Abstract
Fibrillar protein aggregates are a hallmark of a range of human disorders, from prion diseases to dementias, but are also encountered in several functional contexts. Yet, the fundamental links between protein assembly mechanisms and their functional or pathological roles have remained elusive. Here, we analyze the aggregation kinetics of a large set of proteins that self-assemble by a nucleated-growth mechanism, from those associated with disease, over those whose aggregates fulfill functional roles in biology, to those that aggregate only under artificial conditions. We find that, essentially, all such systems, regardless of their biological role, are capable of self-replication. However, for aggregates that have evolved to fulfill a structural role, the rate of self-replication is too low to be significant on the biologically relevant time scale. By contrast, all disease-related proteins are able to self-replicate quickly compared to the time scale of the associated disease. Our findings establish the ubiquity of self-replication and point to its potential importance across aggregation-related disorders.
All pathological protein aggregates self-replicate in vitro on a disease-relevant time scale.
INTRODUCTION
The self-assembly of proteins into ordered, homo-molecular filaments is a process associated with a range of currently incurable human disorders, from prion diseases, through sickle \ anemia, to Alzheimer’s disease (1, 2). In such disorders, proteins that are normally monomeric form aggregates, such as the highly stable amyloid fibrils, with often deleterious effects for the organism (1). However, filamentous protein self-assembly also plays important roles in a functional context, including in the formation of cytoskeletal structures, such as the polymerization of actin (3). Even amyloid fibrils, which, unlike actin, are generally highly resistant to depolymerization, are encountered in functional contexts throughout nature, from structural elements in bacterial biofilms, to long-term memory formation in marine organisms (4). In addition to these functional and disease-associated assemblies, the aggregated state, in the form of amyloid, has been proposed to constitute a general, stable conformation for a large number of proteins (5). However, many proteins only reach this state upon perturbation, such as shaking, heating, or extreme pH conditions, which either helps the system to overcome the large energy barrier that prevents their conversion into amyloid under physiological conditions, or reduces the height of the barrier. With current advances in the theoretical descriptions of aggregation and the increasing accuracy of biophysical measurements in recent years, it is now possible to identify the aggregation mechanisms of many of these proteins (6, 7). Combining the data from dozens of other works, we here deduce the mechanism of aggregation for a range of peptides and proteins to elucidate mechanistic commonalities and differences. While various other types of protein aggregates exist, including well-defined macromolecular assemblies, such as virus capsids, or hierarchical assemblies, such as intermediate filaments (8), we focus here on those proteins that aggregate into homo-molecular, filamentous aggregates, via a nucleated polymerization mechanism, primarily amyloids.
RESULTS
The presence of a self-replication mechanism leads to fundamentally different aggregation behavior
Amyloid fibrils are highly elongated structures made up of many thousands of monomers, but typically very few monomers in diameter, and are therefore approximated as linear aggregates. In descriptions of linear self-assembly, the underlying processes naturally fall into two categories: growth processes, which increase the size of existing aggregates, and nucleation and multiplication processes, which generate new aggregates (9). The first category, growth, usually proceeds by addition of monomers from solution to growth-competent aggregate ends, increasing their length but leaving the total number of aggregates unchanged. The second category, processes that increase the number of aggregates, can be further classified into primary nucleation processes, which only involve monomeric protein and are independent of the concentration of aggregates, and secondary processes (or multiplication processes), which do involve existing aggregates. Primary nucleation can take the form of homogeneous nucleation in solution or heterogeneous nucleation on an interface, whereas secondary processes include the fragmentation of fibrils and the catalysis of nucleation from monomers on the surface of existing fibrils in secondary nucleation. Primary nucleation is always a necessary first step in the formation of aggregates from purely monomeric proteins; by contrast, the presence of secondary processes is not required to fully convert a system to its aggregated state.
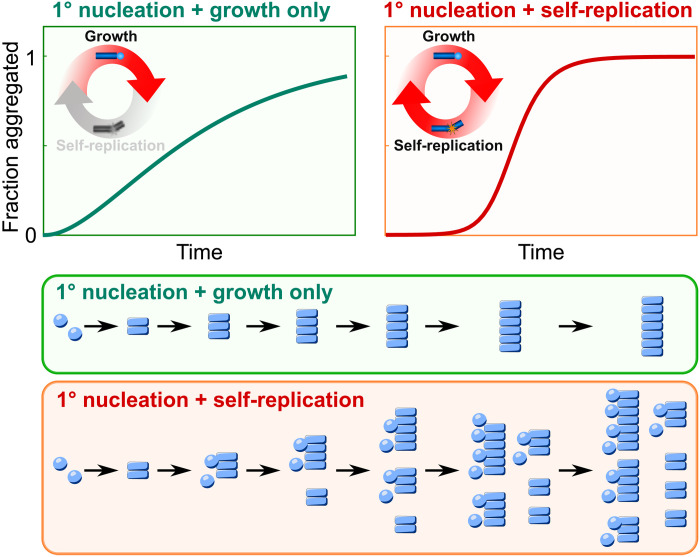
The aggregation behavior of systems with and without secondary processes is fundamentally different (Fig. 1). When secondary processes are active, existing aggregates self-replicate, that is, they create new aggregates autocatalytically, thereby accelerating the overall rate of elongation, which, in turn, speeds up the production of even more aggregates through secondary processes in an iterative manner. This autocatalytic feedback loop means that new aggregates amplify very rapidly once they have reached a limiting concentration. The parent fibril from a single primary nucleation event can thus produce many child fibrils through self-replication, and the aggregate mass increases exponentially with time (see Eq. 2). By contrast, in the absence of secondary processes, each aggregate needs to be initiated by a primary nucleation event, which is independent of the amount of fibrils present. The aggregate mass thus increases more gradually and with polynomial scaling in time (see Eq. 1). The integrated rate laws describing aggregation under constant monomer conditions are given by
| (1) |
in the absence of self-replication and
| (2) |
in the presence of self-replication, where M(t) is the mass concentration of aggregated protein at time t, the parameters κ and λ are defined as and , kprim and ksec are the rates of primary nucleation and secondary processes, respectively, k+ is the rate constant of growth, and m is the concentration of monomer (10). The rate of secondary processes ksec can have contributions from both secondary nucleation and fragmentation, and this rate can vary with monomer concentration in different ways, depending on the specific mechanism. More details can be found in Meisl et al. (7, 9).
Fig. 1. Effect of self-replication.
Illustration of the kinetic curves of aggregate concentration over time without (left) and with self-replication (right), along with a schematic of the reaction in both cases. When aggregation proceeds via nucleation and growth only, without self-replication, each primary nucleation event gives rise to only one fibril, and the aggregate concentration increases gradually. By contrast, when self-replication is present, here illustrated in the form of secondary nucleation, each primary nucleation event gives rise to many fibrils. The positive feedback loop of self-replication leads to exponential growth of aggregate mass and kinetic curves with a much more sudden increase and steeper transition.
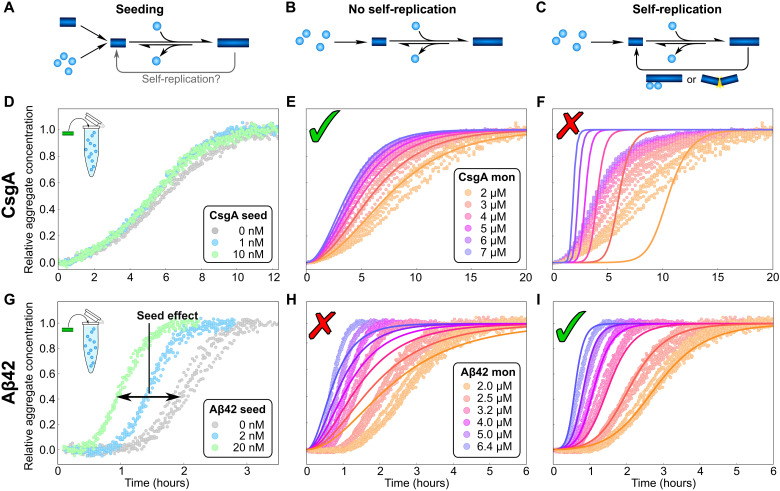
When secondary processes are present, the ability of aggregates to self-replicate in an autocatalytic manner can make such systems extremely sensitive to the introduction of seed fibrils (Fig. 2G). This property is exploited in several amplification assays, which can amplify a single replication-competent aggregate to macroscopically detectable levels (11–13). However, in many biological contexts, this susceptibility to amplify small fluctuations of aggregate concentrations may not be a desirable property, making the speed of the formation, as well as the spatial distribution of new aggregates, difficult to control. In the context of disease, the amplification of spontaneously formed or transferred seed aggregates may be a key step that allows pathology to persist and spread. The effect of small fluctuations in seed concentration on the macroscopic behavior is demonstrated in Fig. 2 (D and G) for two proteins CsgA and Aβ42. CsgA is an Escherichia coli protein that forms functional amyloids during biofilm formation (14, 15) and does not display any significant secondary processes. This is evidenced by the fact that measurements of its aggregation kinetics are well described by a model including only growth and primary nucleation (Fig. 2E) and that the addition of small concentrations of preformed seeds has no effect on its aggregation behavior (Fig. 2D). Aβ42 is one of the main proteins whose aggregation is associated with Alzheimer’s disease. Its aggregation mechanism contrasts with that of CsgA as it is dominated by the secondary nucleation of monomers on the surface of existing fibrils (16). Measurements of its aggregation kinetics cannot be described by a model that does not include a secondary process (Fig. 2H). Moreover, Aβ42 aggregation is very sensitive to the addition of preformed aggregates; even seed concentrations that are orders of magnitude lower than the concentrations of monomeric protein in solution lead to a significant change in the aggregation kinetics (Fig. 2G). With these two proteins exemplifying the two distinct behaviors, we now set out to investigate the universality of self-replication and potential correlations of its presence with the role the protein aggregates play in biology.
Fig. 2. Aggregation kinetics with and without self-replication.
(A to C) Illustration of the aggregation mechanism used in producing the fits. (D and G) Aggregation kinetics of CsgA, 5 μM monomer (D), and Aβ42, 2 μM monomer (G) (52), with increasing concentrations of preformed seeds, monitored by thioflavin T fluorescence, a reporter of the mass of aggregates formed. While the CsgA behavior is not significantly affected by the presence of seeds, a large effect can be seen for Aβ42 aggregation, even when the seed concentration is up to three orders of magnitude below that of the monomeric protein. (E, F, H, and I) Aggregation kinetics of CsgA (E and F) (53) and Aβ42 (H and I) (54) at a range of monomer concentrations in the absence of seeds. The solid lines are global fits of the integrated rate laws in the absence (E and H) and presence (F and I) of secondary processes. In (F), a significant contribution of secondary nucleation is enforced to illustrate the misfit. Data are recorded in triplicates at each concentration; all points are shown.
The ability to self-replicate in vitro is a general characteristic of aggregating proteins
Recent advances in chemical kinetics have allowed us to link the macroscopic aggregation kinetics (Fig. 2) to the underlying molecular mechanisms through integrated rate laws for a range of aggregating proteins (7, 17). While the detailed equations depend on the specific aggregation mechanisms, they have several fundamental properties in common. At a given monomer concentration, the aggregation curves are predominantly determined by two parameters: λ, a measure for the rate at which primary nucleation contributes new aggregates, and κ, a measure for the rate at which secondary processes contribute new aggregates (see also Eqs. 1 and 2). The relative magnitude of these two rates determines which one of the two processes dominates the overall production of new aggregates. By application of our kinetic analysis framework (7), we determined the values of λ and κ for seven functional amyloids, nine pathological amyloids, and eight that do not form amyloid in a biological context (not counting mutants or fragments of the same protein). To have the most representative measure of the intrinsic aggregation mechanism of the proteins and avoid bias, we applied stringent selection criteria (see Materials and Methods) and, in particular, only used aggregation data under quiescent conditions. Shaking and agitation are commonly used to induce aggregation but can considerably alter the mechanism (16) by promoting fragmentation and inducing aggregation through shearing (18). By excluding such datasets, we avoid an artificial bias toward fragmentation-dominated mechanisms. An example of the analysis performed for all these proteins is shown in Fig. 2, for both the functional amyloid CsgA and the disease-associated Aβ42 peptide. We find that almost all of these systems, regardless of their biological role, display the ability to self-replicate via a secondary process when aggregating in vitro. The only exceptions to this are some of the functional aggregates, namely, actin, CsgA, and FapC.
The results of our analysis of these data are summarized in Fig. 3, which shows a rate diagram for amyloid forming proteins. The aggregation mechanisms are quantified by the rate at which new aggregates are produced via a primary nucleation pathway, λ, and the rate at which they are produced via a pathway involving secondary processes, κ. We find that the vast majority of biological protein aggregates, whether functional or disease associated, are able to self-replicate and are thus located in the upper left hand section of the plot in Fig. 3. Thus, the question arises whether the presence of secondary processes in protein aggregation is the default state, or if only the subset of proteins that is prone to aggregation in biological systems, either in a disease context or as functional assemblies, is biased toward self-replication. To answer this question, we included proteins whose aggregation does not occur in a biological context but can be triggered by harsh conditions such as low pH and high temperature in vitro. These systems also, without exception, display the ability to self-replicate. Thus, we conclude that, if a protein can be made to form filamentous aggregates, the ability to self-replicate appears to be the default state. Only a few functional assemblies appear to have evolved to suppress this property to such a degree that it is no longer readily observable on the time scales of in vitro experiments.
Fig. 3. Rate diagrams of aggregation mechanisms show that self-replication is ubiquitous.
The rate at which new aggregates are produced by secondary pathways, κ, is plotted against the rate at which primary pathways produce new aggregates, λ. On the dashed line, the rates of the two processes are equal. It separates systems dominated by primary nucleation (bottom right corner) from systems dominated by a secondary process (top left corner). In primary nucleation-dominated systems, when secondary processes are too slow, only an upper bound for the rate of secondary pathways can be obtained, and similarly, when primary nucleation is so slow that seeding is required, only an upper bound for the primary rate can be obtained. These cases are here illustrated by elongated points. Proteins are split into three classes: those forming pathological amyloids (bottom left), those forming functional amyloids (bottom right), and those that do not generally form amyloid under physiological conditions (top right). Labels are shown above or to the right of the corresponding data point. Note: Aβ42 (V18S + A21S) is classed as a nonbiological amyloid because it is an artificial mutant of Aβ42, itself pathological, designed specifically to try to affect secondary nucleation [see Thacker et al. (28)]. Details on all proteins are given in the Supplementary Materials.
As outlined, there are two distinct molecular mechanisms that can give rise to self-replication in vitro: fragmentation of aggregates and secondary nucleation of monomers on the surface of existing aggregates. Fragmentation, as it does not require any specific molecular interactions, may appear an obvious candidate for a default mechanism of self-replication: It tends to be significant under agitation (16, 19, 20) but has also been found to be important under quiescent conditions in some systems such as yeast prions (21). Fragmentation induced by the proteasome has also been proposed as an important process of self-replication in cell culture (22). However, more unexpectedly, secondary nucleation on the surface of existing aggregates has in fact been established as the main mechanism of self-replication under quiescent conditions in many amyloid forming proteins, where this process has been studied in detail (16, 23–27). For the Aβ42 peptide, even attempts to specifically abolish the ability to secondary nucleate via targeted mutations were unsuccessful. The mutants displayed significant changes in fibril morphology, but self-replication still proceeded by secondary nucleation (28) (see also Fig. 3; Aβ42 V18S + A21S). Secondary nucleation is not exclusive to protein aggregation and appears in many other contexts, such as crystal growth, where it is often a result of strain and local defects (29–31). While our data are too limited to clearly establish the specific mechanism of self-replication for most amyloid-forming systems, the ubiquitous presence of self-replication, alongside the fact that assembly of most of these structures induces strained conformations (32), makes for an intriguing correlation. In addition to fragmentation, secondary nucleation in particular, not just self-replication in general, may thus be a key process in the formation of many filamentous aggregates.
We further note that the specific values of the rates λ and κ fall into a relatively narrow range here, which likely reflects the experimental limitations of measuring aggregation kinetics: To be observable with standard techniques on an experimentally feasible time scale, the conditions and concentrations will be adjusted to result in aggregation over the course of minutes to hours. However, while the absolute rates are biased by the experimental limitations, the dominance of secondary over primary processes remains a robust finding.
The time scales of self-replication correlate with biological roles
To further investigate the importance of our findings in the context of the respective biological systems in which these proteins are found to aggregate, we investigated the potential significance of self-replication at relevant biological time scales and concentrations. While secondary processes, such as fragmentation and fibril-catalyzed nucleation, conceivably proceed in a similar manner in biological systems as they do in vitro, the process of primary nucleation is likely to differ more significantly. In vitro, air-water interfaces (33) or the surface of the reaction vessel may serve as heterogeneous primary nucleation sites, whereas in vivo nucleator proteins for functional aggregates (34) or lipid membranes for disease-associated ones (35) may trigger primary nucleation. To assess the potential role of self-replication in a biological context, we therefore focus exclusively on the rate constant of self-replication obtained from kinetic analysis. The aim of this comparison is to answer the question whether the intrinsic self-replication propensity, as measured in vitro, correlates in any way with the roles that protein aggregation plays in living systems. In general, aggregation is slowed in living systems, for example, through the action of chaperones or of active removal processes (36, 37). Thus, our analysis investigates if, despite these in vivo effects, the intrinsic self-replication propensity can be a predictor of disease association.
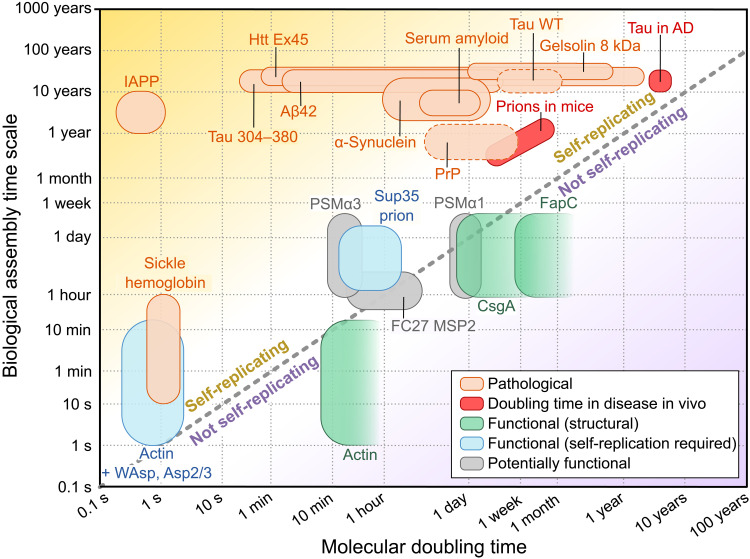
For the subset of functional and disease-associated proteins analyzed in Fig. 3, for which a set of aggregation data at varying monomer concentrations is available, we determined the rate constants and reaction orders. Thus, we were able to evaluate the time to double the number of aggregates through self-replication, t2 = ln (2)/κ, at the protein concentrations encountered in the respective in vivo environment. We then compared this time scale to the characteristic time scale of the in vivo process in which aggregation takes place, such as the time for biofilm maturation for CsgA or the disease duration for the prion protein (PrP; see Fig. 4).
Fig. 4. Time scales of self-replication and relevant in vivo time scale.
Bottom right half: The time scale of self-replication is longer than the relevant in vivo time scale, making self-replication unlikely to be able to contribute to the aggregation kinetics in vivo. Top left half: By contrast, the time scale of self-replication is much shorter than the relevant in vivo time scale, making a contribution of self-replication to the process likely. The time scales of self-replication were computed using the fitted rate constants and the protein concentrations in their respective in vivo environments, the latter being the main source of uncertainty. For two systems, prions in mice and tau in Alzheimer’s disease (AD), the self-replication rates have been determined in the in vivo system directly, requiring no further computation or knowledge of the in vivo concentrations (red). For FapC, CsgA, and actin, the experimental aggregation kinetics are described well with a mechanism that does not include self-replication; therefore, we can only obtain a lower limit on the time scale for those proteins. Dashed lines for PrP and tau denote mild shaking conditions (for details, see Materials and Methods).
We find that all disease-associated proteins, without exception, show self-replication time scales that are much shorter than the time scales of the associated disease. Therefore, all disease-associated aggregates have the intrinsic ability to replicate sufficiently quickly for self-replication to be a relevant mechanism in disease progression. Rates of self-replication measured in the relevant system in vivo are very rare, but we were recently able to determine them in two systems, prions in mice (38) and tau in Alzheimer’s disease (39). These rates are included here alongside the rates calculated from in vitro measurements to serve both as a comparison to the in vitro numbers and as validation of our conclusions on the potential importance of self-replication. While replication is somewhat slowed compared to the in vitro behavior of the same protein, both systems are still clearly in a regime dominated by the self-replication process.
By contrast, the functional assemblies cluster much closer to the threshold at which self-replication becomes too slow to be relevant on the biological assembly time scale (diagonal dashed line in Fig. 4). For those functional assemblies that have been established to fulfill structural roles, FapC, CsgA, and actin, the archetypal biological structural element, the self-replication time scale exceeds that of the relevant in vivo process, suggesting that self-replication is too slow to be relevant in vivo. The time scales of self-replication appear to be just long enough to have no significant contribution to the aggregation process on the relevant biological time scale. This reduction of self-replication only by the minimal amount necessary is further indication in support of the idea that self-replication is ubiquitous and has to be selected against by evolution if undesirable. The two functional systems for which self-replication is likely to be a desirable property, yeast prions (Sup35) and actin in the presence of a branching agent such as Wiskott-Aldrich syndrome protein (WAsp) and actin related protein complex (Arp2/3), are indeed situated in the region of the plot where self-replication is significant. Specifically, the biological role of yeast prions is believed to involve the transfer of the prion between yeast cells, thus requiring self-replication of the aggregates on time scales relevant for yeast reproduction. In actin assembly, if branching, a secondary process, is required, it can be induced in actin in a controlled manner by other molecules such as WAsp (see Fig. 4) (40). Last, the phenol soluble modulins (PSMs), expressed by Staphylococcus bacteria, and the merozoite surface protein, FC27 MSP2, expressed by the malaria parasite, form amyloid fibrils under native conditions in vitro, but the specific biological roles of these assemblies and thus whether one would expect their self-replication to be a desirable property have yet to be established.
Thus, while there appears to be some evidence for an evolutionary pressure to prevent self-replication if aggregates are involved in certain functions, it is unclear how this is achieved. Intriguingly, modification of some structural functional amyloids, e.g., by removal of regions from the sequence, can lead to increased rates of self-replication: CsgA and FapC consist of multiple copies of imperfect repeats of 20 to 35 residues (14, 41). Each repeat is predicted to fold into a β-hairpin conformation, which forms a self-contained element in a β-helix structure (42, 43) and thus constitutes a readily available building block for the efficient construction of an amyloid fibril. Stepwise removal of these repeats increases the tendency of the fibrils to fragment (44). Thus, accumulation of repeats within the protein sequence, at least in these bacterial amyloids, appears to suppress self-replication over primary nucleation. Beyond this observation, a further investigation of the proteins analyzed here for patterns in their sequence did not reveal any clear features associated with the propensity to self-replicate (see fig. S23) (45). The ubiquity of self-replication may prevent a specific sequence feature from being identified across the diverse set of proteins analyzed here. As the set of proteins with known aggregation mechanisms increases, further stratification of the data to investigate the presence of features important for self-replication in different classes of proteins may become possible.
DISCUSSION
In conclusion, our results provide insights into both the widespread presence of self-replication in amyloid formation and the potential importance of this process in disease. Almost all aggregating proteins studied have the ability to self-replicate, although the propensity to do so appears to have been deselected for by evolution in some, but not all, proteins that have evolved to aggregate in a functional context. In light of these results, we propose that for certain functional roles, such as structural support, a crucial aspect in determining whether a protein is suitable for the formation of functional structures may be its propensity to self-replicate. In addition to making the system susceptible to fluctuations in initial seed concentration, secondary nucleation would lead to the formation of new fibrils along the length of existing fibrils, making the control over the location of these new fibrils difficult. Similarly, a high propensity to fragment is unlikely to be desirable in a structural context.
By contrast, the ability of aggregates to self-replicate appears to be a central prerequisite for their involvement in disease. Prion disease, the archetypal protein aggregation disease, requires the self-replication of its disease-causing aggregates to propagate between individuals (46). Moreover, in many model organisms of neurodegenerative disease, it has been shown that the introduction of seeds can trigger the formation of new aggregates (47, 48), leading to the description of several such diseases as prion-like (49). In recent work, we established the replication mechanism of prions in mice and found that it is in fact consistent with the self-replication mechanism of PrP aggregates in vitro (38). In another study, we showed that the relative difference in the replication rate of two strains of α-synuclein aggregates produced in vitro was mirrored in the survival times of mice infected with these strains (50). Last, in recent work, we established that self-replication is the rate-limiting step of the accumulation of tau aggregates in Alzheimer’s disease (39). In light of these clear connections between the in vitro mechanisms and the mechanisms active in disease, our finding of a marked correlation between replication time scales and disease association indicates that self-replication, by the simple mechanisms that act on purified proteins in vitro, should be considered a key factor in the pathology of a wide range of aggregation-related disorders.
MATERIALS AND METHODS
Choice of data for kinetic analysis
A large number of publications exist that contain largely qualitative data of the aggregation of proteins. To also be suitable for interpretation in the context of a quantitative kinetic analysis as we present it here, a number of conditions need to be met. In particular, we excluded the following:
1) experiments that did not measure a reporter of the mass of protein aggregates formed;
2) datasets that showed aggregation that was so fast that nucleation takes place fully in the dead time of the experiment (evident by a lack of positive curvature of the kinetic curves);
3) datasets in which for any other reason the purity of the sample was questionable; and
4) kinetic experiments that were performed under significant agitation [as that biases the system toward fragmentation (16)]. Brief shaking before measurement, e.g., 5 s every 10 min as for gelsolin (51), was deemed acceptable.
The two exceptions to the last point are the data for full-length tau and PrP, both of which were obtained under mild shaking. The presence of shaking may lead to a slight underestimation of the time scale of self-replication. However, we are confident that for both of these systems, self-replication is still sufficiently rapid to be relevant in vivo, because we have in fact obtained measures for the time scale of replication directly from data in living systems.
The biggest experimental problem is the presence of small amounts of preformed aggregates at the start of the experiment. While impurities other than the protein of interest are usually no longer an issue after being removed by standard purification protocols, seed aggregates can easily reform after purification due to improper handling during sample preparation that induces nucleation. The presence of such seeds can lead to a significant shortening of the lag phase in systems dominated by secondary processes. As this might lead to misinterpretation of these systems as lacking secondary processes, this was a major concern in our analysis, and the analyzed datasets were carefully inspected to minimize the risk of such a situation. In the light of the overwhelming evidence for secondary processes, together with the fact that seed impurities would bias the analysis toward a more primary nucleation-dominated conclusion, we conclude that this source of error is unlikely to be significant in the datasets selected here.
Protein purification, assay conditions, and biological role
Please refer to the original publications for the detailed methods of protein purification and conditions of the aggregation assay. In the Supplementary Materials, we give the original source for every dataset used an overview of the biological roles of each of the studied proteins and the rationale behind the choice of relevant biological time scale.
Fitting of kinetic data
The aggregation kinetics were fitted using the AmyloFit interface at www.amylofit.com (7). All data were fitted with a model that includes the processes of primary nucleation, elongation, and secondary nucleation, given by the equations
| (3) |
where the definitions of the parameters are
where M(t) is the mass concentration of aggregates; m0 is the monomer concentration at the beginning of the aggregation reaction; kn, k+, and k2 are the rate constants of primary nucleation, elongation, and secondary nucleation, respectively; and nc and n2 are the reaction orders of primary and secondary nucleation, respectively. In some special cases, the two-step nature of secondary nucleation (for some datasets of Aβ and tau) or of elongation (for α-synuclein) had to be taken into account explicitly [see Meisl et al. (7, 9) for details on the refinement of the model in those cases]. The details for all proteins can be found in the Supplementary Materials.
When data at a range of monomer concentrations are available, the models can be fitted to extract both the reaction orders and the rate constants. These parameters can then be used to calculate the rates λ and κ also at monomer concentrations not directly measured in the experiment. When data at only one monomer concentration are available, only λ and κ can be determined, but the values of the reaction orders and reaction rate constants cannot be determined separately. Thus, only those proteins for which data at a range of monomer concentrations have been measured can be used to calculate relevant time scales at in vivo concentrations, reflected in the fact that Fig. 4 contains only a subset of the proteins shown in Fig. 3. For each system, we determined either the rate constants and reaction orders, k+kn, k+k2, nc, and n2 (if data at a range of monomer concentrations were available) or only the rates λ and κ (if there were insufficient data to determine the reaction orders and rate constants separately). When data at multiple concentrations were available, we chose an intermediate concentration to calculate λ and κ for Fig. 3.
For systems not displaying any detectable secondary processes (e.g., CsgA), we set a conservative upper bound for the rate of self-replication as 10% that of primary nucleation, i.e., κ ≤ 10λ. In those cases where no self-replication was detectable in experiments, it was also not possible to determine the reaction order of the secondary process. Therefore, to calculate a bound for the self-replication rate at in vivo concentrations, we used n2 = 0. This choice was made to ensure that the quoted rate is still an upper bound on the true rate of self-replication; as the monomer concentrations in the in vitro experiments are higher than those encountered in vivo, extrapolation of the rates determined in experiment to in vivo concentrations will yield the maximal rate of self-replication for the minimal choice of reaction order, n2. As this only concerns systems that are in the lower right region of Fig. 4, an upper bound on the self-replication rate (corresponding to a lower bound on the doubling time) is all that is needed.
A similar problem is encountered for systems where primary nucleation is so slow that to observe aggregation on experimentally accessible time scales seeded experiments have to be used (e.g., α-synuclein). In those cases, we obtain an upper bound for the rate of primary nucleation by calculating the initial rate of nucleation from secondary processes acting on the seeds and set the primary rate to be equal to this (i.e., the upper bound for primary nucleation is that it produces as many nuclei as secondary processes at the beginning of the seeded reaction). Expressing this in terms of the rate constants gives , which can be used to show that , where m0 and M0 denote the initial monomer and fibril concentrations, respectively. As primary nucleation is not considered in Fig. 4, an estimate of the reaction order is not required.
Calculation of time scales
The biologically relevant time scale for diseases was chosen to be the approximate time from diagnosis/symptom onset to death, with the exception of sickle cell anemia, for which the relevant time scale was chosen to be the speed of onset of a sickle cell crisis. For the functional proteins, biologically relevant time scale is the time scale over which the associated process takes place, i.e., the time scale of biofilm assembly (CsgA and FapC), of colony spreading (PSMα), of cytoskeleton assembly (actin), of yeast cell multiplication (Sup35 prions), and of formation time of the merozoite form of the malaria parasite (FC27 MSP2). The molecular doubling time was computed using the rate constants and reaction orders determined in our fits of in vitro data along with estimates of the in vivo relevant concentrations of the aggregating proteins. As kinetics at a range of different protein concentrations was available only for a subset of the data analyzed, the time scales at in vivo relevant concentrations could be determined only for this subset of the data, thus not all proteins shown in Fig. 3 could be included in Fig. 4. Detailed numbers and references are given in tables S1 to S3.
Acknowledgments
We thank our colleague and friend the late C. Dobson for insightful comments and helpful discussions. We also thank M. Sunde, R. Anders, and C. Adda for expert input.
Funding: This work was supported by the Danish Council for Independent Research|Natural Sciences (FNU-11-113326) (M.A.), Peterhouse College, Cambridge (T.C.T.M.), Sidney Sussex College, Cambridge (G.M.), the European Research Council grant number 669237 (D.K. and G.M.), and a Herchel Smith Research Studentship (C.K.X.).
Author contributions: G.M. and C.K.X. performed literature data extraction, analysis, and visualization. J.D.T. performed the CsgA experiments. G.M. wrote the manuscript. All authors edited the manuscript.
Competing interests: G.M. is a scientific consultant for Wren Therapeutics. S.L. is a cofounder and employee of Wren Therapeutics. T.P.J.K. is a cofounder of and consultant for Wren Therapeutics. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Materials and Methods
Supplementary Text
Figs. S1 to S23
Tables S1 to S3
References
REFERENCES AND NOTES
- 1.Chiti F., Dobson C. M., Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Oosawa F., Asakura S., Hotta K., Imai N., Ooi T., G-F transformation of actin as a fibrous condensation. Aust. J. Polit. Sci. 37, 323–336 (1959). [Google Scholar]
- 4.Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W., Functional amyloid–from bacteria to humans. Trends Biochem. Sci. 32, 217–224 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Knowles T. P. J., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Knowles T. P. J., Waudby C. A., Devlin G. L., Cohen S. I. A., Aguzzi A., Vendruscolo M., Terentjev E. M., Welland M. E., Dobson C. M., An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Meisl G., Kirkegaard J. B., Arosio P., Michaels T. T. C., Vendruscolo M., Dobson C. M., Linse S., Knowles T. P. J., Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 11, 252–272 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bas J. G., Pieters E., van Eldijk M. B., Nolte R. J. M., Mecinović J., Natural supramolecular protein assemblies. Chem. Soc. Rev. 45, 24–39 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Meisl G., Rajah L., Cohen S. A. I., Pfammatter M., Saric A., Hellstrand E., Buell A. K., Aguzzi A., Linse S., Vendruscolo M., Dobson C. M., Knowles T. P. J., Scaling behaviour and rate-determining steps in filamentous self-assembly. Chem. Sci. 8, 7087–7097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen S. I. A., Vendruscolo M., Welland M. E., Dobson C. M., Terentjev E. M., Knowles T. P. J., Nucleated polymerization with secondary pathways. I. Time evolution of the principal moments. J. Chem. Phys. 135, 065105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arosio P., Knowles T. P. J., Linse S., On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 17, 7606–7618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atarashi R., Moore R. A., Sim V. L., Hughson A. G., Dorward D. W., Onwubiko H. A., Priola S. A., Caughey B., Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat. Methods 4, 645–650 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Pfammatter M., Andreasen M., Meisl G., Taylor C. G., Adamcik J., Bolisetty S., Sanchez-Ferrer A., Klenerman D., Dobson C. M., Mezzenga R., Knowles T. P. J., Aguzzi A., Hornemann S., Absolute quantification of amyloid propagons by digital microfluidics. Anal. Chem. 89, 12306–12313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J., Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Hammer N. D., Chapman M. R., The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 283, 21530–21539 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S. I. A., Linse S., Luheshi L. M., Hellstrand E., White D. A., Rajah L., Otzen D. E., Vendruscolo M., Dobson C. M., Knowles T. P. J., Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. 110, 9758–9763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen S. I. A., Vendruscolo M., Dobson C. M., Knowles T. P. J., From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 421, 160–171 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Dunstan D. E., Hamilton-Brown P., Asimakis P., Ducker W., Bertolini J., Shear flow promotes amyloid-β fibrilization. Protein Eng. Des. Sel. 22, 741–746 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Kundel F., Hong L., Falcon B., McEwan W. A., Michaels T. C. T., Meisl G., Esteras N., Abramov A. Y., Knowles T. J. P., Goedert M., Klenerman D., Measurement of tau filament fragmentation provides insights into prion-like spreading. ACS Chem. Neurosci. 9, 1276–1282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sang J. C., Meisl G., Thackray A. M., Hong L., Ponjavic A., Knowles T. P. J., Bujdoso R., Klenerman D., Direct observation of murine prion protein replication in vitro. J. Am. Chem. Soc. 140, 14789–14798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Chen L.-J., Wang H.-Y., Wang Y.-Q., Perrett S., Deletion of a Ure2 C-terminal prion-inhibiting region promotes the rate of fibril seed formation and alters interaction with Hsp40. Protein Eng. Des. Sel. 24, 69–78 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sang J. C., Hidari E., Meisl G., Ranasinghe R. T., Spillantini M. G., Klenerman D., Super-resolution imaging reveals α-synuclein seeded aggregation in SH-SY5Y cells. Commun. Biol. 4, 613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrone F. A., Hofrichter J., Eaton W. A., Kinetics of sickle hemoglobin polymerization. II. a double nucleation mechanism. J. Mol. Biol. 183, 611–631 (1985). [DOI] [PubMed] [Google Scholar]
- 24.Meisl G., Yang X., Hellstrand E., Frohm B., Kirkegaard J. B., Cohen S. I. A., Dobson C. M., Linse S., Knowles T. P. J., Differences in nucleation behavior underlie the contrasting aggregation kinetics of the aβ40 and aβ42 peptides. Proc. Natl. Acad. Sci. 111, 9384–9389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaspar R., Meisl G., Buell A. K., Young L., Kaminski C. F., Knowles T. P. J., Sparr E., Linse S., Secondary nucleation of monomers on fibril surface dominates α-synuclein aggregation and provides autocatalytic amyloid amplification. Q. Rev. Biophys. 50, E6 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez Camargo D. C., Chia S., Menzies J., Mannini B., Meisl G., Lundqvist M., Pohl C., Bernfur K., Lattanzi V., Habchi J., Cohen S. I. A., Knowles T. P. J., Vendruscolo M., Linse S., Surface-catalyzed secondary nucleation dominates the generation of toxic IAPP aggregates. Front. Mol. Biosci. 8, 757425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez Camargo D. C., Sileikis E., Chia S., Axell E., Bernfur K., Cataldi R. L., Cohen S. I. A., Meisl G., Habchi J., Knowles T. P. J., Vendruscolo M., Linse S., Proliferation of tau 304-380 fragment aggregates through autocatalytic secondary nucleation. ACS Chem. Nerosci. 12, 4406–4415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thacker D., Sanagavarapu K., Frohm B., Meisl G., Knowles T. P. J., Linse S., The role of fibril structure and surface hydrophobicity in secondary nucleation of amyloid fibrils. Proc. Natl. Acad. Sci. 117, 25272–25283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anwar J., Khan S., Lindfors L., Secondary crystal nucleation: Nuclei breeding factory uncovered. Angew. Chem. Int. Ed. 54, 14681–14684 (2015). [DOI] [PubMed] [Google Scholar]
- 30.J. F. Richardson, J. H. Harker, J. R. Backhurst, Chapter 15 - crystallisation, in Chemical Engineering (Fifth Edition), J. F. Richardson, J. H. Harker, J. R. Backhurst, Eds. (Chemical Engineering Series, Butterworth-Heinemann, 5 ed., 2002), pp. 827–900. [Google Scholar]
- 31.Törnquist M., Michaels T. C. T., Sanagavarapu K., Yang X., Meisl G., Cohen S. I. A., Knowles T. P. J., Linse S., Secondary nucleation in amyloid formation. Chem. Commun. 54, 8667–8684 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Close W., Neumann M., Schmidt A., Hora M., Annamalai K., Schmidt M., Reif B., Schmidt V., Grigorieff N., Fändrich M., Physical basis of amyloid fibril polymorphism. Nat. Commun. 9, 699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pham C. L. L., Rey A., Lo V., Soulès M., Ren Q., Meisl G., Knowles T. P. J., Kwan A. H., Sunde M., Self-assembly of mpg1, a hydrophobin protein from the rice blast fungus that forms functional amyloid coatings, occurs by a surface-driven mechanism. Sci. Rep. 6, 25288 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammer N. D., Schmidt J. C., Chapman M. R., The curli nucleator protein, csgb, contains an amyloidogenic domain that directs csga polymerization. Proc. Natl. Acad. Sci. 104, 12494–12499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galvagnion C., Buell A. K., Meisl G., Michaels T. C. T., Vendruscolo M., Knowles T. P. J., Dobson C. M., Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat. Chem. Biol. 11, 229–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson T. B., Meisl G., Knowles T. P. J., Goriely A., The role of clearance mechanisms in the kinetics of pathological protein aggregation involved in neurodegenerative diseases. J. Chem. Phys. 154, 125101 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Frankel R., Törnquist M., Meisl G., Hansson O., Andreasson U., Zetterberg H., Blennow K., Frohm B., Cedervall T., Knowles T. P. J., Leiding T., Linse S., Autocatalytic amplification of Alzheimer-associated aβ42 peptide aggregation in human cerebrospinal fluid. Commun. Biol. 2, 365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meisl G., Kurt T., Condado-Morales I., Bett C., Sorce S., Nuvolone M., Michaels T. C. T., Heinzer D., Avar M., Cohen S. I. A., Hornemann S., Aguzzi A., Dobson C. M., Sigurdson C. J., Knowles T. P. J., Scaling analysis reveals the mechanism and rates of prion replication in vivo. Nat. Struct. Mol. Biol. 28, 365–372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meisl G., Hidari E., Allinson K., Rittman T., De Vos S. L., Sanchez J. S., Xu C. K., Duff K. E., Johnson K. A., Rowe J. B., Hyman B. T., Knowles T. P. J., Klenerman D., In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease. Sci. Adv. 7, eabh1448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgs H. N., Blanchoin L., Pollard T. D., Influence of the C terminus of Wiskott-Aldrich syndrome protein (wasp) and the arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Dueholm M. S., Petersen S. V., Sønderkær M., Larsen P., Christiansen G., Hein K. L., Enghild J. J., Nielsen J. L., Nielsen K. L., Nielsen P. H., Otzen D. E., Functional amyloid in Pseudomonas. Mol. Microbiol. 77, 1009–1020 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Rouse S. L., Matthews S. J., Dueholm M. S., Ecology and biogenesis of functional amyloids in Pseudomonas. J. Mol. Biol. 430, 3685–3695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian P., Boomsma W., Wang Y., Otzen D. E., Jensen M. H., Lindorff-Larsen K., Structure of a functional amyloid protein subunit computed using sequence variation. J. Am. Chem. Soc. 137, 22–25 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Rasmussen C. B., Christiansen G., Vad B. S., Lynggaard C., Enghild J. J., Andreasen M., Otzen D., Imperfect repeats in the functional amyloid protein fapc reduce the tendency to fragment during fibrillation. Protein Sci. 28, 633–642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohan M. C., Shinn M. K., Lalmansingh J. M., Pappu R. V., Uncovering non-random binary patterns within sequences of intrinsically disordered proteins. J. Mol. Biol. 434, 167373 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). [DOI] [PubMed] [Google Scholar]
- 47.Luk K. C., Kehm V., Carroll J., Zhang B., O’Brien P., Trojanowski J. Q., Lee V. M.-Y., Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clavaguera F., Akatsu H., Fraser G., Anthony Crowther R., Frank S., Hench J., Probst A., Winkler D. T., Reichwald J., Staufenbiel M., Ghetti B., Goedert M., Tolnay M., Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. 110, 9535–9540 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meisl G., Knowles T. P. J., Klenerman D., The molecular processes underpinning prion-like spreading and seed amplification in protein aggregation. Curr. Opin. Neurobiol. 61, 58–64 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Lau A., So R. W. L., Lau H. H. C., Sang J. C., Ruiz-Riquelme A., Fleck S. C., Stuart E., Menon S., Visanji N. P., Meisl G., Faidi R., Marano M. M., Schmitt-Ulms C., Wang Z., Fraser P. E., Tandon A., Hyman B. T., Wille H., Ingelsson M., Klenerman D., Watts J. C., α-synuclein strains target distinct brain regions and cell types. Nat. Neurosci. 23, 21–31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solomon J. P., Yonemoto I. T., Murray A. N., Price J. L., Powers E. T., Balch W. E., Kelly J. W., The 8 and 5 kda fragments of plasma gelsolin form amyloid fibrils by a nucleated polymerization mechanism, while the 68 kda fragment is not amyloidogenic. Biochemistry 48, 11370–11380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiffert T., Meisl G., Flagmeier P., De S., Dunning C. J. R., Frohm B., Zetterberg H., Blennow K., Portelius E., Klenerman D., Dobson C. M., Knowles T. P. J., Linse S., Increased secondary nucleation underlies accelerated aggregation of the four-residue n-terminally truncated aβ42 species aβ5–42. ACS Chem. Nerosci. 10, 2374–2384 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Andreasen M., Meisl G., Taylor J. D., Michaels T. C. T., Levin A., Otzen D. E., Chapman M. R., Dobson C. M., Matthews S. J., Knowles T. P. J., Physical determinants of amyloid assembly in biofilm formation. mBio 10, e02279-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X., Meisl G., Frohm B., Thulin E., Knowles T. P. J., Linse S., On the role of sidechain size and charge in the aggregation of aβ42 with familial mutations. Proc. Natl. Acad. Sci. 115, E5849–E5858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M., α-Synuclein in Lewy bodies. Nature 388, 839–840 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Meisl G., Yang X., Dobson C. M., Linse S., Knowles T. P. J., Modulation of electrostatic interactions to reveal a reaction network unifying the aggregation behaviour of the aβ42 peptide and its variants. Chem. Sci. 8, 4352–4362 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dominguez R., Holmes K. C., Actin structure and function. Annu. Rev. Biophys. 40, 169–186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oda T., Iwasa M., Aihara T., Maéda Y., Narita A., The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Fujii T., Iwane A. H., Yanagida T., Namba K., Direct visualization of secondary structures of f-actin by electron cryomicroscopy. Nature 467, 724–728 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Lieleg O., Claessens M. M. A. E., Bausch A. R., Structure and dynamics of cross-linked actin networks. Soft Matter 6, 218–225 (2010). [Google Scholar]
- 61.Sirangelo I., Tavassi S., Irace G., Tryptophanyl contributions to apomyoglobin fluorescence resolved by site-directed mutagenesis. Biochim. Biophys. Acta 1476, 173–180 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Sirangelo I., Malmo C., Casillo M., Mezzogiorno A., Papa M., Irace G., Tryptophanyl substitutions in apomyoglobin determine protein aggregation and amyloid-like fibril formation at physiological ph. J. Biol. Chem. 277, 45887–45891 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Vilasi S., Sarcina R., Maritato R., De Simone A., Irace G., Sirangelo I., Heparin induces harmless fibril formation in amyloidogenic w7fw14f apomyoglobin and amyloid aggregation in wild-type protein in vitro. PLOS ONE 6, e22076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barnhart M. M., Chapman M. R., Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sánchez R., Martínez J., Castro A., Pedrosa M., Quirce S., Rodríguez-Pérez R., Gasset M., The amyloid fold of gad m 1 epitopes governs ige binding. Sci. Rep. 6, 32801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castellanos M., Torres-Pardo A., Rodríguez-Pérez R., Gasset M., Amyloid assembly endows gad m 1 with biomineralization properties. Biomolecules 8, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kakkar V., Månsson C., de Mattos E. P., Bergink S., van der Zwaag M., van Waarde M. A. W. H., Kloosterhuis N. J., Melki R., van Cruchten R. T. P., Al-Karadaghi S., Arosio P., Dobson C. M., Knowles T. P. J., Bates G. P., van Deursen J. M., Linse S., van de Sluis B., Emanuelsson C., Kampinga H. H., The S/T-rich motif in the dnajb6 chaperone delays polyglutamine aggregation and the onset of disease in a mouse model. Mol. Cell 62, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Kar K., Jayaraman M., Sahoo B., Kodali R., Wetzel R., Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat. Struct. Mol. Biol. 18, 328–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pilkington E. H., Xing Y., Wang B., Kakinen A., Wang M., Davis T. P., Ding F., Ke P. C., Effects of protein corona on iapp amyloid aggregation, fibril remodelling, and cytotoxicity. Sci. Rep. 7, 2455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen L., Khurana R., Coats A., Frokjaer S., Brange J., Vyas S., Uversky V. N., Fink A. L., Effect of environmental factors on the kinetics of insulin fibril formation: Elucidation of the molecular mechanism. Biochemistry 40, 6036–6046 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Stroobants K., Kumita J. R., Harris N. J., Chirgadze D. Y., Dobson C. M., Booth P. J., Vendruscolo M., Amyloid-like fibrils from an α-helical transmembrane protein. Biochemistry 56, 3225–3233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasecke F., Miti T., Perez C., Barton J., Schölzel D., Gremer L., Grüning C. S. R., Matthews G., Meisl G., Knowles T. P. J., Willbold D., Neudecker P., Heise H., Ullah G., Hoyer W., Muschol M., Origin of metastable oligomers and their effects on amyloid fibril self-assembly. Chem. Sci. 9, 5937–5948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Konno T., Multistep nucleus formation and a separate subunit contribution of the amyloidgenesis of heat-denatured monellin. Protein Sci. 10, 2093–2101 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Low A., Chandrashekaran I. R., Adda C. G., Yao S., Sabo J. K., Zhang X., Soetopo A., Anders R. F., Norton R. S., Merozoite surface protein 2 of Plasmodium falciparum: Expression, structure, dynamics, and fibril formation of the conserved n-terminal domain. Biopolymers 87, 12–22 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Somé A. F., Bazié T., Zongo I., Serge Yerbanga R., Nikiéma F., Neya C., Taho L. K., Ouédraogo J.-B., Plasmodium falciparum msp1 and msp2 genetic diversity and allele frequencies in parasites isolated from symptomatic malaria patients in Bobo-Dioulasso, Burkina Faso. Parasit. Vectors 11, 323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adda C. G., Murphy V. J., Sunde M., Waddington L. J., Schloegel J., Talbo G. H., Vingas K., Kienzle V., Masciantonio R., Howlett G. J., Hodder A. N., Foley M., Anders R. F., Plasmodium falciparum merozoite surface protein 2 is unstructured and forms amyloid-like fibrils. Mol. Biochem. Parasitol. 166, 159–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Si K., Giustetto M., Etkin A., Hsu R., Janisiewicz A. M., Miniaci M. C., Kim J.-H., Zhu H., Kandel E. R., A neuronal isoform of cpeb regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell 115, 893–904 (2003). [DOI] [PubMed] [Google Scholar]
- 78.Alarcon J. M., Hodgman R., Theis M., Huang Y.-S., Kandel E. R., Richter J. D., Selective modulation of some forms of schaffer collateral-ca1 synaptic plasticity in mice with a disruption of the cpeb-1 gene. Learn. Mem. 11, 318–327 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keleman K., Krüttner S., Alenius M., Dickson B. J., Function of the drosophila cpeb protein orb2 in long-term courtship memory. Nat. Neurosci. 10, 1587–1593 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Bajakian T. H., Cervantes S. A., Soria M. A., Beaugrand M., Kim J. Y., Service R. J., Siemer A. B., Metal binding properties of the N-terminus of the functional amyloid orb2. Biomolecules 7, 57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Periasamy S., Chatterjee S. S., Cheung G. Y. C., Otto M., Phenol-soluble modulins in staphylococci: What are they originally for? Commun. Integr. Biol. 5, 275–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheung G. Y. C., Joo H.-S., Chatterjee S. S., Otto M., Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 38, 698–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang R., Braughton K. R., Kretschmer D., Bach T.-H. L., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., Peschel A., De Leo F. R., Otto M., Identification of novel cytolytic peptides as key virulence determinants for community-associated mrsa. Nat. Med. 13, 1510–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Laabei M., David Jamieson W., Yang Y., van den Elsen J., Toby A., Jenkins A., Investigating the lytic activity and structural properties of staphylococcus aureus phenol soluble modulin (psm) peptide toxins. Biochim. Biophys. Acta 1838, 3153–3161 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Wang R., Khan B. A., Cheung G. Y. C., Bach T.-H. L., Jameson-Lee M., Kong K.-F., Queck S. Y., Otto M., Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwartz K., Syed A. K., Stephenson R. E., Rickard A. H., Boles B. R., Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLOS Pathog. 8, e1002744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tayeb-Fligelman E., Tabachnikov O., Moshe A., Goldshmidt-Tran O., Sawaya M. R., Coquelle N., Colletier J.-P., Landau M., The cytotoxic Staphylococcus aureus psmα3 reveals a cross-α amyloid-like fibril. Science 355, 831–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salinas N., Colletier J.-P., Moshe A., Landau M., Extreme amyloid polymorphism in Staphylococcus aureus virulent psmα peptides. Nat. Commun. 9, 3512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zaman M., Andreasen M., Cross-talk between individual phenol-soluble modulins in Staphylococcus aureus biofilm enables rapid and efficient amyloid formation. eLife 9, e59776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pitkänen P., Westermark P., Cornwell G. G. III, Murdoch W., Amyloid of the seminal vesicles. A distinctive and common localized form of senile amyloidosis. Am. J. Pathol. 110, 64–69 (1983). [PMC free article] [PubMed] [Google Scholar]
- 91.Linke R. P., Joswig R., Murphy C. L., Wang S., Zhou H., Gross U., Rocken C., Westermark P., Weiss D. T., Solomon A., Senile seminal vesicle amyloid is derived from semenogelin I. J. Lab. Clin. Med. 145, 187–193 (2005). [DOI] [PubMed] [Google Scholar]
- 92.Sharma N., Vishwanath S., Patel B. K., Recombinant human semenogelin-1 (sg1) and sg1 (1-159) form detergent stable amyloid like aggregates in vitro. Protein Pept. Lett. 23, 87–96 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Lilja H., Abrahamsson P. A., Lundwall A., Semenogelin, the predominant protein in human semen: Primary structure and identification of closely related proteins in the male accessory sex glands and on the spermatozoa. J. Biol. Chem. 264, 1894–1900 (1989). [PubMed] [Google Scholar]
- 94.Frohm B., DeNizio J. E., Lee D. S. M., Gentile L., Olsson U., Malm J., Åkerfeldt K. S., Linse S., A peptide from human semenogelin I self-assembles into a pH-responsive hydrogel. Soft Matter 11, 414–421 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Pettersson T., Konttinen Y. T., Maury C. P. J., Treatment strategies for amyloid a amyloidosis. Expert Opin. Pharmacother. 9, 2117–2128 (2008). [DOI] [PubMed] [Google Scholar]
- 96.Lu J., Yu Y., Zhu I., Cheng Y., Sun P. D., Structural mechanism of serum amyloid a-mediated inflammatory amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 111, 5189–5194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu J., Zhu H., Guo J.-t., de Beer F. C., Kindy M. S., Expression of mouse apolipoprotein saa1.1 in ce/j mice: Isoform-specific effects on amyloidogenesis. Lab. Invest. 80, 1797–1806 (2000). [DOI] [PubMed] [Google Scholar]
- 98.Srinivasan S., Patke S., Wang Y., Ye Z., Litt J., Srivastava S. K., Lopez M. M., Kurouski D., Lednev I. K., Kane R. S., Col W., Pathogenic serum amyloid A 1.1 shows a long oligomer-rich fibrillation lag phase contrary to the highly amyloidogenic non-pathogenic SAA2.2. J. Biol. Chem. 288, 2744–2755 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sipe J. D., Carreras I., Gonnerman W. A., Cathcart E. S., de Beer M. C., de Beer F. C., Characterization of the inbred ce/j mouse strain as amyloid resistant. Am. J. Pathol. 143, 1480–1485 (1993). [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L., Lashuel H. A., Colón W., From hexamer to amyloid: Marginal stability of apolipoprotein SAA2.2 leads to in vitro fibril formation at physiological temperature. Amyloid 12, 139–148 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Ye Z., Poueymiroy D. B., Javier Aguilera J., Srinivasan S., Wang Y., Serpell L. C., Colón W., Inflammation protein SAA2.2 spontaneously forms marginally stable amyloid fibrils at physiological temperature. Biochemistry 50, 9184–9191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Noguchi C. T., Schechter A. N., Sickle hemoglobin polymerization in solution and in cells. Annu. Rev. Biophys. Biophys. Chem. 14, 239–263 (1985). [DOI] [PubMed] [Google Scholar]
- 103.Eaton W. A., Hofrichter J., Sickle cell hemoglobin polymerization. Adv. Protein Chem. 40, 63–279 (1990). [DOI] [PubMed] [Google Scholar]
- 104.Ferrone F. A., Hofrichter J., Eaton W. A., Kinetics of sickle hemoglobin polymerization. I. studies using temperature-jump and laser photolysis techniques. J. Mol. Biol. 183, 591–610 (1985). [DOI] [PubMed] [Google Scholar]
- 105.Kushnirov V. V., Kochneva-Pervukhova N. V., Chechenova M. B., Frolova N. S., Ter-Avanesyan M. D., Prion properties of the sup35 protein of yeast Pichia methanolica. EMBO J. 19, 324–331 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.True H. L., Lindquist S. L., A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477–483 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Liebman S. W., Chernoff Y. O., Prions in yeast. Genetics 191, 1041–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sant’Anna R., Fernández M. R., Batlle C., Navarro S., de Groot N. S., Serpell L., Ventura S., Characterization of amyloid cores in prion domains. Sci. Rep. 6, 34274–34274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Glover J. R., Kowal A. S., Schirmer E. C., Patino M. M., Liu J.-J., Lindquist S., Self-seeded fibers formed by sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997). [DOI] [PubMed] [Google Scholar]
- 110.Wickner R. B., [URE3] as an altered ure2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994). [DOI] [PubMed] [Google Scholar]
- 111.Coschigano P. W., Magasanik B., The ure2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol. Cell. Biol. 11, 822–832 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang Y., Li H., Zhu L., Zhou J.-M., Perrett S., Amyloid nucleation and hierarchical assembly of ure2p fibrils: Role of asparagine/glutamine repeat and nonrepeat regions of the prion domain. J. Biol. Chem. 279, 3361–3369 (2004). [DOI] [PubMed] [Google Scholar]
- 113.Ferguson N., Berriman J., Petrovich M., Sharpe T. D., Finch J. T., Fersht A. R., Rapid amyloid fiber formation from the fast-folding ww domain fbp28. Proc. Natl. Acad. Sci. U.S.A. 100, 9814–9819 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eaton W. A., Hofrichter J., Ross P. D., Tschudin R. G., Becker E. D., Comparison of sickle cell hemoglobin gelation kinetics measured by nmr and optical methods. Biochem. Biophys. Res. Commun. 69, 538–547 (1976). [DOI] [PubMed] [Google Scholar]
- 115.Dommershuijsen L. J., Heshmatollah A., Darweesh S. K. L., Koudstaal P. J., Arfan Ikram M., Kamran Ikram M., Life expectancy of Parkinsonism patients in the general population. Parkinsonism Relat. Disord. 77, 94–99 (2020). [DOI] [PubMed] [Google Scholar]
- 116.Wilhelm B. G., Mandad S., Truckenbrodt S., Kröhnert K., Schäfer C., Rammner B., Koo S. J., Claßen G. A., Krauss M., Haucke V., Urlaub H., Rizzoli S. O., Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science 344, 1023–1028 (2014). [DOI] [PubMed] [Google Scholar]
- 117.Zanetti O., Solerte S. B., Cantoni F., Life expectancy in Alzheimer’s disease (ad). Arch. Gerontol. Geriatr. 49, 237–243 (2009). [DOI] [PubMed] [Google Scholar]
- 118.Lue L. F., Kuo Y. M., Roher A. E., Brachova L., Shen Y., Sue L., Beach T., Kurth J. H., Rydel R. E., Rogers J., Soluble amyloid beta peptide concentration as a predictor of synaptic change in Alzheimer’s disease. Am. J. Pathol. 155, 853–862 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carlsson A. E., Actin dynamics: From nanoscale to microscale. Annu. Rev. Biophys. 39, 91–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.H. Lodish, A. Berk, S. L. Zipursky, Molecular Cell Biology (The Dynamics of Actin Assembly, New York: W. H. Freeman, 4 ed., 2000), Section 18.2. [Google Scholar]
- 121.Grüring C., Heiber A., Kruse F., Ungefehr J., Gilberger T.-W., Spielmann T., Development and host cell modifications of Plasmodium falciparum blood stages in four dimensions. Nat. Commun. 2, 165 (2011). [DOI] [PubMed] [Google Scholar]
- 122.Gilson P. R., Nebl T., Vukcevic D., Moritz R. L., Sargeant T., Speed T. P., Schofield L., Crabb B. S., Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 5, 1286–1299 (2006). [DOI] [PubMed] [Google Scholar]
- 123.Schmidt E.-K., Atula S., Tanskanen M., Nikoskinen T., Notkola I.-L., Kiuru-Enari S., Causes of death and life span in Finnish gelsolin amyloidosis. Ann. Med. 48, 352–358 (2016). [DOI] [PubMed] [Google Scholar]
- 124.Kulakowska A., Drozdowski W., Sadzynski A., Bucki R., Janmey P. A., Gelsolin concentration in cerebrospinal fluid from patients with multiple sclerosis and other neurological disorders. Eur. J. Neurol. 15, 584–588 (2008). [DOI] [PubMed] [Google Scholar]
- 125.Yu C.-C., Zendzian-Piotrowska M., Charmas M., Długołęcka B., Baranowski M., Górski J., Bucki R., Change in blood gelsolin concentration in response to physical exercise. Biol. Sport 30, 169–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Foroud T., Gray J., Ivashina J., Conneally P. M., Differences in duration of Huntington’s disease based on age at onset. J. Neurol. Neurosurg. Psychiatry 66, 52–56 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Macdonald D., Tessari M. A., Boogaard I., Smith M., Pulli K., Szynol A., Albertus F., Lamers M. B. A. C., Dijkstra S., Kordt D., Reindl W., Herrmann F., Allister G. M., Fischer D. F., Munoz-Sanjuan I., Quantification assays for total and polyglutamine-expanded Huntingtin proteins. PLOS ONE 9, e96854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baldo B., Sajjad M. U., Cheong R. Y., Bigarreau J., Vijayvargia R., Lean C. M., Perrier A. L., Seong I. S., Halliday G., Petersén Å., Kirik D., Quantification of total and mutant huntingtin protein levels in biospecimens using a novel alphaLISA assay. eNeuro 5, ENEURO.0234–ENEU18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fonseca V. A., Defining and characterizing the progression of type 2 diabetes. Diabetes Care 32 ( suppl 2), S151–S156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jaikaran E. T. A. S., Nilsson M. R., Clark A., Pancreatic beta-cell granule peptides form heteromolecular complexes which inhibit islet amyloid polypeptide fibril formation. Biochem. J. 377, 709–716 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pocchiari M., Puopolo M., Croes E. A., Budka H., Gelpi E., Collins S., Lewis V., Sutcliffe T., Guilivi A., Delasnerie-Laupretre N., Brandel J.-P., Alperovitch A., Zerr I., Poser S., Kretzschmar H. A., Ladogana A., Rietvald I., Mitrova E., Martinez-Martin P., de Pedro-Cuesta J., Glatzel M., Aguzzi A., Cooper S., Mackenzie J., van Duijn C. M., Will R. G., Predictors of survival in sporadic Creutzfeldt-Jakob disease and other human transmissible spongiform encephalopathies. Brain 127, 2348–2359 (2004). [DOI] [PubMed] [Google Scholar]
- 132.Vallabh S. M., Nobuhara C. K., Llorens F., Zerr I., Parchi P., Capellari S., Kuhn E., Klickstein J., Safar J. G., Nery F. C., Swoboda K. J., Geschwind M. D., Zetterberg H., Arnold S. E., Minikel E. V., Schreiber S. L., Prion protein quantification in human cerebrospinal fluid as a tool for prion disease drug development. Proc. Natl. Acad. Sci. 116, 7793–7798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kizaki H., Omae Y., Tabuchi F., Saito Y., Sekimizu K., Kaito C., Cell-surface phenol soluble modulins regulate Staphylococcus aureus colony spreading. PLOS ONE 11, e0164523 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ahbap E., Kara E., Sahutoglu T., Basturk T., Koc Y., Sakaci T., Sevinc M., Akgol C., Ucar Z. A., Kayalar A. O., Bayraktar F., Ozagari A. A., Unsal A., Outcome of 121 patients with renal amyloid a amyloidosis. J. Res. Med. Sci. 19, 644–649 (2014). [PMC free article] [PubMed] [Google Scholar]
- 135.Lachmann H. J., Goodman H. J. B., Gilbertson J. A., Ruth Gallimore J., Sabin C. A., Gillmore J. D., Hawkins P. N., Natural history and outcome in systemic AA amyloidosis. N. Engl. J. Med. 356, 2361–2371 (2007). [DOI] [PubMed] [Google Scholar]
- 136.Biasucci L. M., Liuzzo G., Grillo R. L., Caligiuri G., Rebuzzi A. G., Buffon A., Summaria F., Ginnetti F., Fadda G., Maseri A., Elevated levels of c-reactive protein at discharge in patients with unstable angina predict recurrent instability. Circulation 99, 855–860 (1999). [DOI] [PubMed] [Google Scholar]
- 137.Sack G. H. Jr., Serum amyloid A – a review. Mol. Med. 24, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hofrichter J., Ross P. D., Eaton W. A., Kinetics and mechanism of deoxyhemoglobin s gelation: A new approach to understanding sickle cell disease. Proc. Natl. Acad. Sci. U.S.A. 71, 4864–4868 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Diggs L. W., Sickle cell crises: Ward Burdick Award contribution. Am. J. Clin. Pathol. 44, 1–19 (1965). [Google Scholar]
- 140.P. R. Sarma, Red cell indices, in Clinical Methods: The History, Physical, and Laboratory Examinations, H. K. Walker, W. D. Hall, J. W. Hurst, Eds. (Butterworths, 3 ed., 1990). [PubMed] [Google Scholar]
- 141.Minois N., Frajnt M., Wilson C., Vaupel J. W., Advances in measuring lifespan in the yeast saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 102, 402–406 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Váchová L., Čáp M., Palková Z., Yeast colonies: A model for studies of aging, environmental adaptation, and longevity. Oxid. Med. Cell. Longev. 2012, 601836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khan T., Kandola T. S., Wu J., Venkatesan S., Ketter E., Lange J. J., Gama A. R., Box A., Unruh J. R., Cook M., Halfmann R., Quantifying nucleation in vivo reveals the physical basis of prion-like phase behavior. Mol. Cell 71, 155–168.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Supplementary Text
Figs. S1 to S23
Tables S1 to S3
References