMD2.G and the target vector using Transporter 5 transfection reagent (Polysciences) with a 3:1 (volume:mass) ratio of transfection reagent to plasmid DNA. The media was replaced 8 hours post-transfection and the supernatant was collected at 48- and 72-hours post-transfection. The supernatants from both time points were pooled and filtered through a 0.45μm syringe filter before being aliquoted and frozen at -80°C until use. Virus was titrated in MDA-MB-231 cells to determine the concentration of viral particles used to calculate multiplicity of infection (MOI).
Lentiviral transduction
MDA-MB-231 or SUM159 cells were transduced with lentivirus using a MOI of 4. Lentivirus was added to cells in media containing 8 μg/ml polybrene (Sigma-Aldrich) and centrifuged at 1500rpm for 1 hour at room temperature. The media was replaced 24 hours later and supplemented with either 2 μg/ml puromycin or 5 μg/ml blasticidin, or both, depending on the required selection antibiotics. For YTHDC1 loss of function experiments, MDA-MB-231 cas9 or SUM159 cells were transduced with lentivirus carrying either sgRNA or shRNA, respectively. MDA-MB-231 cas9 YTHDC1 knockout (KO) cells were selected using puromycin for 4 days whereas SUM159 YTHDC1 knockdown (KD) cells were selected with puromycin for 2 days before experiments were performed. For rescue experiments, cells were transduced with the SMAD3 overexpression vector and selected for 7 days with blasticidin. The cells were then replated into 6-well plates and transduced with sgRNA or shRNA and selected with puromycin and blasticidin for either 4 or 2 days, respectively, before being used in further experiments.
Western Blotting
Following trypsinization, cells were washed once with PBS and pelleted by centrifugation at 800 rpm for 5 minutes. The cell pellet was lysed with RIPA buffer (Sigma-Aldrich) supplemented with HaltTM protease and phosphatase inhibitors (Thermo Fisher Scientific) and incubated on ice for 20 minutes. The lysate was cleared by centrifugation and protein concentration was determined using the Bio-Rad Protein Assay Kit (Bio-Rad). For each sample, 20 μg of protein was separated using a 10% SDS-PAGE gel and transferred onto a PVDF membrane (Thermo Fisher Scientific). Membranes were blocked with 5% skim milk in PBST and incubated with primary antibody. Goat anti-mouse-HRP (ab6789, Abcam) or anti-rabbit-HRP (ab6721, Abcam) were used as secondary antibodies. Substrate was added to the blots and imaging performed on a ChemiDoc MP (Bio-Rad) imager. Primary antibodies used for Western blotting were: YTHDC1 (A305-096A, Bethyl Laboratories), β-Actin (3700S, Cell Signaling Technologies), SMAD2/3 (3102S, Cell Signaling Technologies), SNAI1 (3895S, Cell Signaling Technologies), CDH1 (PA5-80457, Thermo Fisher Scientific), HDAC1 (sc-7872, Santa Cruz Biotechnology), Flag (F3165, Sigma-Aldrich), and GAPDH (10494-1-AP, Proteintech).
Transwell migration and invasion assays
Transwell chambers with 8 μm pore size in 24-well plates (3422, Corning) were used for both migration and invasion assays. For invasion, 30 μg matrigel (356234, Corning) diluted in serum free media was used to coat the top of each Transwell and was left to solidify at 37°C for 2 hours before the cells were layered onto the gel. MDA-MB-231 or SUM159 cells were seeded in 200 μl serum free media above the Transwell membrane at the indicated cell concentration. 750 μL media with 10% FBS as a chemoattractant was added to the bottom of the well and cells were incubated for the indicated duration. For TGF-β experiments, the top and bottom layers of Transwells were supplemented with 5 ng/ml TGF-β (100-21, Peprotech).
Animal experiments
NOD-SCID IL2Rγ-null (NSG, Jackson 005557) mice were originally purchased from the Jackson Laboratory and bred in accordance to the Institutional Animal Care and Use Committee (IACUC) protocols approved by City of Hope. All mice were grouped housed on a 12 hour:12 hour, light:dark cycle with food and water ad libitum. For each experiment, similar aged (6-9 weeks) mice were randomly assigned to experimental groups.
Mammary fat pad engraftment of cells
MDA-MB-231 and SUM159 were transduced with pLenti-CMV-Puro-LUC (17477, Addgene) to generate luciferase expressing cells and selected with 2 μg/ml of puromycin for 4 days. Cells were then either transduced with YTHDC1 or SMAD3 overexpressing lentivirus or YTHDC1 sgRNA/shRNA lentivirus and their respective controls. Cells resuspended in 100μL PBS were injected into the 4th mammary fat pad of NSG mice. Tumors were measured with calipers twice weekly and tumor volume was calculated using the formula: (width2 × length) / 2. All mice were euthanized at the same time when the primary tumor volume exceeded 1500 mm3 or when tumors showed signs of ulceration.
Bioluminescence imaging of lung metastases
Both the left and right lungs were removed from each mouse and laid out on a petri dish. 50 μL of luciferin substrate (LUCK-2G, Goldbio) was pipetted onto each lung prior to imaging on a Lago X Imaging System (Spectral Instruments). Radiance (watts per square meter per steradian) was measured from the images using Aura Imaging Software (Spectral Instruments) by utilizing an identical selection size for each lung in all images.
CK18 immunohistochemistry staining of lung samples
Dissected lung tissue was incubated in 4% formaldehyde for 48 hours at room temperature, dehydrated using ethanol, and embedded in a paraffin block. Sections of tissue were made and mounted onto glass slides. The sections were deparaffinized with two changes of xylene and then rehydrated by incubation in decreasing concentrations of ethanol (100%, 95%, 70%, and 50%). Antigen retrieval was performed by boiling the slides in Tris-EDTA buffer pH 9.0 in a pressure cooker for 10 minutes. Endogenous peroxidase activity was blocked with 3% H2O2 and the slides were rinsed with PBS followed by blocking with 10% FBS in PBS for 1 hour at room temperature. The slides were stained with anti-human CK18 antibody (ab133263, Abcam, 1:100 dilution) for one hour at room temperature. Following two PBS washes, slides were incubated with goat anti-rabbit HRP secondary antibody (ab6721, Abcam, 1:200 dilution) at room temperature for 30 minutes. Slides were washed three times with PBS and DAB substrate was added to visualize human CK18. Slides were imaged using an Olympus BX61 imaging system.
Nuclear/cytoplasmic separation of cells
One million cells were incubated in 100 μl hypotonic buffer (10mM Tris pH 7.4, 1.5mM MgCl2, 10mM KCl, 1% NP-40) on ice for 10 minutes to separate intact nuclei from cytoplasmic contents. The nuclei were pelleted by centrifugation at 5,000 × g at 4°C for 10 minutes and the supernatant containing the cytoplasmic fraction was collected. The nuclear pellet was washed once with 100 μl hypotonic buffer, spun down, and resuspended in 100 μl hypotonic buffer. RNA was isolated from half of the cytoplasmic and nuclear fractions using Trizol, and the other half of the fraction was incubated with 16.7 μl 4x Laemli buffer for protein extraction.
RNA extraction from cells for RT-qPCR, RNA-seq, and m6A-seq
Cells were trypsinized, transferred to 1.5 ml tubes, and pelleted by centrifugation. 1 ml of Trizol (Qiagen) was added to each sample and the cells were lysed by vortexing. After incubation at room temperature for 10 minutes, 200 μl of chloroform was added and the solution was thoroughly mixed by shaking. Then, the tubes were centrifuged at 12,000 × g for 15 minutes at 4°C and 500 μl of the aqueous phase was transferred to another 1.5 ml tube. An equal amount of isopropanol was added followed by thorough mixing. The tubes were centrifuged at 12,000 × g for 15 minutes at 4°C and the supernatant was discarded. The pellet was washed with 1 ml of 75% ethanol and then pelleted by centrifugation at 12,000 × g for 5 minutes at 4°C. The supernatant was carefully discarded, and the pellet was left to air dry. Nuclease-free water (Thermo Fisher Scientific) was then used to resuspend the RNA pellet and its concentration was determined using a NanoDrop One (Thermo Fisher Scientific).
mRNA isolation
PolyATtract® mRNA Isolation Systems kit (Promega) was used to isolate mRNA from purified total RNA following manufacturer's instructions.
m6A-IP for m6A-RT-qPCR and m6A-seq
Purified mRNA was precipitated with 1:10 volume of 3M sodium acetate (pH 5.2) and 2.5 volumes of 100% ethanol at -80°C overnight. The next day, the mRNA was centrifuged at 15,000 × g for 15 minutes at 4°C and the supernatant was discarded. The pellet was washed with 1mL 75% ethanol and centrifuged at 15,000 × g for 5 minutes at 4°C. The supernatant was discarded and the pellet was left to air dry before being resuspended in nuclease-free water. The concentration was determined using a NanoDrop One (Thermo Fisher Scientific) and adjusted to 1 ug/ul with nuclease-free water. The mRNA was divided into 18uL aliquots in thin-walled PCR tubes. 2 μL of 10× fragmentation buffer (100mM ZnCl2 in 100mM Tris-HCl pH 7.0) was added and tubes were incubated at 94°C for exactly 4 minutes. Immediately, 2 μl of 500mM EDTA was added to each tube and put on ice. The fragmented mRNA was then purified by precipitation as outlined above before immunoprecipitation. At least 100 ng of fragmented RNA was retained as the input fraction. 50 μl of Protein A/G magnetic bead slurry (Thermo Fisher Scientific) were added to a 1.5 ml tube and washed twice with 1 ml of cold RNase-free PBS. Then, 5 μg of anti-m6A antibody (202003, Synaptics Systems) was incubated with the beads for 3 hours at 4°C. The beads were washed 3× with cold PBS and then once with IP buffer (10mM Tris-HCl pH 7.4, 750 mM NaCl and 0.5% (vol/vol) Igepal CA-630). A total of 10 μg of fragmented mRNA was incubated with the beads overnight at 4°C. The next day, the beads were washed 5× with IP buffer. To elute bound RNA, 100 μl of elution buffer (6.67mM m6A salt (Sigma-Aldrich) in IP buffer) was incubated with the beads for 1 hour at 4°C and the supernatant containing immunoprecipitated RNA was collected. The elution was repeated with another 100 μl of elution buffer. The eluted RNA was purified using the RNeasy Mini Kit (Qiagen) and eluted to a final volume of 15 μl. For m6A-RT-qPCR, 3 μl of immunoprecipitated eluate was used and 1% (1 ng) of the input amount used in each RT reaction. For m6A-seq, an Agilent Bioanalyzer 2100 was used to determine the size and quality of both IP and input samples prior to RNA library construction using KAPA Stranded mRNA-Seq Kit (Illumina Platforms) (KR0960m Kapa Biosystems) and sequenced on an Illumina Hiseq2500.
Crosslinking and Immunoprecipitation (CLIP)-RT-qPCR and RIP-seq
MDA-MB-231 cells overexpressing flag-tagged-YTHDC1 or its mutants were grown to 80% confluence in 15cm tissue culture dishes. Cells were washed with cold PBS and UV-crosslinked 3× at 120,000 μJ/cm2 using a 254nm lamp. Nuclear isolation buffer (320mM sucrose 10mM Tris-HCl pH 7.5, 5mM MgCl2, 1% Triton X-100 vol/vol) was added onto the dish and the cells were detached with a cell scraper. The cells were incubated on ice for 20 minutes on ice followed by pelleting of nuclei by centrifugation at 1500 × g for 15 minutes at 4°C. The pellet was resuspended in RIP buffer (150mM NaCl, 25mM Tris-HCl pH 7.4, 5mM EDTA, 0.5mM DTT, and 0.5% Tergitol NP40) containing HaltTM Protease (Thermo Fisher Scientific) and RiboLock (Thermo Fisher Scientific) RNase inhibitors and sonicated using a Qsonica Q800R3 sonicator to isolate nuclear contents. The lysate was centrifuged at 15,000 × g for 10 minutes at 4°C and the supernatant was pre-cleared by incubation with protein A/G beads for 1 hour at 4°C. The amount of protein in each sample was quantified using the Bio-Rad Protein Assay Kit (Bio-Rad). For each immunoprecipitation reaction, 5 μg of anti-flag antibody (F3165, Sigma-Aldrich) or normal mouse IgG (12-371, Millipore) was conjugated to 50 μl Protein A/G magnetic beads (Thermo Fisher Scientific) for 4 hours at 4°C. The beads were washed twice with RIP buffer and incubated with supernatant containing 500 μg of protein overnight at 4°C. Meanwhile, supernatant containing 50 μg of protein (10% input) was saved for each RNA or protein input sample. The Trizol extraction method described above was used to purify input RNA, which was resuspended in a final volume of 50 μl using RNase free water. Laemli buffer was added to input protein and denatured for 5 minutes at 95°C. Both RNA and protein input samples were kept at -80°C until needed. The next day, the immunoprecipitation reactions were washed 3x with RIP buffer and resuspended in 100 μl PBS. For isolation of immunoprecipitated protein, 20 μl of beads were transferred to another 1.5 ml tube and 6.7 μl of 4x Laemli buffer was added and the sample was incubated at 95°C for 5 minutes. The remaining beads were then treated with DNase 1 (Thermo Fisher Scientific) followed by Proteinase K (Thermo Fisher Scientific) to remove unwanted DNA and uncrosslinked protein. The tube was then placed on a magnetic stand and the supernatant containing the immunoprecipitated protein fraction was transferred to a new tube and frozen at -80°C. Trizol was added to the remaining 80 μl of beads and RNA was isolated as described above and resuspended in a final volume of 15 μl. For CLIP-RT-qPCR, 3 μl of immunoprecipitated eluate was used as the IP group and 1% of the input amount (5 μl) used in the input group for each RT reaction. RNA used in RIP-seq was first rRNA depleted followed by library construction using KAPA Stranded mRNA-Seq Kit (Illumina Platforms) (KR0960m Kapa Biosystems) with 10 cycles of PCR amplification and sequenced on an Illumina Hiseq2500.
RNA-seq and data analysis
RNA from whole cells were polyA selected and RNAseq libraries were prepared using KAPA RNA HyperPrep Kit with RiboErase (HMR) (Illumina Platforms) (Kapa Biosystems), using 12 cycles of PCR amplification. Libraries were purified using AxyPrep Mag PCR Clean-up kit (Thermo Fisher Scientific). Sequencing was performed on an Illumina® HiSeq 2500 (Illumina) instrument. Nuclear or cytoplasmic fractions were rRNA-depleted and libraries were prepared similar to whole cell RNA. Data analysis of polyA RNA-seq of whole cells entailed mapping raw reads to a human transcriptome built with GENCODE v33 annotations 43 using STAR (version 2.7.2b) 44 and the expression of genes, including read counts and transcript per million (TPM), were calculated by RSEM (version 1.2.31) 45. Batch effect normalization was performed via the ComBat-seq function of sva (version 3.40) 46 and removed genes with read counts <5 in at least 50% of the samples prior to using DESeq2 (version 1.32.0) 47 to calculate the dispersion and fold change following YTHDC1 KO in MDA-MB-231 cells. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA) analyses were performed by utilizing clusterProfiler4 (version 4.0.2) 48. For calculation of nuclear/cytoplasmic ratio of RNA following YTHDC1 KO, RNA-seq of nuclear and cytoplasmic fractions were analyzed with RUVSeq (version 1.26.0) 49 to remove any unwanted batch effects. The ratio of ratios test model was employed by DESeq2 to identify potential genes that are differentially retained in the nucleus in the YTHDC1 KO group. A transcript was considered as nuclear retained when it has a nuclear to cytoplasmic log2 fold change of 0.5 or higher when comparing sgB vs sgNS.
RIP-seq analysis
Raw reads from RIP-seq were mapped to the transcriptome and quantified using the same data processing procedures described for RNA-seq above. To identify YTHDC1 target transcripts, we kept transcripts with read counts ≥ 5 in at least 50% of the samples, performed batch effect correction using RUVSeq, and calculated the dispersion and fold change of transcripts upon YTHDC1 knockdown in MDA-MB-231 cells by DESeq2 with shrinkage estimators. The transcripts with adjusted P-value ≤ 0.1 and fold change ≥ 2 were recognized as YTHDC1 bound targets.
m6A-seq analysis
Raw reads from m6A-seq were mapped to the human transcriptome annotations (GENCODE v33) using STAR with the parameters -outFilterMismatchNoverReadLmax 0.04 and --outFilterMultimapNmax 20. Potential m6A peaks were identified using exomePeak (version 2.16) 50 with default parameters. Significant m6A peaks were defined as having a fold enrichment ≥ 1 and FDR ≤ 0.05.
RT-qPCR for gene expression analysis
QuantiTect Reverse Transcription Kit (Qiagen) was used to synthesize cDNA for RT-qPCR, RIP-RT-qPCR and m6A-RT-qPCR using manufacturers protocols. For RT-qPCR, 500 ng of total RNA was used for each reaction. The cDNA samples were diluted 1:10 using nuclease-free water before qPCR analysis. For each qPCR reaction, 5 μl of Applied Biosystems PowerUp SYBR Green Master Mix (Thermo Fisher Scientific), 3 μl nuclease-free water, 1 μl mix of forward and reverse primers (5 μM), and 1 μl of cDNA was used in a 384-well format. The qPCR was run on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific) using the standard cycling mode. β-actin was used as a loading control for gene expression analysis. A list of qPCR primers used is provided in Table S2.
Immunofluorescence imaging
Cells seeded in 24-well plates were fixed with 4% formaldehyde at room temperature for 15 minutes and then permeabilized with 1% Triton-X100 in PBS for 15 minutes at room temperature. Wells were blocked by incubating with 10% FBS in PBS for one hour at room temperature and cells were stained with mouse anti-vimentin (1:200, MA511883, Thermo Fisher Scientific) overnight at 4°C. The cells were washed with PBS and stained with Alexa Fluor 488 goat anti-mouse secondary antibody (1:500, A11001, Thermo Fisher Scientific). After three PBS washes, cells were stained with DAPI and imaged on an Olympus BX61 imaging system. Cell aspect ratio was calculated using ImageJ. The threshold function was used on vimentin images to convert them to a binary image containing the outline of cells. Aspect ratio was determined using the analyze particles function, which was calculated by dividing by the length by the width of a cell.
Fluorescence in situ hybridization
Custom Stellaris® FISH Probes were designed against the SMAD3 coding sequence by utilizing the Stellaris® RNA FISH Probe Designer (Biosearch Technologies, Inc.) available online at www.biosearchtech.com/stellarisdesigner (version 4.2). Samples were hybridized with the SMAD3 Stellaris RNA FISH Probe set labeled with Quasar® 570 dye (Biosearch Technologies, Inc.), following the manufacturer's instructions. Briefly, cells were fixed in 4% formaldehyde, permeabilized with 70% ethanol and incubated with probe overnight. The samples were mounted onto slides with Fluoroshield® medium containing DAPI (F6057, Sigma-Aldrich) and imaged using a Carl Zeiss LSM880 confocal laser scanning microscope.
Dual luciferase assay
HEK293T cells were transfected with firefly luciferase fused to either WT or mutant SMAD3 3'UTR plasmids and a renilla luciferase plasmid using Effectene Transfection Reagent (Qiagen) according to the manufacturer's protocol. Two days later, cells were then transduced with lentivirus carrying YTHDC1 shRNA as described above.
Three days after lentiviral transduction, cells were trypsinized and the Dual-Luciferase Reporter Assay System (Promega) was used for detection according to manufacturer's instructions. Briefly, lysate from 10,000 cells was pipetted into each well of a 96-well white bottom plate. Then, firefly luciferase substrate was added, and the signal was quantified on a GloMax® Navigator Microplate Luminometer (Promega). Lastly, a solution that quenches firefly luciferase activity and contains a substrate for renilla luciferase was added and the signal was quantified again. The firefly luciferase signal was normalized using the renilla luciferase signal.
Statistical analyses
Statistical analyses of experimental data were calculated with Prism Version 8 (GraphPad Software). The log-rank test was used to determine significance for survival plots. For comparison between two samples, the Student's t-test was employed. One-way ANOVA with Dunnett's test was used for comparisons between more than two samples. For comparison of growth curves and primary tumor sizes, Two-way ANOVA with Tukey's multiple comparisons test was used. P-values less than 0.05 were considered significant.
Data availability
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE193156.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
Research reported in this publication includes work performed by City of Hope shared resources supported by the National Cancer Institute of the NIH under award number P30 CA033572.
Author Contributions
Brandon Tan: Conceptualization, contributed resources, designed experiments, performed experiments, statistical analyses, interpreted data, and wrote the manuscript. Keren Zhou and Lu Yang: Bioinformatics and statistical analyses and edited the manuscript. Wei Liu, Emily Prince, Ying Qing, Yangchan Li, Li Han, Xi Qin, Rui Su, Sheela Pangeni Pokharel, Chao Shen, Wei Li, and Zhicong Zhao: Designed experiments, performed experiments, interpreted data, contributed resources to the study, and edited the manuscript. Andrew Small, Kitty Wang, and Keith Leung contributed resources to the study and edited the manuscript. Zhenhua Chen, Zheng Zhang, and Xiaolan Deng: Interpreted data and critical editing of the manuscript. Chun-Wei Chen, and Binghui Shen: Conceptualization, interpreted data, funding support, and supervision. Jianjun Chen: Conceptualization, designed experiments, interpreted data, wrote the manuscript, supervision, and funding support.
Financial support
This work was supported in part by the National Institutes of Health (NIH) grants T32 CA186895 (B.T.), R01 CA243386 (J.C.), R01 CA214965 (J.C.), R01 CA236399 (J.C.), R01 CA271497 (J.C.), R01 DK124116 (J.C), R01CA085344 (B.H.S), The Margaret Early Medical Research Trust (R.S.), and The Simms/Mann Family Foundation (J.C.). J.C. is a Leukemia & Lymphoma Society (LLS) Scholar.
Abbreviations
- ALKBH5
AlkB Homolog 5, RNA Demethylase
- AML
Acute myeloid leukemia
- CK18
Cytokeratin 18
- CPTAC
Clinical Proteomic Tumor Analysis Consortium
- CRISPR
clustered regularly interspaced short palindromic repeats
- EMT
Epithelial to mesenchymal transition
- EPHA1
EPH Receptor A1
- FTO
FTO Alpha-Ketoglutarate Dependent Dioxygenase
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- HPRT1
Hypoxanthine Phosphoribosyltransferase 1
- IL11
Interleukin 11
- m6A
N6-methyladenosine
- METTL3
Methyltransferase 3, N6-Adenosine-Methyltransferase Complex Catalytic Subunit
- METTL14
Methyltransferase 14, N6-Adenosine-Methyltransferase Subunit
- MOI
Multiplicity of infection
- KO
knockout
- KD
knockdown
- PCA
Principal component analysis
- SMAD3
SMAD Family Member 3
- SNAI1
Snail Family Transcriptional Repressor 1
- SRSF3
Serine and Arginine Rich Splicing Factor 3
- TCGA
The Cancer Genome Atlas
- TNBC
triple negative breast cancer
- TGF-β
Transforming Growth Factor Beta
- TGFBR1
Transforming Growth Factor Beta Receptor 1
- WT
Wild type
- YTHDC1
YTH Domain Containing 1
- YTHDF2
YTH N6-Methyladenosine RNA Binding Protein 2
References
- 1. SEER*Explorer: An interactive website for SEER cancer statistics [https://seer.cancer.gov/explorer/]. Accessed on December 8th 2021.
- 2.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR. et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–14. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR. et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarzynska JM. Two faces of TGF-beta1 in breast cancer. Mediators Inflamm. 2014;2014:141747. doi: 10.1155/2014/141747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–9. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169:1187–200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 11.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhtar J, Renaud Y, Albrecht S, Ghavi-Helm Y, Roignant JY, Silies M. et al. m(6)A RNA methylation regulates promoter- proximal pausing of RNA polymerase II. Mol Cell. 2021;81:3356–67. doi: 10.1016/j.molcel.2021.06.023. e6. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Meyer KD, Jaffrey SR. Rethinking m(6)A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 2017;33:319–42. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 2018;14:e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81:3368–85. doi: 10.1016/j.molcel.2021.07.024. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife. 2017. 6. [DOI] [PMC free article] [PubMed]
- 20.Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang H, Weng H, Deng X, Chen J. RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annual Review of Cancer Biology. 2020;4:221–40. [Google Scholar]
- 22.Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer cell. 2020;37:270–88. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L. et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722:144076. doi: 10.1016/j.gene.2019.144076. [DOI] [PubMed] [Google Scholar]
- 25.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Einstein JM, Perelis M, Chaim IA, Meena JK, Nussbacher JK, Tankka AT. et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol Cell. 2021;81:3048–64. doi: 10.1016/j.molcel.2021.06.014. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X. et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer cell. 2021;39:958–72. doi: 10.1016/j.ccell.2021.04.017. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng Y, Wei J, Yu F, Xu H, Yu C, Wu Q, A Critical Role of Nuclear m6A Reader YTHDC1 in Leukemogenesis by Regulating MCM Complex-Mediated DNA Replication. Blood. 2021. [DOI] [PMC free article] [PubMed]
- 29.Zhang B, Shao X, Zhou J, Qiu J, Wu Y, Cheng J. YT521 promotes metastases of endometrial cancer by differential splicing of vascular endothelial growth factor A. Tumour Biol. 2015. [DOI] [PubMed]
- 30.Hirschfeld M, Zhang B, Jaeger M, Stamm S, Erbes T, Mayer S. et al. Hypoxia-dependent mRNA expression pattern of splicing factor YT521 and its impact on oncological important target gene expression. Mol Carcinog. 2014;53:883–92. doi: 10.1002/mc.22045. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Yi Y, Miao Y, Long W, Long T, Chen S. et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019;79:5785–98. doi: 10.1158/0008-5472.CAN-18-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, Scherer PE. Adipocyte-derived endotrophin promotes malignant tumor progression. J Clin Invest. 2012;122:4243–56. doi: 10.1172/JCI63930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA. et al. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer discovery. 2014;4:702–15. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 34.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y. et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–9. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 36.He X, Tan L, Ni J, Shen G. Expression pattern of m(6)A regulators is significantly correlated with malignancy and antitumor immune response of breast cancer. Cancer Gene Ther. 2021;28:188–96. doi: 10.1038/s41417-020-00208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiuan E, Inala A, Wang S, Song W, Youngblood V, Chen J. et al. Host deficiency in ephrin-A1 inhibits breast cancer metastasis. F1000Res. 2020;9:217. doi: 10.12688/f1000research.22689.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Science advances. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Gong Q, Lin Z, Wang Y, Li M, Wang L. et al. Emerging Roles of SRSF3 as a Therapeutic Target for Cancer. Front Oncol. 2020;10:577636. doi: 10.3389/fonc.2020.577636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L. et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256–9. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A. et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–8. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J. et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D73. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform. 2020;2:lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (N Y) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y. et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–81. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.
Data Availability Statement
The data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE193156.