Abstract
Recent studies have mapped key genetic changes in colorectal cancer (CRC) that impact important pathways contributing to the multistep models for CRC initiation and development. In parallel with genetic changes, normal and cancer tissues harbor epigenetic alterations impacting regulation of critical genes that have been shown to play profound roles in the tumor initiation. Cumulatively, these molecular changes are only loosely associated with heterogenous transcriptional programs, reflecting the heterogeneity in the various CRC molecular subtypes and the paths to CRC development. Studies from mapping molecular alterations in early CRC lesions and use of experimental models suggest that the intricate dependencies of various genetic and epigenetic hits shape the early development of CRC via different pathways and its manifestation into various CRC subtypes. We highlight the dependency of epigenetic and genetic changes in driving CRC development and discuss factors affecting epigenetic alterations over time and, by extension, risk for cancer.
Keywords: colorectal cancer, epigenetics, aging, classification, subtyping, DNA methylation, consensus molecular subtypes
Introduction
Colorectal cancer (CRC), which involves cancers occurring in the colon and rectum, is the third most prevalent cancer by incidence and mortality worldwide. In fact, it is estimated that by 2030 there will be globally >2.2 million new cases and 1.1 million deaths due to CRC [1]. CRC is a heterogenous group of tumors in the colon and rectum that can be sub-grouped based on distinct molecular patterns, including pathological parameters, genomic alterations, and gene-expression patterns. This heterogeneity in the molecular patterns of CRC further reflects differences in the molecular evolution of CRC from the early stages of tumor initiation to its clinical presentation, including different molecular drivers of carcinogenesis, routes to tumor progression, factors contributing to the risk of cancer development, and potentially even cell of origin. Recent genome-wide sequencing projects and development of experimental models have revealed the importance of the interaction of genetic and epigenetic alterations in the early stages of CRC molecular evolution [2–6]. This review will discuss recent advances in the molecular genetics of early CRC evolution and examine the role early epigenetic changes may have in creating the appropriate gene-expression profile for genetic driver mutations to promote CRC tumor initiation and progression. Within this context, we strive to answer the following questions: Which key physiological and environmental factors contribute to epigenetic changes? How do these epigenetic changes play a role in tumor initiation and increasing cancer risk by genetic driver mutations?
Molecular heterogeneity of CRC
CRC carcinogenesis occurs due to the progressive accumulation of different combinations of molecular changes in the form of genetic and epigenetic insults during aging that inactivate tumor suppressor genes and activate proto-oncogenes [7, 8]. Classifying CRC based on these molecular changes, which is still an ongoing process, has immense value for understanding essential events in CRC evolution and their clinical implications. Developments in molecular and genomic technologies have continually refined the classification of CRC into various subtypes over the years. Recent integrated molecular characterizations of CRC, such as The Cancer Genome Atlas (TCGA) project, have revealed important relationships among the genetic, epigenetic, and transcriptional changes [9]. Earlier molecular classifications were primarily based on genetic and epigenetic parameters, while newer classifications have also incorporated transcriptional signatures for more robust clinical implications [10–12]. Below we discuss the relationships between the molecular features underlying both the conventional classification and the recent gene-expression-based classification, which serve in further understanding the genetic and epigenetic basis for CRC evolution.
Conventional classification
CRC is primarily classified based on the following genetic and epigenetic molecular features, which are important determinants of the alternate routes of genomic instability in CRC development.
Chromosomal instability
The chromosomal instability (CIN) subtype is the most prevalent feature accounting for 80%–85% of all CRC cases and is characterized by sporadic genomic instability. It is defined by abnormalities in either the chromosomal structure resulting in loss of heterozygosity (LOH) or somatic copy number alterations (SCNA; aneuploidy or polyploidy) associated with a specific set of tumor suppressor and proto-oncogene loci [13–15]. A hallmark of 70%–80% of CIN cases is loss of the tumor suppressor and Wnt pathway negative regulator APC, which occurs by sporadic mutation in one allele of APC followed by loss of the other APC allele due to flaws in chromosome segregation [16, 17]. Other genes that are typically affected include TP53, KRAS, and PIK3CA. LOH in chromosome 18q containing tumor suppressor genes SMAD2, SMAD4, and DCC is also common as these genes are transcriptional mediators of the transforming growth factor (TGF)-β signaling pathway, which regulates cell growth, differentiation, and apoptosis, and promotes MYC activation [9, 16, 18]. CRC with CIN is more frequently observed in the distal colon [17].
Microsatellite instability
CRC classified as microsatellite instability (MSI) form the other major type of genomic instability aside from CIN. MSI develops due to inactivation of the mismatch repair genes and accounts for 15%–20% of CRC. Twenty percent of MSI cases are hereditary and develop due to germline mutations in the mismatch repair genes, specifically MLH1, MSH2, MSH6, and PMS2. The remaining 80% of these cases are sporadic and develop due to spontaneous epigenetic silencing of the MLH1 gene by promoter hypermethylation in the CpG-island methylator phenotype (CIMP) [14, 19]. Rather than affecting whole or parts of chromosomes, MSI is defined by a high frequency of replication errors, specifically an accumulation of insertions and/or deletions, in repetitive mononucleotide and dinucleotide DNA sequences referred to as microsatellites in exon regions of tumor suppressor genes, resulting in loss of function hypermutation [18]. MSI tumors have been subclassified into three groups based on the number of microsatellites associated: (i) high MSI (MSI-H), (ii) low MSI (MSI-L), and (iii) microsatellite stable (MSS). Frequently targeted genes for mutation in MSI tumors include BRAF (which is mutated in >8% of all CRC cases), ACVR2A, TGFBR2, MSH3, and MSH6 along with Wnt pathway regulators RNF43, RNF213, and ZNRF3 [20, 21]. LOH and mutations in APC, TP53, and KRAS are less frequent in MSI tumors than in CIN tumors, with loss of APC dropping from 80% down to ∼50% in MSI cases [9]. CRC with MSI status is more frequently observed in the proximal colon [17]. In contrast to the MSI tumors, CIN tumors are non-hypermutated and MSS.
A more recent and less well-studied class of MSI that is different from the classical MSI is the elevated microsatellite alterations at selected tetranucleotide repeats (EMAST), which occurs due to mismatch repair dysfunction resulting from loss of function of MSH3 [22]. Herein, MSH3 loss of function is caused by nuclear to cytoplasmic export of MSH3 in response to oxidative stress and pro-inflammatory cytokine interleukin (IL)-6 [23]. Unlike MSI due to MLH1 loss, which occurs in ∼15% of all CRC cases, EMAST is observed to be prevalent in 60% of CRC [24]. MSH3 dysfunction can also contribute to defects in double strand DNA repair by homologous-recombination, leading to the CIN phenotype [25]. Thus, microenvironmental changes, such as those that cause oxidative stress or inflammatory exposure of colon cells, may lead to this alternate form of MSI, i.e. EMAST, which may play a role in genetic events that lead to a substantial number of CIN CRC.
CpG-island methylator phenotype
CIMP tumors are characterized by a very high frequency of promoter gene methylation [26, 27]. In general, ∼50% of gene promoters in humans harbor 200–800 base-pair stretches of nucleotides called CpG-islands that are unusually enriched with the dinucleotide sequence containing cytosine followed by guanine (termed CpG-residues). CpG-islands have important roles in the regulation of gene expression, and methylation of the CpG-islands in promoters causes silencing of these genes [26]. Widespread hypermethylation involves methylation of the promoter regions of many tumor suppressor genes leading to their epigenetic silencing and is present in 20%–30% of CRC cases, both MSI and CIN [28, 29]. CIMP classification of CRC represents the spectrum of CRC with differing frequency of hypermethylated CpG-island promoters [30, 31]. Based on the degree of methylation at a group of genes/loci, clinically cancers with the highest frequency of hypermethylation of these marker loci are classified into CIMP-high (CIMP-H), those with intermediate frequency are classified into CIMP-low (CIMP-L), and those lacking hypermethylation of the marker loci are classified as CIMP-negative (CIMP-). In general, the CIMP-H cancers have variably been associated with poor outcomes [32, 33] with the CIMP-H being a negative indicator of overall survival, whereas MSI-H is a positive indicator [34, 35]. Recent pooled analysis suggests that CIMP-H CRC harboring BRAF mutation but with MSI-L status have the highest disease-associated mortality [36]. CIMP-H is prevalent in tumors in the proximal colon with MSI-H and BRAF mutation, whereas CIMP-L tends to occur in the context of KRAS mutations in the distal colon; however, there are also wide heterogeneities in these co-occurrences and their corresponding impact on disease outcomes as CIMP-H can also occur in CIN (MSI-L/MSS) tumors in the proximal colon with higher frequency of BRAF mutations, which tends to have worse survival [36, 37]. These incidence patterns of CIMP in relation to other molecular features suggest that CIMP is an important and early premalignant pathological pathway particularly involved in BRAF-induced CRC development.
Transcriptional subtypes and their relation to conventional subtypes
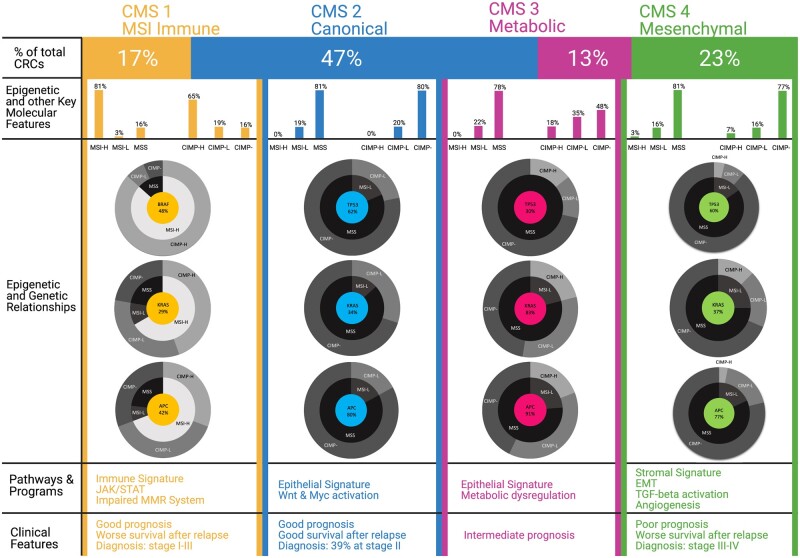
The conventional classification of CRC discussed above provides the critical reference point for understanding molecular drivers of CRC and guiding therapeutic and response dynamics. However, it does not encompass the diversity of CRC phenotypes and contribution of transcriptional landscape to these molecular genetic phenotypes. An integrated transcriptome-based classification using approaches from six independent studies have classified CRC into the consensus molecular subtypes (CMS), which has helped better understand biological features and molecular properties of the various CRC subtypes [38]. Based on the CMS clustering approach, 80%–90% of CRC falls into one of the four major transcriptional subgroups termed CMS1–4 and the remaining are heterogeneous cases exhibiting “mixed or indeterminate” gene-expression patterns with varying features of these subtypes [21, 38, 39]. The most striking outcome of the CMS classification is the separation of CRC into subtypes representing the following major molecular pathways that also associate with the molecular genetics and epigenetic phenotypes as well as the site of colon at which the CRC originates [40, 41]: (1) CMS1 represents the MSI immune subtype characterized by diffuse immune infiltrate with concomitant immune evasion signatures. (2) CMS2, also known as the canonical subtype, has a strong epithelial signature with activation of Wnt and MYC targets. (3) CMS3 is characterized by deregulation of metabolic signature pathways. (4) CMS4 is characterized by signatures of increased epithelial-to-mesenchymal transition (EMT), activation of TGF-β signaling, extracellular matrix remodeling, angiogenesis, and the complement-mediated inflammatory system (Figure 1).
Figure 1.
Epigenetic features of the various CRC CMS subtypes. Mutation frequencies of key genes (APC, KRAS, BRAF, TP53) and the MSI status and CIMP status of CRC samples in the TCGA dataset (185 samples) available on cBioPortal is related to the consensus molecular subtypes (CMS). Each transcriptional subtype shows distinct molecular and clinical features within the CMS classification. CMS1 (MSI immune) cancers show a high MSI and CIMP status, BRAF mutations, diffuse immune infiltration, and good prognosis but worse survival after relapse. On the other hand, CMS2–4 cancers show an MSS and CIMP-low/negative status. CMS2 (canonical) cancers display a high level of CIN, loss of APC and TP53 mutations, and activation of the Wnt and MYC signaling pathways. CMS3 (metabolic) cancers are characterized by their low level of CIN, overrepresentation of KRAS mutations, and dysregulation of metabolic pathways, including carbohydrate and fatty acid oxidation. Similar to CMS2, CMS4 (mesenchymal) cancers display a high level of CIN, loss of APC and TP53 mutations, and diffuse stromal infiltration along with worse survival overall and after relapse. Figure generated using BioRender. MSI, microsatellite instability; MSS, microsatellite stable; CIMP, CpG-island methylator phenotype; CIN, chromosomal instability; MMR, mismatch repair; EMT, epithelial-to-mesenchymal transition.
Although the CMS subtypes provide a transcriptional basis for the biological features of CRC and relate it to outcomes, it paints a complex picture regarding the conventional molecular subtypes mentioned above, with only CMS1 showing the highest association with the conventional classification in that these tumors are highly enriched for the CIMP-H, MSI-H, and low CIN hypermutator cases (Figure 1). The remaining CRC cases showing CIN and MSS are distributed among the CMS2–4 subtypes. Reflecting the heterogeneous nature of CRC, key genetic mutations that are known drivers of CRC do not show distinct association with the CMS subtypes. The only exceptions to this finding are CRC with mutations in BRAF or KRAS, which are distributed among CMS1 and CMS3, respectively (Figure 1). Such diversity in a transcriptional landscape that is very loosely associated with genetic mutations most likely hints at different cells of origin, which in turn depends on the epigenetic state of the cells, or same cells of origin but with different acquired epigenetic alterations in response to environmental cues. CRC with the BRAF and KRAS mutations, derived from the serrated or conventional adenoma-to-carcinoma pathways (discussed further in detail later), are also represented by distinct epigenetic subtypes that may represent basic differences in transcriptional programs involved in these cancers. The gene-expression signatures of BRAF- and KRAS-mutated tumors typifying the CMS subtypes have been observed in adenomas, the precursors of CRC, indicating that these gene-expression patterns are early changes during CRC evolution that occur before acquisition of all of the relevant cancer-driver mutations [32, 42] (Figure 2). These observations in early adenomas are suggestive again that a significant component of the transcriptional programs contributing to the CMS types may be driven more so by the cell of origin or the epigenetic state of the cells that give rise to tumor clones than by the driver mutations occurring later during adenoma-to-carcinoma conversion.
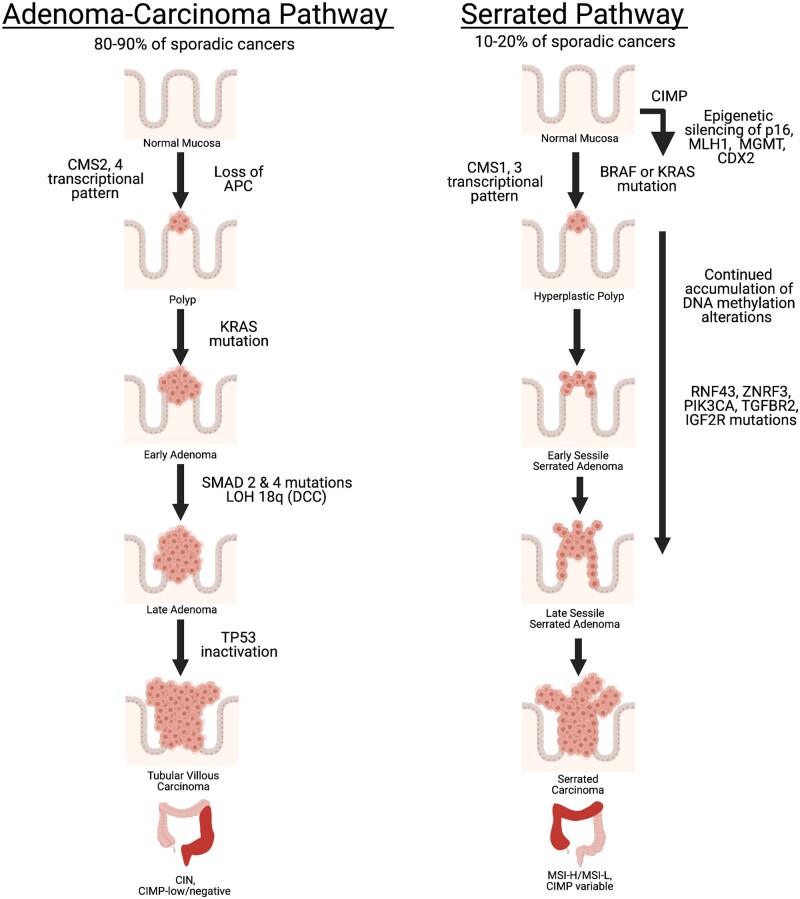
Figure 2.
Molecular dependencies of adenoma–carcinoma and serrated pathways. In the classical adenoma–carcinoma pathway, activation of both the Wnt pathway via APC loss and MEK–ERK pathway via the KRAS oncogene followed by tumor suppressor inactivation of SMAD4 and TP53 develops microsatellite stable (MSS), CpG-island methylator phenotype (CIMP)-low/negative cancers with chromosomal instability (CIN). In the serrated pathway, activation of the MEK–ERK pathway occurs via the KRAS or BRAF oncogenes in parallel with CIMP-high related DNA methylation accumulation. This involves methylation of important genes such as CDKN2A/p16, MLH1, and CDX2. Methylation at the MLH1 promoter results in high microsatellite instability (MSI-H) cancers, while cases lacking MLH1 methylation result in low microsatellite instability (MSI-L) cancers. Other DNA repair genes, such as MGMT, also get affected by promoter methylation. Mutations in various genes most likely occur later during progression of the serrated lesions, leading to dysregulation of the major cancer pathways, including due to inactivation of the RNF43–ZNRF3 axis for Wnt pathway activation. Consensus molecular subtypes (CMS) expression phenotypes may be an early event occurring at the early lesion stage and may have important roles in the path to tumor development. Figure generated using BioRender.
The distinction of CRC into different CMS expression types with no underlying mutational drivers, first, strengthens observations made over the years that CRC is a very heterogenous group of tumors and, second, indicates that although CRC is initiated by common driver events, the manifestation of the expression phenotypes may be subject to various other factors, such as the cell of origin, tissue location, and the role of factors epigenetically predisposing to tumorigenesis, such as aging, inflammation, environmental exposures, metabolic factors, and microbiome [40]. Gaining a clear understanding of the drivers of CMS expression phenotypes warrants further analyses by inclusion of these various other factors.
Genetic and epigenetic contributions to the major pathways of CRC development
Genome-wide exome sequencing studies have revealed a plethora of mutations and epigenetic alterations that are involved in genome maintenance along with the Wnt, MEK–ERK, PI3K, TGF-β, and p53 pathways to be the primary mediators of CRC development [9, 43]. A key insight this model provides is to view tumor initiation as a consequence of the dysregulation of a limited set of these four to five pathways. As discussed in detail below, epigenetic mediated gene silencing plays parallel, if not similar, roles in dysregulation of these pathways and there is a close interdependency between the genetic and epigenetic mechanisms in driving CRC initiation. In addition, the differences in the spectrum of mutations and epigenetic alterations for CRC initiation at various sites along the colon axis starkly reflect different dependencies on disruption of these above pathways (Figure 2). These alterations are early events also observed in the adenomas, and thus closely reflect the major routes to CRC development described below (Figure 2).
Classical adenoma–carcinoma pathway
The majority (50%–70%) of CRC cases arise via the classical adenoma–carcinoma pathway that begins as an adenomatous precursor lesion protruding from the epithelial lining [8]. The incidence and risk for advanced adenomas and carcinomas increase with age, with ∼25% of men in the USA by age 50 years showing adenomas [44–46]. The incidence rate of adenomas in women is 2- to 3-fold lower than that of men [44]. Approximately 10% of these adenomas in both men and women progress to carcinomas when unchecked, indicating various tissue and immune-microenvironment-based controls that prevent development of carcinomas [47]. The pathological and molecular features of a sporadically occurring adenoma–carcinoma pathway closely mimic the CRC arising in the familial adenomatous polyposis syndrome. In most cases of the adenoma–carcinoma sequence, the development of aberrant crypt foci in the initial stages of CRC development involves unregulated activation of the Wnt pathway via an inactivating APC mutation. Next, the founder cells acquire further mutations in KRAS, SMAD4, and TP53 [9, 48, 49] followed by copy number alterations and additional epigenetic changes during development of a benign adenomatous polyp to transition to an advanced premalignant polyp with high-grade dysplastic foci and finally to an invasive carcinoma [50] (Figure 2). A cornerstone of the adenoma–carcinoma pathway is the development of early Wnt independency combined with mutations activating MEK–ERK signaling and inactivating cell-cycle check-point pathways [51]. Wnt independency occurs mainly due to APC mutation, which is found to be inactivated in 75% of CRC cases arising via the conventional adenoma–carcinoma pathway [52–54], and the subsequent dysregulation of MEK–ERK signaling and cell-cycle check points is primarily due to the high likelihood of APC mutations to co-occur with KRAS mutations and/or TP53 mutations. When this co-occurrence is present during progression of the adenoma–carcinoma sequence, APC tends to be the primary mutation and is followed by KRAS mutation, TP53 mutation, or both [55] (Figure 2). The importance of Wnt activation in this progression is exemplified using genetically engineered mouse models which show that sustained activation of the Wnt pathway by loss of Apc function is a key driver of the tumorigenic property as restoration of Apc completely reverses the tumor phenotype by reestablishing normal colon stem-cell differentiation [56]. However, in human CRC, although Wnt activation by APC mutations sufficiently activates Wnt signaling [57], multiple other mutations in the Wnt pathway are observed in the adenomas in both familial adenomatous polyposis patients [58] and sporadic adenomas [59, 60], and the resulting sporadic carcinomas [9]. These mutations include important Wnt pathway genes, such as LRP1B, SOX9, FAT4, TCF7L2, FBXW7, ARID1A, CTNNB1, and AXIN2 (Table 1). Thus, these additional genetic defects in the Wnt pathway suggests dependencies on mechanisms for sustained activation of the Wnt pathway in addition to that from loss of APC function.
Table 1.
Summary of the various gene mutations in CRC characterized in the TCGA [9] and DFCI [175] sequencing projects
| Gene | Sample size (#) | Proximal colon cancer (%) | Distal colon cancer (%) | Rectal cancer (%) | Pathway | Function |
|---|---|---|---|---|---|---|
| MLH1 (P) | 25 | 59.6 | 25.2 | 15.2 | DNA mismatch repair | MMR after DNA replication; forms heterodimer with PMS2 |
| MLH3 (P) | 22 | 68.2 | 14.4 | 17.4 | DNA mismatch repair | MMR after DNA replication; competes against PMS2 to from heterodimer with MLH1 |
| MSH2 (P) | 12 | 48.8 | 23.2 | 28.1 | DNA mismatch repair | MMR after DNA replication; binds to MSH6 or MSH3 to form heterodimer |
| MSH3 (P) | 20 | 54.3 | 36.8 | 8.9 | DNA mismatch repair | MMR after DNA replication; binds with MSH2 to form heterodimer that recognizes insertion-deletion loops |
| MSH6 (P) | 27 | 62.5 | 16.9 | 20.5 | DNA mismatch repair | MMR after DNA replication; binds MSH2 to form heterodimer |
| PMS1 (P) | 16 | 69.5 | 30.5 | 0 | DNA mismatch repair | MMR after DNA replication; forms heterodimer with MLH1 |
| PMS2 (P) | 18 | 49.6 | 31.4 | 19.0 | DNA mismatch repair | MMR after DNA replication; forms heterodimer with MLH1 |
| POLE (P) | 47 | 49.2 | 36.2 | 14.6 | DNA mismatch repair | Involved in chromosomal DNA replication, recombination, and DNA repair via base and nuclear excision pathways |
| APC | 361 | 29.7 | 32.0 | 38.3 | Wnt signaling | Tumor suppressor; regulator of Wnt pathway and involved in cell cycle and cell adhesion |
| AXIN1 | 23 | 63.6 | 20.1 | 16.3 | Wnt signaling | Tumor suppressor; component of β-catenin destruction complex |
| AXIN2 | 49 | 55.1 | 26.9 | 18.1 | Wnt signaling | Tumor suppressor; component of β-catenin destruction complex |
| CTNNB1 | 36 | 49.6 | 31.4 | 19.0 | Wnt signaling | Oncogene; makes protein product β-catenin ligand of Wnt pathway |
| TCF7L2 | 44 | 34.8 | 37.3 | 27.9 | Wnt signaling | Tumor suppressor; TF for many genes by altering the chromatin structure around those genes, suppresses transcription of CTNBB1 |
| ARID1A (P) | 67 | 43.6 | 34.4 | 22.0 | Wnt signaling | Tumor suppressor; TF for many genes by altering the chromatin structure around those genes, suppresses transcription of CTNBB1 |
| FBXW7 | 86 | 35.5 | 30.7 | 33.7 | Wnt signaling | Tumor suppressor; involved in the ubiquitination and degradation of the cell-cycle regulators |
| RNF213 (P) | 80 | 59.3 | 22.5 | 18.2 | Wnt signaling | Tumor suppressor; ubiquitin ligase that acts on frizzled receptors in Wnt pathway |
| RNF43 (P) | 72 | 81.7 | 9.6 | 8.7 | Wnt signaling | Tumor suppressor; ubiquitin ligase that acts on frizzled receptors in Wnt pathway |
| ZNRF3 (P) | 38 | 68.6 | 21.1 | 10.3 | Wnt signaling | Tumor suppressor; ubiquitin ligase that acts on frizzled receptors in Wnt pathway |
| SOX9 | 62 | 41.2 | 25.4 | 33.4 | Wnt signaling | Tumor suppressor; TF for intestinal stem-cell differentiation; promotes ubiquitination and degradation of β-catenin |
| FAM123B/WTX | NA | NA | NA | NA | Wnt signaling | Tumor suppressor; promotes ubiquitination and degradation of β-catenin, stabilizes Axin2 |
| KLF5 | 14 | 38.9 | 27.6 | 33.5 | Wnt signaling | Oncogene; plays a critical role in β-catenin activation by increasing interaction with TCF4 |
| TGFBR1 (P) | 8 | 42.0 | 0 | 58.0 | TGF-β signaling | Tumor suppressor; component of TGF-β receptor |
| TGFBR2 (P) | 29 | 60.3 | 20.8 | 18.8 | TGF-β signaling | Tumor suppressor; component of TGF-β receptor |
| SMAD2 | 32 | 47.4 | 21.4 | 31.2 | TGF-β signaling | Tumor suppressor; downstream signaling molecule for TGF-β signaling, when phosphorylated forms SMAD2-SMAD4 heterodimer to modify transcription of TGF-β target genes |
| SMAD3 (P) | 21 | 39.2 | 30.9 | 29.9 | TGF-β signaling | Tumor suppressor; downstream signaling molecule for TGF-β signaling, when phosphorylated forms SMAD3-SMAD4 heterodimer to modify transcription of TGF-β target genes |
| SMAD4 | 73 | 39.8 | 34.1 | 26.1 | TGF-β signaling | Tumor suppressor; downstream signaling molecule for TGF-β signaling, when phosphorylated forms SMAD2/3-SMAD4 heterodimer to modify transcription of TGF-β target genes |
| ACVR2A (P) | 59 | 73.6 | 19.6 | 6.8 | TGF-β signaling | Activin receptor type 2A; component of activin receptor; complex phosphorylates SMAD2/3 |
| ACVR1B (P) | 29 | 40.7 | 31.8 | 27.5 | TGF-β signaling | Activin receptor type 1B; component of activin receptor; complex phosphorylates SMAD2/3 |
| ATM | 64 | 53.9 | 29.5 | 16.6 | TGF-β signaling | Tumor suppressor; kinase that phosphorylates multiple targets including TGF-β receptor to activate TGF-β signaling and p53 to activate DNA damage pathways |
| ERBB2 (HER 2) (P) | 38 | 45.6 | 32.5 | 21.9 | MAPK signaling | Oncogene; EGFR; activates oncogenic Ras–Raf–MEK–ERK signaling pathway |
| ERBB3 (HER3) (P) | 36 | 51.0 | 11.6 | 37.5 | MAPK signaling | Oncogene; EGFR; activates oncogenic Ras–Raf–MEK–ERK signaling pathway |
| ERBB4 (HER4) (P) | 37 | 42.2 | 40.1 | 17.7 | MAPK signaling | Oncogene; EGFR: activates oncogenic Ras–Raf–MEK–ERK signaling pathway |
| KRAS (D) | 173 | 39.3 | 28.7 | 31.9 | MAPK signaling | Oncogene; component of oncogenic Ras–Raf–MEK–ERK signaling pathway |
| NRAS (P) | 27 | 31.0 | 36.2 | 32.9 | MAPK signaling | Oncogene; component of oncogenic Ras–Raf–MEK–ERK signaling pathway |
| BRAF (P) | 127 | 71.9 | 21.9 | 6.1 | MAPK signaling | Oncogene; component of oncogenic Ras–Raf–MEK–ERK signaling pathway |
| IGF1 (P) | 4 | 56.6 | 0 | 43.4 | PI3K signaling | Oncogene; ligand for IGF1R; Regulates cell proliferation |
| IGF2 (P) | 4 | 100.0 | 0 | 0 | PI3K signaling | Oncogene; ligand for IGF1R; Regulates cell proliferation |
| IGF1R (P) | 26 | 72.4 | 12.5 | 15.1 | PI3K signaling | Oncogene; RTK receptor phosphorylates ISR1 and ISR2 |
| IRS1 (P) | 47 | 44.8 | 14.1 | 41.1 | PI3K signaling | Oncogene; once phosphorylated, activates PI3K-AKT/mTOR pathway and Ras–Raf–MEK–ERK signaling pathway |
| IRS2 (P) | 15 | 35.0 | 24.8 | 40.2 | PI3K signaling | Oncogene; once phosphorylated, activates PI3K-AKT/mTOR pathway and Ras–Raf–MEK–ERK signaling pathway |
| PIK3CA (P) | 132 | 50.1 | 35.6 | 14.3 | PI3K-signaling | Oncogene; triggers PI3K-Akt/mTOR pathway |
| PIK3R1 (P) | 30 | 50.2 | 38.2 | 11.6 | PI3K signaling | Tumor suppressor; regulates PI3CA gene |
| mTOR (P) | 50 | 47.4 | 16.3 | 36.3 | PI3K signaling | Oncogene; kinase activating PI3K-AKT/mTOR pathway |
| AKT1 (P) | 11 | 81.3 | 0 | 18.7 | PI3K signaling | Triggers PI3K-Akt/mTOR pathways that have a central regulatory role in promoting cell growth and proliferation and inhibiting apoptosis |
| PTEN (P) | 51 | 51.6 | 28.0 | 20.4 | PI3K signaling | Tumor suppressor; negatively regulates PI3K-ATK/mTOR pathway |
| TP53 (p53) (D) | 320 | 24.2 | 36.3 | 39.5 | p53 signaling | Tumor suppressor; regulates cell cycle and activates DNA damage pathways |
| MYC (D) | 12 | 59.6 | 25.2 | 15.2 | MYC signaling | Oncogene; codes for family of TFs |
The most frequent mutations were obtained from cBioPortal and compiled according to the weighted incidence in the various sections of the colon and rectum. Key pathways associated with the genes are listed. A weighted incidence was calculated by assuming an equal representation of each section of the colon and rectum in the sample size and accounting for both the overrepresentation of proximal CRC and underrepresentation of distal and rectal CRC in the original data. Gene names followed by "D" in parenthesis have higher tendency to be mutated in distal colon cancers while those followed by "P" in parenthesis tend to be mutated in proximal colon cancers.
CRC, colorectal cancer; MMR, mismatch repair; TF, transcription factor.
Another modality through which the Wnt pathway is activated is epigenetic silencing of Wnt pathway negative regulators. Epigenetic changes, especially DNA methylation occurring at promoters that mediate gene silencing, are very early changes observed in the adenomas [61–64]. Many of the genes affected by promoter DNA methylation in cancers are already methylated in early adenomas and in the normal tissue adjacent to the cancers (field defect) [65, 66]. The negative regulator genes of the Wnt pathway that are affected by epigenetic inactivation are often important in developmental and differentiation pathways and have also been found to be methylated during aging [67–71]. As such, these additional modalities of Wnt activation, the methylation mediated inactivation of Wnt pathway negative regulators and supplementary mutations in other Wnt pathway genes aside from APC, may thus synergize with APC inactivation to maintain Wnt activation at multiple levels. These additional modalities of Wnt activation might be relevant in the context of the spectrum of APC mutations that activate the Wnt pathway to different levels [52]. This is especially relevant while considering observations that tumors with mutations in APC can still be responsive to Wnt ligands that suppress external Wnt signaling [72]. In the intestinal epithelial microenvironment, the antagonistic gradient of Wnt (stem-cell activating) and BMP signaling (differentiation inducing) may thus necessitate the need for multiple cell intrinsic mechanisms for Wnt pathway activation in addition to the APC mutation for selection of clones with stronger Wnt activation. Herein, additional mutations affecting the Wnt pathway, and especially epigenetic alterations, may provide this further impetus to early Wnt independency. Understanding the spectrum of molecular changes affecting the Wnt pathway in CRC will be critical in designing strategies for targeting its activation in cancer treatment.
Serrated pathway
About 15%–20% of CRC cases arise through the serrated pathway wherein the precursor lesions exhibit the distinguished serrated (saw-tooth-like) morphology in the epithelial glands [73, 74]. These early lesions present as hyperplastic polyps and sessile or traditional serrated adenomas, which arise sporadically or in the context of serrated polyposis syndrome. In stark contrast to the tubular adenomas in the adenoma–carcinoma pathway, in which APC inactivation is a founder mutation for Wnt activation leading to the formation of the early tubular adenoma lesions, sessile serrated adenomas tend to significantly lack APC mutations [58, 59, 75]. Instead, mutations in KRAS, or more commonly BRAF, activate the RAS–RAF–MEK–ERK–MAPK axis as these are the earliest mutations observed in sessile serrated adenomas and its precursor hyperplastic polyps [76–84] (Figure 2). In mouse models, these mutations by themselves are not sufficient to trigger intestinal tumorigenesis, except over prolonged periods (∼1 year) in the small intestine [85] or colon [4]. Further, it has been shown that oncogenic KRAS and BRAF mutations trigger oncogene-induced senescence (OIS) via the p16-RB and ATM–ATR-mediated DNA damage response pathways [86–88]. Therefore, since OIS is one of the earliest tumor suppressor mechanisms that is triggered in response to activation of the MEK–ERK pathway by oncogenic mutations in KRAS or BRAF, founder cells have to first overcome OIS for tumor initiation to occur [85, 89, 90]. Although, in principle, oncogenic BRAF can induce hyperplasia, which may accompany induction of OIS [85], these hyperplastic cells continue to proliferate while accumulating further genetic and epigenetic alterations, and develop into adenomas and then progress to carcinomas [4]. In these mouse models, progression to carcinomas requires additional genetic and epigenetic changes that intensify MAPK signaling, Wnt pathway activation, and inactivation of cell-cycle and senescence pathways by epigenetic silencing of CDKN2A/p16 [85].
Analysis of the molecular changes in the human serrated polyps has shown that early genetic and epigenetic alterations play profound roles in overcoming the OIS response and promoting early Wnt pathway activation in the absence of APC mutations, which forms the basis for progression of hyperplastic polyps to serrated lesions to carcinomas. One of the major regulators of OIS is the product of CDKN2A/p16, which is silenced by DNA methylation during progression to advanced serrated lesions [91]. The CIMP-H, which includes a high frequency of methylation in the CDKN2A/p16 promoter, evolves during the progression from hyperplastic polyps to sessile serrated adenomas [92] (Figure 2). Analyses of individuals with serrated polyposis, i.e. incidence of multiple sessile serrated adenomas simultaneously, have identified germline alterations in genes including ATM, RBL1, XAF1, PIF1, TELO2, and RNF43, which are involved in OIS through roles in DNA repair, cell cycle, apoptosis, genome stability, and Wnt regulation [93]. Herein, inactivating mutations in RNF43 (RING-type E3 ubiquitin ligase) is an important cancer-driver mutation, not only in the familial serrated polyposis cases [93, 94], but also in the sporadically occurring BRAF-mutant MSI-H CRC [20, 94–97]. Mechanistically, RNF43 functions as a negative regulator of the Wnt pathway by ubiquitination of Wnt receptors and targeting them for degradation in colon stem cells [98]. Interestingly, although inactivation of RNF43 may play a role in suppressing OIS induction [93], its loss in the serrated pathway is more likely to function in activating the Wnt pathway. RNF43 mutations have been observed in BRAF-mutant sessile or traditional serrated adenomas, but not in their KRAS-mutant counterparts [99], suggesting that the alternate modalities of MEK–ERK pathway activation may determine the method by which the Wnt pathway is activated.
Furthermore, various features of the RNF43 mutations highlight the importance of early CIMP development in driving RNF43 mutations. This finding comes from observations that while CRCs arising in the familial serrated polyposis patients have BRAF mutation and are predominantly MSS, the sporadic CRC arising via the serrated pathway are predominantly MSI and CIMP-H. Particularly, the MSI-related DNA repair defects in the mismatch repair pathway are associated with development of RNF43 mutations in the sporadic cases as the RNF43 mutations identified in the sporadically occurring early serrated polyps and CRC derived via the serrated pathway harbor truncating mutations in mononucleotide repeats [94]. These mutations are a signature of the MSI phenotype due to loss of MLH1, which in the CIMP-H cases most frequently occurs due to DNA methylation of MLH1 promoter [100] (Figure 2). In contrast to the sporadic serrated pathway, the familial serrated lesions with BRAF mutations that are MSS, i.e. without defects in the mismatch repair pathway, have been shown to harbor non-repeat tract mutations in RNF43. These studies have highlighted the importance of inactivating RNF43 in the progression of the sporadic, BRAF-mutant, MSI-H, CIMP-H, and/or MLH1-methylated CRC arising via the serrated pathway. Particularly, these studies indicate that RNF43 mutations occur via the hypermutator pathway due to silencing of MLH1 by promoter methylation in the CIMP-H context.
Distinct modes and roles for Wnt pathway activation in the two major CRC types
The importance of Wnt pathway activation as an early genetic dependency for progression of the serrated pathway is further highlighted by the occurrence of genetic alterations involving RSPO fusions in adenomas and CRC arising from the serrated pathway [94, 97, 101]. These RSPO fusions lead to overexpression of the secreted factor R-spondin, which amplifies Wnt signaling by negatively regulating RNF43 and ZNRF3. Recent analysis of a panel of CRC-specific mutations discriminates the traditional serrated adenoma lesions from sessile serrated adenoma lesions as those that carry more frequent mutations in the Wnt pathway, interestingly indicating different dependencies on Wnt pathway mutations within the serrated pathway [99]. These studies firmly show that BRAF-driven CRC arising from the serrated pathway has an early dependency on Wnt activation by inactivating the RNF43–ZNRF3 axis. Therefore, mutations in components of the Wnt pathway have very early roles in the serrated pathway, similar to the role of APC mutations in driving the adenoma–carcinoma pathway in CRC. However, a major distinction in this parallelism is that Wnt-pathway-activating APC mutations have precursor roles in the adenoma–carcinoma pathway, while in the serrated pathway, the Wnt-pathway-activating mutations mentioned above most likely occur after BRAF mutations. In this sense, disruption of the RNF43–ZNRF3 axis in the serrated pathway may serve similar, but not identical, functions to the APC mutations do in the adenoma–carcinoma pathway, which may have implications for the therapeutic targeting of the Wnt pathway in different CRC subtypes.
In addition to dependency on the acquisition of mutations in RNF43 via promoter DNA methylation mediated silencing of MLH1, epigenetic defects in the regulation of the Wnt and differentiation pathways may also have early and profound roles in tolerating the increased MEK–ERK signaling by oncogenic BRAF. For example, CDX2 has been shown to be downregulated in CIMP-H CRC and is preferentially methylated in this CRC subtype [102–106]. In mouse models, inactivating Cdx2 in the context of oncogenic BRAF (BRAFV600E, mouse-human hybrid version of the V600E oncogene) leads to serrated lesions and tumor development [107]. Cdx2 loss in BRAFV600E inducible colon organoids allows overcoming senescence upon induction of BRAFV600E [5]. In these studies, ex vivo aging of mouse colon organoids involves evolution of promoter DNA hypermethylation, which facilitates activation of the Wnt pathway and predisposes transformation by oncogenic BRAF. Further, simultaneous inactivation of genes subject to frequent epigenetic silencing in human CIMP-H CRC, such as CDX2, SFRP4, SOX17, and CDKN2A, in freshly isolated organoids results in rapid induction of tumorigenesis upon BRAFV600E. Otherwise, in the same model, transformation takes ∼5 months after BRAFV600E induction, during which time DNA methylation alterations occur across the genome, including promoter methylation of these same genes [5]. Thus, the clinical genetics observations of CRC, and experimental observations from both in vivo mouse studies and ex vivo models, suggest that initiation of the serrated route has high reliance on epigenetic inactivation of regulators of the Wnt and senescence pathways, which allows oncogenic BRAF mutations to effectively drive tumorigenesis.
The key difference between the adenoma–carcinoma pathway and serrated pathway to CRC development may consequently rely on the nature of Wnt activation signals via accumulation of either mutations or sufficient epigenetic alterations by the founder cells. The extent and nature of the epigenetic alterations observed in CRC arising from these two pathways suggest different dependencies between the mutations and epigenetic alterations during CRC development. The fact that downstream activation of MEK–ERK signaling by BRAF shows a preference for the serrated pathway is intriguing and begs the question as to why this is the case. These insights into the molecular genetics and epigenetics of CRC development via the two major pathways, which account for the majority of CRC cases, will help refine strategies for early diagnosis and prognosis that can use a range of genetic markers, such as RNF43, ZNRF3, and RSPO fusions, in addition to methylation-based markers, such as CDX2, CDKN2A, and various Wnt regulator genes. Further, key differences in Wnt pathway activation could be exploited for therapeutic intervention with Wnt inhibitors that act at different levels of the Wnt pathway in combination with other targeted MEK–ERK and epigenetic inhibitors.
Factors contributing to epigenetic changes in cells of the gastrointestinal tract
The DNA methylation alterations at promoter and other regulatory elements occur during various normal physiological processes as these epigenetic mechanisms orchestrate gene-expression changes in response to metabolic and environmental signals, including those derived from intracellular and extracellular environments. In totality, these epigenetic alterations manifest as age-related methylation changes and may reflect the cumulative impact of exposure to various environmental signals [108]. Individual exposures that directly affect the colon are important in understanding how the ensuing epigenetic changes may alter gene expression and predispose us to CRC. Below we discuss some of these factors that lead to accumulation of epigenetic changes and are known risk factors for CRC (Figure 3).
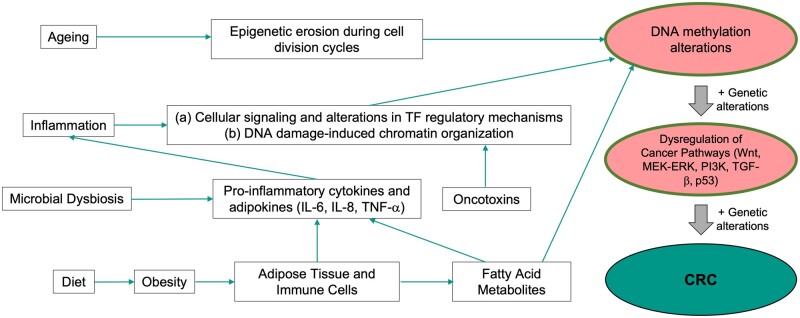
Figure 3.
Various factors contributing to DNA methylation alterations and linked to the risk for CRC development. Various physiological and environmental risk factors (e.g. microbiome, obesity and diet, inflammation, and aging) contribute to the early acquisition of epigenetic alterations over time, eventually leading to cancer initiation following genetic driver mutations. These factors together are linked to the various macro-environmental exposures that contribute to the age-related DNA methylation changes. CRC, colorectal cancer; TF, transcription factor.
Aging
The normal maintenance and regeneration of tissues during mammalian growth are associated with the accumulation of epigenetic changes involving both hypermethylation and hypomethylation of CpG-residues. Globally, the human genome contains ∼28 million CpG-sites unevenly distributed across the genome [109]. Of these CpG-sites, a minor subset (∼7%) are enriched in CpG-islands, which in normal cells are protected from DNA methylation [110]. About 75% of the remaining CpG-sites, scattered in the genic and non-genic regions of the genome, are normally methylated in somatic cells. During aging, substantial proportions of these non-CpG-island-associated CpG dinucleotides undergo demethylation, tending to occur at specific sites and regions of the genome [111]. These hypomethylation changes impact various aspects of genome structure and integrity, leading to deregulation in transcription and genome stability, which are important for cancer initiation and progression. On the other hand, the CpG-sites in CpG-islands, ∼50% of which are associated with gene promoter regions and otherwise normally unmethylated, gain methylation during aging [69, 112, 113]. This age-related hypermethylation occurs in CpG-sites that are associated with genes involved in development and differentiation [69, 114, 115]. Normally these genes are maintained in a repressive state by the H3K27me3 chromatin modification in progenitor cells, which in very primitive stem cells, like embryonic stem cells, co-occur with the active H3K4me3 modification termed the bivalent chromatin mark [116, 117]. Furthermore, the target sites associated with age-related hypermethylation are enriched for specific transcription factors. For example, during aging, leukocyte DNA undergoes hypermethylation at sites that are enriched for genes. These genes are predicted to be regulated by groups of transcription factors involved in biological functions related to development [118]. This phenomenon pans across species, including humans, dogs, and bats, which all show age-related methylation changes at regions related to developmental processes [118–121]. Importantly, these same regions show distinct and amplified hypermethylation in human cancers, indicating common mechanisms, and that the age-related methylation changes may induce tumorigenesis [5, 67]. The acquisition of age-related hypermethylation of multiple genes in colon cancer is important for studying colon cancer development. As mentioned earlier, methylation of genes such as CDX2, SFRP4, SOX17, CDKN2A, MLH1, and SFRP1 are observed during normal aging [5] and are important in the development of CRC. We have shown that inactivation of some of these genes predisposes colon stem cells to tumor initiation [5]. Herein, it is possible that subclones of cells that have acquired promoter hypermethylation simultaneously in many of such genes during aging may be more prone to undergo tumorigenesis in the context of a pre-existing genetic mutation or acquired genetic mutations.
The mechanisms by which age-related hypermethylation and hypomethylation accumulate are not clear [122]. Aging involves organ growth and development until adulthood, and later organ and tissue maintenance in the context of constant tissue damage and replenishment by stem-cell divisions and tissue repair. The constant cell division in the context of environmental exposures (such as pollutants, cigarette smoke, microbial exposures including infections as well as normal flora) and physiological exposures (such as obesity and hormonal changes) is an important component of aging. Multiple studies have shown that constant cell divisions lead to global methylation changes involving genome-wide hypomethylation at intergenic and intragenic regions, along with hypermethylation at the CpG-island promoter elements [123–125]. Specifically, hypomethylation has been identified as a feature of mitotic cell division, occurring at CpG-residues in specific sequence contexts and in the late replicating DNA [125]. The replicating regions during the late S phase constitute the heterochromatic regions of the genome that are heavily methylated and replicate in large replication factories consisting of the enzyme machinery for DNA replication as well as the DNA methyltransferase (DNMT)1 [126]. DNMT1 is purported to be a catalytically slow enzyme [127] and due to the large numbers of CpG-residues in the late replicating DNA that have to be methylated during the S phase at each cell division, it is hypothesized that gradually certain residues are not faithfully methylated. Such passive loss of methylation during subsequent cell-division cycles may result in progressive hypomethylation. By contrast, regions that get hypermethylated, which are mainly in the promoter and other regulatory regions, may potentially acquire hypermethylation due to multiple cellular pathways that impinge upon transcription factor expression changes and DNA-damage-induced chromatin reorganization. The acquisition of methylation at promoter CpG-sites at these genes that are normally regulated by polycomb group proteins (primarily establishing and maintaining the H3K27me3 mark) is correlated with the number of stem-cell divisions in different organs, and accordingly can be used to determine the mitotic age of a tissue [128]. These studies have suggested that cancerous and pre-cancerous lesions can be discriminated from normal tissues by estimating the mitotic age of tissues, which tends to be higher in the former indicating increased cell divisions. Overall, current models suggest that the aging-related methylation changes arise due to inefficiencies in maintaining the epigenetic information during mitotic cycles in the context of exposures that alter chromatin and cellular signaling pathways. Various factors impacting the epigenome, discussed below, may directly fit into the broad category of such exposures to the stem-cell microenvironment that maintains tissue homeostasis.
Microbiome
The gut microbiome represents an important microenvironmental exposure component essential in normal functioning of the colon and is also a strong modulator of CRC development (Figure 3). Distinct microbial taxonomic subgroups are altered during adenoma-to-carcinoma development [129, 130]. These studies suggest that microbial dysbiosis can occur early in the adenoma stage and that diet especially affects this dysbiosis in a manner such that consumption of red meat is associated with a hostile gut microbiome compared with the high-fiber fruits and vegetables diet. In direct relation to CRC development, individual species of bacterial populations have been shown to express pro-oncogenic genes, such as Bacteroides fragilis toxin (BFT), also called oncotoxins, that promote tumorigenesis [131–136]. In addition to the direct impact on CRC development by such oncotoxins, various studies have shown that the gut microbiome along with its metabolites and toxins can directly interact with epigenetic machinery and reprogram the epigenome [137, 138]. In this context, the microbiome has an important role in modulating epigenetic changes and normal postnatal development of the intestine [139]. In healthy individuals, the normal gut microbiome plays important roles in homeostasis of the intestinal epithelium by directly influencing the transcriptome and preventing inflammation [137, 139]. In contrast, dysbiosis of the gut microbiome is known to cause chronic inflammation, which could alter the epigenome and increases the risk for CRC (Figure 3). As evidence, dysbiosis in the gut due to antibiotic usage leads to an increased risk of CRC [140–142]. While the gut microbiome may cause these changes via directly affecting the immune and cytokine balance in the epithelial microenvironment [143, 144], it also reprograms the epigenome, resulting in gene-expression programs that are pro-oncogenic. Relative proportions of defined microbial species in the gut microbiota of CRC patients have been linked to methylation/demethylation changes at specific regions in the genome of epithelial cells in CRC [145]. Such roles for the CRC-associated microbiome in altering the methylation landscape of the colonic epithelial cells have been observed in fecal transplant studies, wherein the microbiota from the CRC patients when transferred to the colon of mice induced increased aberrant crypt foci and marked changes in the methylation of gene promoters in the recipient gut epithelium of the mice. Some of the genes that were identified to have altered methylation in this model are important in stem-cell functions belonging to the Wnt and Notch pathways [145].
Further, the role for microbial populations in CRC, potentially by modulating the epigenome, is indicated by studies showing that the CIMP-H CRC cases in the proximal colon are enriched in Fusobacterium species [146, 147]. Cancers in the proximal colon and the normal tissues from these patients have been observed to harbor distinct, invasive bacterial aggregates, called biofilms, that are associated with increased proliferation [148]. Transfer of biofilm from tumors or normal tissue of these CRC patients to APCMin mice models induces inflammation and tumor initiation [149]. In experiments comparing genome-wide expression and methylation changes in the small intestines of germ-free and/or conventional microbiota-containing mice, the presence of microbiome is associated with large changes in the epithelial transcriptional programs, of which many genes were associated with methylation changes and were enriched for processes involving intestinal epithelial proliferation and regeneration [138]. Specifically, the important tumor suppressor gene Rb1 was observed to be downregulated in the presence of microbiome. In these studies, the tumor protective vs promoting role of the conventional microbiota is unclear as Rb1 is downregulated. Further, the presence of the BFT has been directly associated with methylation changes in important tumor suppressors, such as Hoxa5, Polg, Runx1, Runx3, CD37, Stx11, Tceb2, Lgr6, Cdx1, and Fut4, in tumors forming in mice [150]. Interestingly, in support of the close relation between the microbiome changes associated with CRC, specific bacterial taxonomic groups, assessed by 16S rRNA sequencing, were associated with the CMS subtypes. For example, the CMS1 subgroup that is enriched for the CIMP-H subgroup is associated with increased colonization by Fusobacterium hwasookii, Porphyromonas gingivalis, Fusobacterium nucleatum, Parvimonas micra, and Peptostreptococcus stomatis; the CMS2 subgroup is enriched for Selenomas and Prevotella species [151]. Overall, there is building evidence for the microbiome influencing DNA methylation during normal epithelial homeostasis as well as especially during tumor development.
Molecularly, there are potentially multiple mechanisms by which a normal microbiome and conditions of dysbiosis affect the host epithelial epigenome. It has been demonstrated that bacterial dysbiosis and the oncotoxins cause inflammation and increased DNA damage. In this context, DNA damage is associated with genome-wide relocalization of epigenetic silencing complexes containing histone modifiers (polycomb repressive proteins) and DNA methyltransferases (DNMT1 and DNMT3B) at CpG-islands [152]. The recruitment of DNMT1 to the CpG-islands is directly mediated by DNA mismatch repair proteins [150, 153]. The affected CpG-islands include promoters of genes such as SFRP4, MLH1, SFRP5, and SOX17, which are known to be methylated in colon cancers and important in tumorigenesis. In addition to the methylation changes at the promoter CpG-islands, exposure to BFT causes genome-wide changes in chromatin accessibility, which is indicative of epigenetic reprogramming involving altered transcriptional programs regulated by changes in enhancer elements [154]. Further, gut microbes produce millimolar amounts of short-chain fatty acid metabolites, such as acetate, propionate, and butyrate, produced as a result of fermentation of dietary fibers [155, 156]. These metabolites are a direct source for acetyl-CoA, which is a key substrate for histone acetyl transferases, and butyrate, which is known to affect histone modifications by directly acting as an inhibitor of histone deacetylases (HDAC) that play an important role in recruiting the DNMTs to specific regions of the genome (Figure 3). In the proximal colon of mice, a Western diet, rich in fat and sucrose containing reduced amounts of fermentable dietary fibers, has reduced SCNAs and altered histone modifications compared with the regular chow diet [157]. The gut microbiome, in combination with an obesity-inducing diet, alters the histone modifications in enhancer regulatory elements of colon epithelial cells [158]. This in turn alters the expression of transcription factors, and the resulting changes in gene expression are similar to gene-expression changes during CRC development. Thus, mechanistically, the microbiome modulates the intestinal epigenome via a multitude of interactions with the host tissue, and the key challenges are to determine the epigenome modulating roles of the individual components, such as the direct effect of oncotoxins, interactions with the epithelial and immune components, inflammation, and the role of microbial metabolites. A cumulative understanding of how the ensuing epigenetic changes impact colon stem-cell maintenance and differentiation is important to understand CRC risk.
Metabolic influences: obesity and diet
Obesity is one of the major risk factors for colon cancers [159–161]. Adipose tissue is enriched in immune cells such as macrophages and secretes pro-inflammatory adipokines (e.g. tumor necrosis factor [TNF]), which results in chronic low-grade systemic inflammation [160, 162]. Several studies have shown that obesity directly influences gene-expression changes with important roles in predisposing to tumorigenesis. Most direct analyses on the effect of obesity on epigenetic alterations come from studies in mice, wherein it has been shown that obesity is directly associated with an increase in pro-oncogenic signals, such as RAS, PI3K, and JNK signaling pathways. Changes in these pathways are brought about by alterations in the histone H3K27-acetylation (H3K27ac) mark at enhancer regulatory regions in the genome, which is an important indicator of active enhancer regulatory elements that control expression of target genes from a distance [163]. Obesity induces significant loss of H3K27ac at enhancer regions that are specific to colon tissue, and the loss of activity of these enhancer elements is similar to that observed in colon cancers [163, 164]. The transcriptomic and epigenomic alterations may result from the direct impact of diet as well as due to obesity-induced metabolic and inflammatory changes. Diet is known to affect metabolism and the availability of different species of long-chain fatty acids, and the latter may directly interact with cell signaling and epigenetic machineries. For example, butyrate is an important product of metabolizing a low-fat diet, which in addition to being an energy resource is also a known modulator of HDAC. Further, obesity-inducing high-fat diets in mice stimulate an increase in Lgr5+ colon stem cells and stem-cell function, via activation of PPAR-δ target genes, which are involved in the oxidation of long-chain fatty acids [165]. Mice fed with an obesity-inducing diet primarily show significant changes in expression of genes involved in metabolic processes, such as lipid metabolism, carbohydrate metabolism, and energy production, and importantly these genes show similar upregulation or downregulation in human CRC. Administration of a high-fat diet involves a shift from butyrate-dependent metabolism, which inhibits intestinal stem/progenitor cell proliferation, to metabolism of the stem cell promoting long-chain fatty acid [166]. Importantly, these studies have shown that, in parallel with metabolic changes, a high-fat diet induces gains and losses of DNA methylation at regulatory elements throughout the genome. These studies have reported that the majority of DNA methylation gains and losses are associated with enhancer regulatory elements, which potentially control cancer-related metabolic genes. Over the long term, these methylation changes are of important consequence as they impact future gene-expression changes. Thus, obesity and high-fat diet are associated with alterations in both histone and DNA methylation changes with important consequences for increasing risk of colon cancer. The mechanisms by which obesity alters DNA methylation are not clear and, as mentioned earlier, may involve direct impact on epigenetic enzymes by the metabolites of fatty acid metabolism or due to inflammatory signals induced by the obesity-associated adipose tissues. In regard to the latter, adipocytes release a number of hormones, as well as attract immune cells that may create an inflammatory environment resulting in high levels of pro-inflammatory cytokines, such as IL-6, IL-1β, and TNF-α, released by the adipocytes as well as the infiltrating immune cells [162]. Such alterations of the inflammatory environment linked to obesity can impact the epigenome, as discussed further in the inflammation section (Figure 3).
Inflammatory microenvironment
Inflammation can occur on its own, due to the gut microbiome, or due to diet and obesity, and it is a very strongly associated factor with the cancer microenvironment. The relationship between inflammation and CRC has been shown to be reciprocal in nature as chronic inflammation increases the risk of cancer initiation and cancer progression often leads to an inflamed microenvironment [167]. Accordingly inflammatory genes, such as IL-1, IL-6, IL-17A, and IL-23, are increased in most sporadic CRC cases [168]. Looking more closely at the relationship between inflammation and CRC, it is well known that patients with chronic inflammation in their bowels are at a higher risk for CRC. Diseases that result in inflammation of the bowels are majorly inflammatory bowel diseases (IBD) such as Crohn’s disease and ulcerative colitis. Meta-analysis of population-based cohort studies has shown that the incidence of CRC is positively correlated with worse IBD severity and longer disease duration with cumulative risks of CRC at 1%, 2%, and 5% after ≤10, ≤20, and >20 years of IBD diagnosis, respectively [169]. Further, obesity also results in chronic low-grade inflammation as well as alterations in the gut microbiome as mentioned above. Chronic inflammation has been shown to speed up the rate of age-related epigenetic changes via aberrant DNA methylation patterns seen in sporadic CRC [170]. A temporal analysis of methylation patterns based on the infection-associated inflammatory response to Helicobacter pylori shows that several inflammation-related genes, such as CXCL2, IL-1b, NOS2, and TNF-α, positively correlate with temporal changes in methylation levels. Further, when H. pylori is administered but the infection-associated inflammatory response is suppressed using the immunosuppressive drug, cyclosporin A, alterations in DNA methylation no longer occur, implying that inflammation itself is inducing the aberrant methylation changes [171].
One mechanism for these aberrant methylation changes may be due to higher expression levels of DNMT1, the enzyme that methylates and silences genes, in inflamed mucosa. Treatment of colon cancer cells with the inflammatory biomarker IL-6 alone results in an increase in DNMT1 expression independently of de novo gene expression as well as an increase in methylation of the promoter regions of tumor suppressor genes, leading to their downregulation. This downregulation in expression can be prevented by pre-incubation with a DNMT1 inhibitor, showing that inflammation-associated changes in DNA methylation patterns may occur through the activity of DNMT1 [172]. Studies have suggested that the mechanism by which inflammation promotes CRC may occur through the methylation of specific genes. In particular, the inflammatory NF-κB and STAT3 signaling pathways play an important role by secreting a large number of pro-inflammatory cytokines into the cell microenvironment (Figure 3). These pro-inflammatory cytokines include IL-6 and TNF, which aid in the methylation of distinct tumor suppressor genes including ITGA4, TFPI2, VIM, SOCS3, p14ARF, p16INK4A, and PTX3 that promote malignant transformation [168].
The cytochrome P450s CYP2E1 and CYP1B1 along with FXR and VDR are also associated with inflammatory conditions and deficiency via methylation, which may allow weakened defenses against carcinogens, resulting in carcinogenesis [167]. Inflammation-induced epigenetic silencing of these genes typically occurs by either of two mechanisms. The first mechanism is hypermethylation of CpG-island promoters, which results in the CIMP. Comparing the methylation rates for seven CpG-island sites in mucosae of individuals who are healthy and individuals infected by H. pylori inducing inflammation, Maekita et al. [173] show that infection significantly increases methylation of CpG-islands to various levels and that these methylation levels of specific CpG-islands were predictive of cancer risk in health individuals. The second mechanism of methylation is a novel epigenetic program identified by Abu-Remaileh et al. [174] using monoalkyl methylation maps of a colitis-induced mouse colon cancer model. This program is characterized by hypermethylation of DNA methylation valleys (DMVs), which leads to epigenetic silencing of DMV-associated genes. Ultimately, inflammation-induced epigenetic silencing via hypermethylation of CpG-island promoters or DMVs aids in the malignant transformation of inflamed mucosa to CRC. Lastly, inflammation has also been shown to promote early epigenetic modifications through histone modification, microRNAs, and lncRNAs; however, we have chosen to focus on DNA methylation specifically for this review.
Conclusion
The long debate as to what constitutes the early dependencies for CRC development is still ongoing. It is becoming clear that the modes of Wnt pathway activation may rely on different mechanisms in the two major pathways of CRC. Although epigenetic changes seem to play important roles in the classical pathway, the serrated pathway appears to be more dependent on early epigenetic changes. Understanding how an array of environmental and physiological factors affect DNA methylation in colon epithelial cells, and defining the gene promoter and regulatory elements involved, will be important in understanding and preventing the risk for CRC in the future.
Authors’ Contributions
S.P. and H.E. conceived and wrote the manuscript. Both authors read and approved the final manuscript.
Funding
This work was supported by the National Cancer Institute and National Institute on Aging at the National Institutes of Health [NIH/NCI R01CA229240; NIH U01AG066101].
Acknowledgements
None.
Conflict of Interest
None declared.
Contributor Information
Sehej Parmar, Cancer Genetics and Epigenetics, Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Hariharan Easwaran, Cancer Genetics and Epigenetics, Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
References
- 1. Arnold M, Sierra MS, Laversanne M. et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683–91. [DOI] [PubMed] [Google Scholar]
- 2. Chan DKH, Buczacki SJA.. Tumour heterogeneity and evolutionary dynamics in colorectal cancer. Oncogenesis 2021;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo M, Peng Y, Gao A. et al. Epigenetic heterogeneity in cancer. Biomark Res 2019;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rad R, Cadinanos J, Rad L. et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell 2013;24:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tao Y, Kang B, Petkovich DA. et al. Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and Braf(V600E)-induced tumorigenesis. Cancer Cell 2019;35:315–28.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yi JM, Dhir M, Van Neste L. et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res 2011;17:1535–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pancione M, Remo A, Colantuoni V.. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int 2012;2012:509348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamagishi H, Kuroda H, Imai Y. et al. Molecular pathogenesis of sporadic colorectal cancers. Chin J Cancer 2016;35:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sawayama H, Miyamoto Y, Ogawa K. et al. Investigation of colorectal cancer in accordance with consensus molecular subtype classification. Ann Gastroenterol Surg 2020;4:528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh MP, Rai S, Pandey A. et al. Molecular subtypes of colorectal cancer: an emerging therapeutic opportunity for personalized medicine. Genes Dis 2021;8:133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ten Hoorn S, de Back TR, Sommeijer DW. et al. Clinical value of consensus molecular subtypes in colorectal cancer: a systematic review and meta-analysis. J Natl Cancer Inst 2021;djab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geigl JB, Obenauf AC, Schwarzbraun T. et al. Defining “chromosomal instability.” Trends Genet 2008;24:64–9. [DOI] [PubMed] [Google Scholar]
- 14. Gupta R, Sinha S, Paul RN.. The impact of microsatellite stability status in colorectal cancer. Curr Probl Cancer 2018;42:548–59. [DOI] [PubMed] [Google Scholar]
- 15. Pino MS, Chung DC.. The chromosomal instability pathway in colon cancer. Gastroenterology 2010;138:2059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Armaghany T, Wilson JD, Chu Q. et al. Genetic alterations in colorectal cancer. Gastrointest Cancer Res 2012;5:19–27. [PMC free article] [PubMed] [Google Scholar]
- 17. Baran B, Mert Ozupek N, Yerli Tetik N. et al. Difference between left-sided and right-sided colorectal cancer: a focused review of literature. Gastroenterology Res 2018;11:264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen HT, Duong H-Q.. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett 2018;16:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nojadeh JN, Sharif Sakhinia BS.. E. Microsatellite instability in colorectal cancer. Excli J 2018;17:159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bond CE, McKeone DM, Kalimutho M. et al. RNF43 and ZNRF3 are commonly altered in serrated pathway colorectal tumorigenesis. Oncotarget 2016;7:70589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong SN. Genetic and epigenetic alterations of colorectal cancer. Intest Res 2018;16:327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carethers JM, Koi M, Tseng-Rogenski SS.. EMAST is a form of microsatellite instability that is initiated by inflammation and modulates colorectal cancer progression. Genes (Basel )2015;6:185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tseng-Rogenski SS, Hamaya Y, Choi DY. et al. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology 2015;148:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boland CR, Goel A.. Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–87.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munakata K, Koi M, Kitajima T. et al. Inflammation-associated microsatellite alterations caused by MSH3 dysfunction are prevalent in ulcerative colitis and increase with neoplastic advancement. Clin Transl Gastroenterol 2019;10:e00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Toyota M, Ahuja N, Ohe-Toyota M. et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999;96:8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hughes LAE, Melotte V, de Schrijver J. et al. The CpG island methylator phenotype: what’s in a name? Cancer Res 2013;73:5858–68. [DOI] [PubMed] [Google Scholar]
- 28. De Palma FDE, D’Argenio V, Pol J. et al. The molecular hallmarks of the serrated pathway in colorectal cancer. Cancers (Basel )2019;11:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simons CCJM, Hughes LAE, Smits KM. et al. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol 2013;24:2048–56. [DOI] [PubMed] [Google Scholar]
- 30. Weisenberger DJ, Siegmund KD, Campan M. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787–93. [DOI] [PubMed] [Google Scholar]
- 31. Hinoue T, Weisenberger DJ, Lange CP. et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2011;22:271–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Sousa E Melo F, Wang X, Jansen M. et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013;19:614–8. [DOI] [PubMed] [Google Scholar]
- 33. Gallois C, Taieb J, Le Corre D. et al. Prognostic value of methylator phenotype in stage III colon cancer treated with oxaliplatin-based adjuvant chemotherapy. Clin Cancer Res 2018;24:4745–53. [DOI] [PubMed] [Google Scholar]
- 34. Phipps AI, Limburg PJ, Baron JA. et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148:77–87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sinicrope FA, Shi Q, Smyrk TC. et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015;148:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phipps AI, Alwers E, Harrison T. et al. Association between molecular subtypes of colorectal tumors and patient survival, based on pooled analysis of 7 international studies. Gastroenterology 2020;158:2158–68.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Samowitz WS, Albertsen H, Herrick J. et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005;129:837–45. [DOI] [PubMed] [Google Scholar]
- 38. Guinney J, Dienstmann R, Wang X. et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thanki K, Nicholls ME, Gajjar A. et al. Consensus molecular subtypes of colorectal cancer and their clinical implications. Int Biol Biomed J 2017;3:105–11. [PMC free article] [PubMed] [Google Scholar]
- 40. Fessler E, Medema JP.. Colorectal cancer subtypes: developmental origin and microenvironmental regulation. Trends Cancer 2016;2:505–18. [DOI] [PubMed] [Google Scholar]
- 41. Lee MS, Menter DG, Kopetz S.. Right versus left colon cancer biology: integrating the consensus molecular subtypes. J Natl Compr Canc Netw 2017;15:411–9. [DOI] [PubMed] [Google Scholar]
- 42. Komor MA, Bosch LJ, Bounova G. et al. ; NGS-ProToCol Consortium. Consensus molecular subtype classification of colorectal adenomas. J Pathol 2018;246:266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Koveitypour Z, Panahi F, Vakilian M. et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci 2019;9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diamond SJ, Enestvedt BK, Jiang Z. et al. Adenoma detection rate increases with each decade of life after 50 years of age. Gastrointest Endosc 2011;74:135–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo S, Gu J, Zhang D. et al. The elderly harbor greater proportions of advanced histology in subcentimeter adenomas: implications for screening colonoscopy approaches. Eur J Gastroenterol Hepatol 2022;34:281–7. [DOI] [PubMed] [Google Scholar]
- 46. Pommergaard H-C, Burcharth J, Rosenberg J. et al. The association between location, age and advanced colorectal adenoma characteristics: a propensity-matched analysis. Scand J Gastroenterol 2017;52:1–4. [DOI] [PubMed] [Google Scholar]
- 47. Stryker SJ, Wolff BG, Culp CE. et al. Natural history of untreated colonic polyps. Gastroenterology 1987;93:1009–13. [DOI] [PubMed] [Google Scholar]
- 48. Lin M-T, Zheng G, Tseng L-H. et al. Multiclonal colorectal cancers with divergent histomorphological features and RAS mutations: one cancer or separate cancers? Hum Pathol 2020;98:120–8. [DOI] [PubMed] [Google Scholar]
- 49. Sylvester BE, Vakiani E.. Tumor evolution and intratumor heterogeneity in colorectal carcinoma: insights from comparative genomic profiling of primary tumors and matched metastases. J Gastrointest Oncol 2015;6:668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cummings OW. Pathology of the adenoma-carcinoma sequence: from aberrant crypt focus to invasive carcinoma. Semin Gastrointest Dis 2000;11:229–37. [PubMed] [Google Scholar]
- 51. Murakami T, Mitomi H, Saito T. et al. Distinct WNT/beta-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol 2015;28:146–58. [DOI] [PubMed] [Google Scholar]
- 52. Christie M, Jorissen RN, Mouradov D. et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/beta-catenin signalling thresholds for tumourigenesis. Oncogene 2013;32:4675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miyoshi Y, Nagase H, Ando H. et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet 1992;1:229–33. [DOI] [PubMed] [Google Scholar]
- 54. Powell SM, Zilz N, Beazer-Barclay Y. et al. APC mutations occur early during colorectal tumorigenesis. Nature 1992;359:235–7. [DOI] [PubMed] [Google Scholar]
- 55. Schell MJ, Yang M, Teer JK. et al. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun 2016;7:11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dow LE, O'Rourke KP, Simon J. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 2015;161:1539–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Korinek V, Barker N, Morin PJ. et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997;275:1784–7. [DOI] [PubMed] [Google Scholar]
- 58. Borras E, San Lucas FA, Chang K. et al. Genomic landscape of colorectal mucosa and adenomas. Cancer Prev Res (Phila) 2016;9:417–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lin SH, Raju GS, Huff C. et al. The somatic mutation landscape of premalignant colorectal adenoma. Gut 2018;67:1299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou D, Yang L, Zheng L. et al. Exome capture sequencing of adenoma reveals genetic alterations in multiple cellular pathways at the early stage of colorectal tumorigenesis. PLoS ONE 2013;8:e53310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bormann F, Rodriguez-Paredes M, Lasitschka F. et al. Cell-of-origin DNA methylation signatures are maintained during colorectal carcinogenesis. Cell Rep 2018;23:3407–18. [DOI] [PubMed] [Google Scholar]
- 62. Hanley MP, Hahn MA, Li AX. et al. Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia. Oncogene 2017;36:5035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koestler DC, Li J, Baron JA. et al. Distinct patterns of DNA methylation in conventional adenomas involving the right and left colon. Mod Pathol 2014;27:145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luo Y, Wong CJ, Kaz AM. et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 2014;147:418–29.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Belshaw NJ, Elliott GO, Foxall RJ. et al. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer 2008;99:136–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Belshaw NJ, Pal N, Tapp HS. et al. Patterns of DNA methylation in individual colonic crypts reveal aging and cancer-related field defects in the morphologically normal mucosa. Carcinogenesis 2010;31:1158–63. [DOI] [PubMed] [Google Scholar]
- 67. Easwaran H, Johnstone SE, Van Neste L. et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res 2012;22:837–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ohm JE, McGarvey KM, Yu X. et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 2007;39:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rakyan VK, Down TA, Maslau S. et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 2010;20:434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schlesinger Y, Straussman R, Keshet I. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 2007;39:232–6. [DOI] [PubMed] [Google Scholar]
- 71. Widschwendter M, Fiegl H, Egle D. et al. Epigenetic stem cell signature in cancer. Nat Genet 2007;39:157–8. [DOI] [PubMed] [Google Scholar]
- 72. Voloshanenko O, Erdmann G, Dubash TD. et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun 2013;4:2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 2007;50:113–30. [DOI] [PubMed] [Google Scholar]
- 74. Langner C. Serrated and non-serrated precursor lesions of colorectal cancer. Dig Dis 2015;33:28–37. [DOI] [PubMed] [Google Scholar]
- 75. Fearon ER, Vogelstein B.. A genetic model for colorectal tumorigenesis. Cell 1990;61:759–67. [DOI] [PubMed] [Google Scholar]
- 76. Boparai KS, Dekker E, Polak MM. et al. A serrated colorectal cancer pathway predominates over the classic WNT pathway in patients with hyperplastic polyposis syndrome. Am J Pathol 2011;178:2700–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carr NJ, Mahajan H, Tan KL. et al. Serrated and non-serrated polyps of the colorectum: their prevalence in an unselected case series and correlation of BRAF mutation analysis with the diagnosis of sessile serrated adenoma. J Clin Pathol 2009;62:516–8. [DOI] [PubMed] [Google Scholar]
- 78. Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol 2011;6:479–507. [DOI] [PubMed] [Google Scholar]
- 79. Fu X, Li L, Peng Y.. Wnt signalling pathway in the serrated neoplastic pathway of the colorectum: possible roles and epigenetic regulatory mechanisms. J Clin Pathol 2012;65:675–9. [DOI] [PubMed] [Google Scholar]
- 80. Kim K-M, Lee EJ, Ha S. et al. Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol 2011;35:1274–86. [DOI] [PubMed] [Google Scholar]
- 81. O'Brien MJ, Yang S, Mack C. et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006;30:1491–501. [DOI] [PubMed] [Google Scholar]
- 82. Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 2011;42:1–10. [DOI] [PubMed] [Google Scholar]
- 83. Velho S, Moutinho C, Cirnes L. et al. BRAF, KRAS and PIK3CA mutations in colorectal serrated polyps and cancer: primary or secondary genetic events in colorectal carcinogenesis? BMC Cancer 2008;8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang S, Farraye FA, Mack C. et al. BRAF and KRAS mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol 2004;28:1452–9. [DOI] [PubMed] [Google Scholar]
- 85. Carragher LA, Snell KR, Giblett SM. et al. V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO Mol Med 2010;2:458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Serrano M, Lin AW, McCurrach ME. et al. Oncogenic RAS provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997;88:593–602. [DOI] [PubMed] [Google Scholar]
- 87. Michaloglou C, Vredeveld LC, Soengas MS. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005;436:720–4. [DOI] [PubMed] [Google Scholar]
- 88. Bartkova J, Rezaei N, Liontos M. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006;444:633–7. [DOI] [PubMed] [Google Scholar]
- 89. Bennecke M, Kriegl L, Bajbouj M. et al. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell 2010;18:135–46. [DOI] [PubMed] [Google Scholar]
- 90. Henriques AFA, Barros P, Moyer MP. et al. Expression of tumor-related Rac1b antagonizes B-Raf-induced senescence in colorectal cells. Cancer Lett 2015;369:368–75. [DOI] [PubMed] [Google Scholar]
- 91. Kriegl L, Neumann J, Vieth M. et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol 2011;24:1015–22. [DOI] [PubMed] [Google Scholar]
- 92. Fernando WC, Miranda MS, Worthley DL. et al. The CIMP phenotype in BRAF mutant serrated polyps from a prospective colonoscopy patient cohort. Gastroenterol Res Pract 2014;2014:374926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gala MK, Mizukami Y, Le LP. et al. Germline mutations in oncogene-induced senescence pathways are associated with multiple sessile serrated adenomas. Gastroenterology 2014;146:520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yan HHN, Lai JCW, Ho SL. et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut 2017;66:1645–56. [DOI] [PubMed] [Google Scholar]
- 95. Giannakis M, Hodis E, Jasmine Mu X. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014;46:1264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jo YS, Kim MS, Lee JH. et al. Frequent frameshift mutations in 2 mononucleotide repeats of RNF43 gene and its regional heterogeneity in gastric and colorectal cancers. Hum Pathol 2015;46:1640–6. [DOI] [PubMed] [Google Scholar]
- 97. Sekine S, Yamashita S, Tanabe T. et al. Frequent PTPRK-RSPO3 fusions and RNF43 mutations in colorectal traditional serrated adenoma. J Pathol 2016;239:133–8. [DOI] [PubMed] [Google Scholar]
- 98. Koo B-K, Spit M, Jordens I. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012;488:665–9. [DOI] [PubMed] [Google Scholar]
- 99. Nakanishi H, Sawada T, Kaizaki Y. et al. Significance of gene mutations in the Wnt signaling pathway in traditional serrated adenomas of the colon and rectum. PLoS ONE 2020;15:e0229262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wong JJ, Hawkins NJ, Ward RL. et al. Methylation of the 3p22 region encompassing MLH1 is representative of the CpG island methylator phenotype in colorectal cancer. Mod Pathol 2011;24:396–411. [DOI] [PubMed] [Google Scholar]
- 101. Seshagiri S, Stawiski EW, Durinck S. et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488:660–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Baba Y, Nosho K, Shima K. et al. Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res 2009;15:4665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dawson H, Galvan JA, Helbling M. et al. Possible role of Cdx2 in the serrated pathway of colorectal cancer characterized by BRAF mutation, high-level CpG Island methylator phenotype and mismatch repair-deficiency. Int J Cancer 2014;134:2342–51. [DOI] [PubMed] [Google Scholar]
- 104. Graule J, Uth K, Fischer E. et al. CDX2 in colorectal cancer is an independent prognostic factor and regulated by promoter methylation and histone deacetylation in tumors of the serrated pathway. Clin Epigenetics 2018;10:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kawai H, Tomii K, Toyooka S. et al. Promoter methylation downregulates CDX2 expression in colorectal carcinomas. Oncol Rep 2005;13:547–51. [PubMed] [Google Scholar]
- 106. Wang Y, Li Z, Li W. et al. Methylation of promoter region of CDX2 gene in colorectal cancer. Oncol Lett 2016;12:3229–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sakamoto N, Feng Y, Stolfi C. et al. BRAFV600E cooperates with CDX2 inactivation to promote serrated colorectal tumorigenesis. Elife 2017;6:e20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fraga MF, Ballestar E, Paz MF. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005;102:10604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Stirzaker C, Taberlay PC, Statham AL. et al. Mining cancer methylomes: prospects and challenges. Trends Genet 2014;30:75–84. [DOI] [PubMed] [Google Scholar]
- 110. Bitzer M, Malek NP, Chapter 42: personalized epigenetics. In: Tollefsbol TO (ed.). Medical Epigenetics. Boston: Academic Press, 2016, 843–58. [Google Scholar]
- 111. Unnikrishnan A, Hadad N, Masser DR. et al. Revisiting the genomic hypomethylation hypothesis of aging. Ann N Y Acad Sci 2018;1418:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ahuja N, Li Q, Mohan AL. et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 1998;58:5489–94. [PubMed] [Google Scholar]
- 113. Maegawa S, Hinkal G, Kim HS. et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res 2010;20:332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Teschendorff AE, Menon U, Gentry-Maharaj A. et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 2010;20:440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xie W, Kagiampakis I, Pan L. et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell 2018;33:309–21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bernstein BE, Mikkelsen TS, Xie X. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315–26. [DOI] [PubMed] [Google Scholar]
- 117. Mikkelsen TS, Ku M, Jaffe DB. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007;448:553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Marttila S, Kananen L, Häyrynen S. et al. Ageing-associated changes in the human DNA methylome: genomic locations and effects on gene expression. BMC Genomics 2015;16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Thompson MJ, vonHoldt B, Horvath S. et al. An epigenetic aging clock for dogs and wolves. Aging (Albany NY) 2017;9:1055–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wang T, Ma J, Hogan AN. et al. Quantitative translation of dog-to-human aging by conserved remodeling of the DNA methylome. Cell Syst 2020;11:176–85.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wilkinson GS, Adams DM, Haghani A. et al. DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat Commun 2021;12:1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sen P, Shah PP, Nativio R. et al. Epigenetic mechanisms of longevity and aging. Cell 2016;166:822–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Berman BP, Weisenberger DJ, Aman JF. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 2011;44:40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hansen KD, Timp W, Bravo HC. et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 2011;43:768–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zhou W, Dinh HQ, Ramjan Z. et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat Genet 2018;50:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Easwaran HP, Schermelleh L, Leonhardt H. et al. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. Embo Rep 2004;5:1181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pradhan S, Bacolla A, Wells RD. et al. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem 1999;274:33002–10. [DOI] [PubMed] [Google Scholar]
- 128. Teschendorff AE. A comparison of epigenetic mitotic-like clocks for cancer risk prediction. Genome Med 2020;12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Feng Q, Liang S, Jia H. et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun 2015;6:6528. [DOI] [PubMed] [Google Scholar]
- 130. Nakatsu G, Li X, Zhou H. et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 2015;6:8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Arthur JC, Perez-Chanona E, Mühlbauer M. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Dejea CM, Fathi P, Craig JM. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359:592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. DeStefano Shields CE, White JR, Chung L. et al. Bacterial-driven inflammation and mutant BRAF expression combine to promote murine colon tumorigenesis that is sensitive to immune checkpoint therapy. Cancer Discov 2021;11:1792–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kumar R, Herold JL, Schady D. et al. Streptococcus gallolyticus subsp. gallolyticus promotes colorectal tumor development. PLoS Pathog 2017;13:e1006440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wang X, Yang Y, Huycke MM.. Commensal-infected macrophages induce dedifferentiation and reprogramming of epithelial cells during colorectal carcinogenesis. Oncotarget 2017;8:102176–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wu S, Rhee K-J, Albesiano E. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 2009;15:1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Ansari I, Raddatz G, Gutekunst J. et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat Microbiol 2020;5:610–9. [DOI] [PubMed] [Google Scholar]
- 138. Pan W-H, Sommer F, Falk-Paulsen M. et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med 2018;10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yu D-H, Gadkari M, Zhou Q. et al. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol 2015;16:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Armstrong D, Dregan A, Ashworth M. et al. The association between colorectal cancer and prior antibiotic prescriptions: case control study. Br J Cancer 2020;122:912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ma W, Chan AT.. Antibiotic use and colorectal cancer: a causal association? Gut 2020;69:1913–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Yang T, Owen JL, Lightfoot YL. et al. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med 2013;19:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Bhatt AP, Redinbo MR, Bultman SJ.. The role of the microbiome in cancer development and therapy. CA Cancer J Clin 2017;67:326–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Sears CL, Pardoll DM.. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011;203:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Sobhani I, Bergsten E, Couffin S. et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci USA 2019;116:24285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Lennard KS, Goosen RW, Blackburn JM.. Bacterially-associated transcriptional remodelling in a distinct genomic subtype of colorectal cancer provides a plausible molecular basis for disease development. PLoS One 2016;11:e0166282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mima K, Cao Y, Chan AT. et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol 2016;7:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Dejea CM, Wick EC, Hechenbleikner EM. et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci USA 2014;111:18321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tomkovich S, Dejea CM, Winglee K. et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 2019;129:1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Maiuri AR, Peng M, Podicheti R. et al. Mismatch repair proteins initiate epigenetic alterations during inflammation-driven tumorigenesis. Cancer Res 2017;77:3467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Purcell RV, Visnovska M, Biggs PJ. et al. Distinct gut microbiome patterns associate with consensus molecular subtypes of colorectal cancer. Sci Rep 2017;7:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. O'Hagan HM, Wang W, Sen S. et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell 2011;20:606–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ding N, Bonham EM, Hannon BE. et al. Mismatch repair proteins recruit DNA methyltransferase 1 to sites of oxidative DNA damage. J Mol Cell Biol 2016;8:244–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Allen J, Hao S, Sears CL. et al. Epigenetic changes induced by bacteroides fragilis toxin. Infect Immun 2019;87:e00447-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 1990;70:567–90. [DOI] [PubMed] [Google Scholar]
- 156. Macfarlane S, Macfarlane GT.. Regulation of short-chain fatty acid production. Proc Nutr Soc 2003;62:67–72. [DOI] [PubMed] [Google Scholar]
- 157. Krautkramer KA, Kreznar JH, Romano KA. et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell 2016;64:982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Qin Y, Roberts JD, Grimm SA. et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol 2018;19:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dong Y, Zhou J, Zhu Y. et al. Abdominal obesity and colorectal cancer risk: systematic review and meta-analysis of prospective studies. Biosci Rep 2017;37:BSR20170945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Friedenreich CM, Ryder-Burbidge C, McNeil J.. Physical activity, obesity and sedentary behavior in cancer etiology: epidemiologic evidence and biologic mechanisms. Mol Oncol 2021;15:790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Renehan AG, Tyson M, Egger M. et al. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 2008;371:569–78. [DOI] [PubMed] [Google Scholar]
- 162. Booth A, Magnuson A, Fouts J. et al. Adipose tissue, obesity and adipokines: role in cancer promotion. Horm Mol Biol Clin Investig 2015;21:57–74. [DOI] [PubMed] [Google Scholar]
- 163. Li R, Grimm SA, Chrysovergis K. et al. Obesity, rather than diet, drives epigenomic alterations in colonic epithelium resembling cancer progression. Cell Metab 2014;19:702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Akhtar-Zaidi B, Cowper-Sal-Lari R, Corradin O. et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 2012;336:736–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Beyaz S, Mana MD, Roper J. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016;531:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kaiko GE, Ryu SH, Koues OI. et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 2016;165:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Cervena K, Siskova A, Buchler T. et al. Methylation-based therapies for colorectal cancer. Cells 2020;9:1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yang Z-H, Dang Y-Q, Ji G.. Role of epigenetics in transformation of inflammation into colorectal cancer. World J Gastroenterol 2019;25:2863–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lutgens MWMD, van Oijen MGH, van der Heijden GJMG. et al. Declining risk of colorectal cancer in inflammatory bowel disease: an updated meta-analysis of population-based cohort studies. Inflamm Bowel Dis 2013;19:789–99. [DOI] [PubMed] [Google Scholar]
- 170. Issa JP, Ahuja N, Toyota M. et al. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res 2001;61:3573–7. [PubMed] [Google Scholar]
- 171. Niwa T, Tsukamoto T, Toyoda T. et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res 2010;70:1430–40. [DOI] [PubMed] [Google Scholar]
- 172. Foran E, Garrity-Park MM, Mureau C. et al. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res 2010;8:471–81. [DOI] [PubMed] [Google Scholar]
- 173. Maekita T, Nakazawa K, Mihara M. et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res 2006;12:989–95. [DOI] [PubMed] [Google Scholar]
- 174. Abu-Remaileh M, Bender S, Raddatz G. et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res 2015;75:2120–30. [DOI] [PubMed] [Google Scholar]
- 175. Giannakis M, Mu XJ, Shukla SA. et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep 2016;15:857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]