Abstract
An estimated 3 billion people lack access to dermatological care globally. Artificial intelligence (AI) may aid in triaging skin diseases and identifying malignancies. However, most AI models have not been assessed on images of diverse skin tones or uncommon diseases. Thus, we created the Diverse Dermatology Images (DDI) dataset—the first publicly available, expertly curated, and pathologically confirmed image dataset with diverse skin tones. We show that state-of-the-art dermatology AI models exhibit substantial limitations on the DDI dataset, particularly on dark skin tones and uncommon diseases. We find that dermatologists, who often label AI datasets, also perform worse on images of dark skin tones and uncommon diseases. Fine-tuning AI models on the DDI images closes the performance gap between light and dark skin tones. These findings identify important weaknesses and biases in dermatology AI that should be addressed for reliable application to diverse patients and diseases.
A diverse, curated clinical dermatology image set can help address AI in dermatology limitations.
INTRODUCTION
Globally, an estimated 3 billion people have inadequate access to medical care for skin disease (1). Even in developed countries, such as the United States, there is a shortage and unequal distribution of dermatologists, which can lead to long wait times for skin evaluation (2). Artificial intelligence (AI) diagnostic and decision support tools in dermatology, which have seen rapid development over the last few years, could help triage lesions or aid nonspecialist physicians in diagnosing skin diseases and identifying potential malignancies (3–5). There are several commercial skin cancer detection algorithms with the CE mark in Europe (6).
Despite the widespread interest in dermatology AI, systematic evaluation of state-of-the-art dermatology AI models on independent real-world data has been limited. Most images used to train and test malignancy identification algorithms use siloed, private clinical image data; the data sources and curating methods are often not clearly described (5). Moreover, limitations in the current training and testing of dermatology AI models may mask potential vulnerabilities. Many algorithms are trained or tested on the International Skin Imaging Collaboration (ISIC) dataset, which contains histopathology-confirmed dermoscopic images of cutaneous malignancies but lacks images of inflammatory and uncommon diseases, or images across diverse skin tones (7–9). Images from online atlases such as Fitzpatrick 17k do not have histopathological confirmation of malignancies and likely have label noise—a subset of 504 images were reviewed by board-certified dermatologists, and only 69% of the images appeared diagnostic of the labeled condition (10). No public skin disease AI benchmarks have images of biopsy-proven malignancy on dark skin (5).
Label noise is also a major concern, as many previously published AI algorithms rely on images labeled by visual consensus—meaning that dermatologists provide labels by reviewing only a digital image without information on follow-up or biopsy confirmation (5). Visual inspection, however, can be unreliable for determining cutaneous malignancies, which often require histopathological confirmation (11).
RESULTS
Diverse Dermatology Images dataset
To ascertain potential biases in algorithm performance in this context, we curated the Diverse Dermatology Images (DDI) dataset—a pathologically confirmed benchmark dataset with diverse skin tones. The DDI was retrospectively selected from reviewing histopathologically proven lesions diagnosed in Stanford Clinics from 2010 to 2020. For all lesions, the Fitzpatrick skin type (FST), a clinical classification scheme for skin tone, was determined using chart review of the in-person visit and consensus review by two board-certified dermatologists. This dataset was designed to allow direct comparison between patients classified as FST V–VI (dark skin tones) and patients with FST I–II (light skin tones) by matching patient characteristics. There were a total of 208 images of FST I–II (159 benign and 49 malignant), 241 images of FST III–IV (167 benign and 74 malignant), and 207 images of FST V–VI (159 benign and 48 malignant) (table S1).
Previously developed dermatology AI algorithms perform worse on dark skin tones and uncommon diseases
We evaluated three algorithms on their ability to distinguish benign versus malignant lesions: ModelDerm [using the application programming interface (API) available at https://modelderm.com/] (12) and two algorithms developed from previously described datasets—DeepDerm (4) and HAM10000 (7). These algorithms were selected on the basis of their popularity, availability, and previous demonstrations of state-of-the-art performance (4, 7, 12).
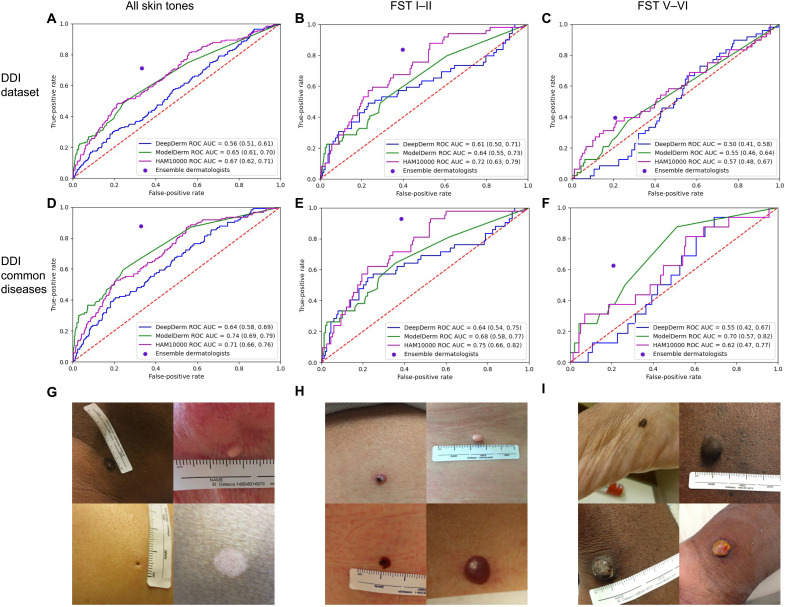
Although all three algorithms had good performance on the original datasets they were trained and tested on, their performance was worse on DDI. In the literature, ModelDerm had a previously reported receiver operator curve area under the curve (ROC-AUC) of 0.93 to 0.94 (12), while DeepDerm achieved an ROC-AUC of 0.88 and HAM10000 achieved an ROC-AUC of 0.92 on its own test data. However, when evaluated on the DDI dataset, ModelDerm had an ROC-AUC of 0.65 [95% confidence interval (CI), 0.61 to 0.70], DeepDerm had an ROC-AUC of 0.56 (0.51 to 0.61), and HAM10000 had an ROC-AUC of 0.67 (95% CI, 0.62 to 0.71) (Fig. 1A and table S2).
Fig. 1. DDI dataset and algorithm performance.
Row 1: Performance of all three AI models and the majority vote of an ensemble of dermatologists on the entire DDI dataset (A), FST I–II (B), and FST V–VI (C). Row 2: Performance across the DDI common diseases dataset with the performance of all algorithms and ensemble of dermatologists on the entire DDI common diseases dataset (D), FST I–II (E), and FST V–VI (F). Row 3: Example images from the entire DDI dataset for all skin tones (G), FST I–II (H), and FST V–VI (I). Photo Credit: DDI dataset, Stanford School of Medicine.
We assessed differential performance between FST I–II and FST V–VI; these two subsets of images were matched for diagnostic class (malignant versus benign) and patient demographics, enabling a direct comparison of performance. Across all three algorithms on the DDI dataset, ROC-AUC performance was better on the subset of Fitzpatrick I–II images (Fig. 1B) compared to Fitzpatrick V–VI (Fig. 1C), with ModelDerm having an ROC-AUC of 0.64 (0.55 to 0.7) (FST I–II) versus 0.55 (0.46 to 0.64) (FST V-VI), DeepDerm having an ROC-AUC of 0.61 (0.50 to 0.71) (FST I–II) versus 0.50 (0.41 to 0.58) (FST V–VI), and HAM10000 having an ROC-AUC of 0.72 (0.63 to 0.79) (FST I–II) versus 0.57 (0.48 to 0.67) (FST V–VI). The use of recently developed robust training methods on the DeepDerm data did not reduce the gap in performance between FST I–II and FST V–VI (table S3) (13–15). The performance on FST III-IV, which was not explicitly matched with FST I–II or FST V–VI, can be found in table S4.
Because detecting malignancy is an important feature of these algorithms and clinical care, we assessed sensitivity to see whether there was differential performance across skin tones (table S2). Across the entire DDI dataset, two algorithms showed statistically significant differential performance in detecting malignancies between FST I–II and FST V–VI: ModelDerm (sensitivity, 0.41 versus 0.12; Fisher’s exact test P = 0.0025) and DeepDerm (sensitivity, 0.69 versus 0.23; Fisher’s exact test P = 5.65 × 10−6). HAM10000 had poor sensitivity across all subsets of the data despite having a sensitivity of 0.68 on its own test set (table S2).
The datasets used to train all three AI models consist mostly of common malignancies, while the DDI dataset also includes uncommon benign and malignant lesions. To assess how this distribution shift could contribute to the model’s performance drop-off, we repeated our analysis on only the common disease split of the DDI dataset. While removing uncommon diseases led to overall improvement in performance in distinguishing between benign versus malignant disease across all algorithms with ModelDerm having an ROC-AUC of 0.74 (CI, 0.69 to 0.79), DeepDerm having an ROC-AUC of 0.64 (95% CI, 0.58 to 0.69), and HAM10000 having an ROC-AUC of 0.71 (0.66 to 0.76), this performance was still lower than performance on each AI algorithm’s original test set (Fig. 1D). DeepDerm and HAM10000 continued to have a difference in performance between FST I–II and FST V–VI even among common diseases (table S2). The performance on the uncommon disease subset can be seen in table S5.
Fine-tuning on diverse data can close performance gap between light and dark skin tones
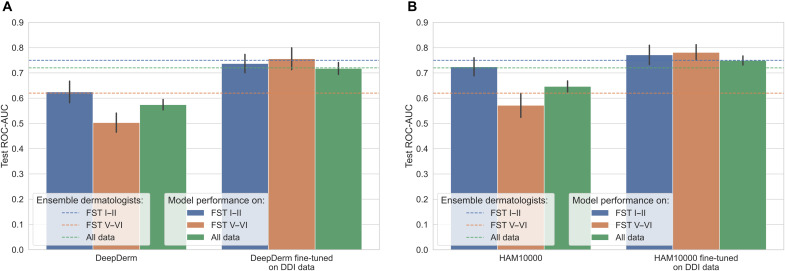
We assessed how using DDI for model fine-tuning could be used to help close the differential performance between FST I–II and FST V–VI for DeepDerm and HAM10000, the two algorithms for which we had access to model weights. Fine-tuning improved overall model performance across all the skin tones. We found that the inclusion of the diverse DDI data with fine-tuning closed the gap in performance between FST I–II and FST V–VI (Fig. 2) for both DeepDerm and HAM10000. After fine-tuning the DeepDerm algorithm on DDI data, we achieved an ROC-AUC of 0.73 (95% CI, 0.70 to 0.77) on FST I–II compared to 0.76 (0.71 to 0.80) for FST V–VI. Similarly, after fine-tuning the HAM10000 algorithm, we achieved ROC-AUCs of 0.77 (0.73 to 0.80) on FST I–II data and 0.78 (0.75 to 0.81) on FST V–VI data. Fine-tuning also made the performance of the algorithms to be equivalent or exceeding that of the dermatologists (Fig. 2). For FST V–VI data, the fine-tuned algorithms were significantly better than dermatologists (P = 9.33 × 10−5 for DeepDerm, P = 5.91 × 10−10 for HAM10000; one-sided t test) and baseline models before fine-tuning (P = 1.71 × 10−10 for DeepDerm, P = 5.72 × 10−9 for HAM10000; one-sided t test). Fine-tuning algorithms only on FST I–II data led to an overall improvement of performance of all skin tones, but the gap in performance between FST I–II and FST V–VI remained significant (P = 3.23 × 10−3, two-sided t test) (fig. S2).
Fig. 2. Algorithm performance after fine-tuning.
Fine-tuned DeepDerm (A) and HAM10000 (B) on the DDI dataset (as described in Materials and Methods) compared to baseline (first three bars in each panel). Fine-tuning closes the gap between FST I–II and FST V–VI performance and leads to overall performance improvement. Ninety-five percent confidence interval is calculated using bootstrapping across the 20 seeds for both baseline and fine-tuned models to allow direct comparison.
Dermatologist consensus labeling can lead to differential label noise across skin tones
Many dermatology AI algorithms rely on data labels generated from dermatologists reviewing images alone, a task that is different from actual clinical practice where in-person assessment and diagnostic tests aid diagnosis. To assess the potential for label noise, we compare dermatologist-generated labels (benign or malignant) versus biopsy-proven labels across DDI. Notably, dermatologist labels generally outperformed algorithms before fine-tuning (Fig. 1). Dermatologist labels were less noisy on images of DDI common diseases compared to the whole DDI dataset (ROC-AUC, 0.82 to 0.72; sensitivity, 0.88 to 0.71; P = 0.0089). Label noise also varied between FST I–II and FST V–VI (sensitivity, 0.72 versus 0.59; Fisher’s exact test P = 8.8 × 10−6), and this difference remained statistically significant when assessing only common disease (sensitivity, 0.93 versus 0.62; Fisher’s exact test P = 0.0096).
DISCUSSION
Dermatology AI algorithms have been envisioned as providing support to nondermatologist specialists or helping with triaging lesions before clinical care (16). Our analyses highlight several key challenges for AI algorithms developed for detecting cutaneous malignancies: (i) State-of-the-art dermatology AI algorithms have substantially worse performance on lesions appearing on dark skin compared to light skin using biopsy-proven malignancies, the gold standard for disease annotation; (ii) as a consequence, there is a substantial drop-off in the overall performance of AI algorithms developed from previously described data when benchmarked on DDI; however, fine-tuning on diverse data can help close performance gaps between skin tones; and (3) there are differences in dermatologist visual consensus label performance, which is commonly used to train AI models, across skin tones and uncommon conditions.
In the current dermatology workflow, there are disparities in skin cancer diagnosis and outcomes, with patients with skin of color getting diagnosed at later stages, leading to increased morbidity, mortality, and costs (17, 18). To alleviate disparities rather than exacerbate them, dermatology datasets used for AI training and testing must include dark skin tones. We release the DDI benchmark as a first step toward this goal and demonstrate how such data can be useful in further evaluation of previously developed algorithms. Because every lesion in DDI is biopsied, we are less likely to have label noise. The dataset also includes ambiguous benign lesions that would have been difficult to label visually but are representative of the type of lesions seen in clinical practice.
Resources such as DDI can show the limitations of current dermatology AI models, which can be sensitive to their training data distributions. ModelDerm was trained on predominantly clinical images, DeepDerm used a mix of clinical and dermoscopic images, while HAM10000 only trained on dermoscopic images. ModelDerm has greater skin tone diversity, since it was trained on a mix of White and Asian patients, while DeepDerm and HAM10000 were trained on predominantly White patients. Unlike DeepDerm and HAM10000, ModelDerm did not show a drop-off in ROC-AUC between FST I–II and FST V–VI on the DDI common diseases dataset. The HAM10000 data are more limited in the number of diagnoses compared to DeepDerm or ModelDerm; however, unlike those datasets, every malignancy in HAM10000 is histopathologically confirmed, meaning there is less label noise (4, 7, 12). This could be one reason why the model trained on HAM10000 achieved the best ROC-AUC performance on the DDI dataset. To assess whether more recent training techniques would improve this disparity, we applied three different robust training methods that are designed to improve algorithmic fairness to train DeepDerm. However, these training regimens did not reduce disparity across the skin tones, further suggesting that the performance limitations lie with the lack of diverse training data, consistent with previous reports in other settings (10). The sensitivity-specificity threshold for all the algorithms was set on the basis of the original training/test data to avoid overfitting and to mimic how thresholds are set for real-world commercial applications. These thresholds may not generalize well to new datasets.
Many previously published algorithms have relied on visual consensus labels from a small group of dermatologists (4, 5, 19). However, dermatologist labels from visual inspection can be noisy since they lack pertinent information used for diagnosis such as clinical history, in-person evaluation, and other diagnostic tests. Consistent with our findings, Hekler et al. (20) showed that a melanoma classifier trained on dermatologist visual consensus labels rather than ground truth pathologic diagnosis performed significantly worse than a classifier trained on ground truth labels. We found that consensus label performance was worse when uncommon diagnoses were included and on FST V–VI skin tones. Biases in photography such as color balancing could be a contributor as could the difficulty of capturing salient features such as erythema in photography of dark skin tones (21). Prior studies have also suggested that the lack of diverse skin tones in dermatology educational materials may also contribute (22–24). Here, we make no claims regarding overall general dermatologists’ performance but illustrate the potential for differences in label noise across skin tones during the consensus labeling process.
Prior to the DDI dataset, there were no publicly available benchmark datasets that included biopsy-proven malignancies in dark skin (5). Although this dataset increases the number of biopsy-proven diagnoses across diverse skin tones, it has limitations because of the nature of biopsy-curated data. Because benign lesions are not regularly biopsied, this dataset is enriched for “ambiguous” lesions that a clinician determined required biopsy; however, these lesions are reflective of the kinds of lesions an algorithm may encounter. This dataset is not comprehensive of all diagnoses in dermatology and is tailored toward algorithms for triaging malignant from benign lesions, a common task in dermatology AI (4, 5, 25). Primary analysis focused on comparing FST I–II and FST V–VI images because they were matched across diagnostic category (benign versus malignant), patient age, sex, and date of photos. While this is not comprehensive of all the potential confounders that could exist when comparing FST I–II and V–VI, our fine-tuning experiments show that the gap in performance can be closed by using diverse data. Although FST is used most commonly for labeling skin tones for images used in AI studies, this scale has limitations and does not capture the full diversity of human skin tones (26).
While the DDI dataset may not be sufficiently large for training models from scratch, we found that differences in performance between FST I–II and FST V–VI in previously state-of-the-art algorithms could be overcome by fine-tuning on diverse images from DDI. Fine-tuning on FST I–II images in DDI alone did not close the gap in performance between FST I–II and FST V–VI, further supporting the need for high-quality, diverse data. The fine-tuned algorithms performed better than the dermatologist labelers on skin tones V–VI, suggesting that future algorithms trained on diverse data could have the potential for providing decision support. Thus, we believe this dataset will not only provide a useful evaluation benchmark but also allow model developers to reduce the performance disparity across skin tones in dermatology AI algorithms.
MATERIALS AND METHODS
Image selection and processing
The Stanford Research Repository was used to identify self-described Black, Hispanic, or Latino patients who received a biopsy or tangential shave (CPT codes: skin biopsy 11100, tangential shave 11102) within the past 10 years starting from 1 January 2010. Per institutional practice, all biopsied lesions undergo clinical photography taken from approximately 6 inches (15.2 cms) from the lesion of interest, typically using a clinic-issued smartphone camera. These clinical images were pulled from the electronic medical record. Each case was represented by a single image. Lesions were characterized by specific histopathologic diagnosis, benign versus malignant status, date of biopsy, and also age, sex, and FST of the patient. A patient’s FST was labeled after assessing the clinical documentation from an in-person visit, demographic photo if available, and the image of the lesion. Two board-certified dermatologists with 1 and 5 years of experience after board certification (R.D. and A.C.) performed a final review of these three pieces of information; discrepancies were adjudicated using a consensus process. Discrepancies were seen in less than 1% of the data; and there was full agreement between the consensus labelers in those cases.
FST I–II patients were generated by matching each lesion diagnosed in the FST V–VI cohort with a lesion with the identical histopathologic diagnosis diagnosed in a patient with FST I–II regardless of race using PowerPath software. Patient demographics were assessed using chart review and included as a matched control if the lesion was diagnosed within the same 3 year period to account for incremental improvements in phone camera technology, and if the patient had the corresponding sex, age within 10 years, and FST type I–II. FST was confirmed in the same manner as the prior cohort; lesions that did not meet FST criteria were excluded. In instances where a matching control strictly meeting the above criteria could not be identified, a lesion with a similar diagnostic category (i.e., matching one type of nonmelanoma skin cancer with another type) in an FST I–II patient meeting the majority of the control criteria would be included instead to preserve the ratio of malignant to benign lesions. During this process, skin lesions associated with patients determined to be FST III–IV were included in the overall FST III–IV group, which represents a convenience sample that was not matched to the FST V–VI group. Figure S3 includes a diagram of sample collection and exclusion.
Diagnosis labels and malignant versus benign labels were determined by a board-certified dermatologist (R.D.) and board-certified dermatopathologist (R.N.) reviewing the histopathologic diagnoses. Any additional tests ordered on pathology was also considered for borderline cases—for example, atypical compound melanocytic proliferation with features of an atypical spindle cell nevus of Reed was labeled as malignant due to diffuse loss of p16 on immunohistochemical stains, while an atypical lymphocytic infiltrate was confirmed to be benign based on negative clonality studies.
Released images
Before image release, an additional review of all photos was done. Additional cropping was done after independent reviews by four board-certified dermatologists as part of the expert determination process of ensuring that all images were deidentified (no full-face images, identifiable tattoos, unique jewelry, or labels with potential identifiers). This was a Stanford Institutional Review Board–approved study: protocol numbers 36050 and 61146.
Dermatologist reader study and quality filtering
Three independent board-certified dermatologists not involved in ground truth or FST labeling with 5, 9, and 27 years of experience after board certification and no previous access to the image data or pathology labels were asked to assess each image in an untimed manner. All the dermatologists practiced within the Stanford system, and none have completed skin of color fellowships. They were asked to assign a photo quality score using a previously developed image quality scale (27). Overall quality scores were tabulated by taking the mean of all three dermatologists. Thirteen images did not meet quality metrics and were removed from analysis, leaving a total of 656 images. We compared the quality score distribution between Fitzpatrick I–II photo and Fitzpatrick V–VI photos using the Mann-Whitney U test, showing no statistically significant difference in quality (scipy.stats package, ipython 7.8.0). In addition, dermatologists were asked to rate whether they thought the presented image showed a benign or malignant process. An ensemble of the three dermatologists’ predictions was created using a majority vote.
AI algorithms
The labels generated by AI algorithms came from three models, labeled “ModelDerm” and “DeepDerm” and “HAM10000.” ModelDerm is a previously described algorithm with an available online API (https://jid2020.modelderm.com/); outputs were generated in December 2020 (12).
DeepDerm is an algorithm developed from using previously described data and similar parameters to the algorithm developed by Esteva et al. (4). The algorithm uses the Inception V3 architecture (23). The data sources include ISIC, two datasets sourced from Stanford hospital, Edinburgh Dermofit Library, and open-access data available on the web, as previously described (4). We do not have permission to share the DeepDerm datasets; we received the raw data directly from the authors. We mix all datasets and randomly generate train (80%) and test splits (20%). Images are resized and cropped to a size of 299 × 299 pixels. During training, we also augment data by randomly rotating and vertically flipping images before the resize and crop operation previously described; the rotation operation involves rotating the image and cropping to the largest upright, inscribed rectangle in the image. We train using the Adam optimization method with a learning rate of 10−4 and with binary cross entropy loss to classify images as malignant/benign (28). We use balanced sampling across benign and malignant images during training.
HAM10000 is an algorithm developed from the previously described, publicly available HAM10000 dataset (7). The algorithm and training methods are identical to the DeepDerm methods, with the only difference being the data used. The HAM10000 dataset consists of 10,015 dermoscopy images; all malignancies are biopsy confirmed (7). The thresholds determined for DeepDerm and HAM10000 were based on the maximum F1 score on the original test set used to train each respective algorithm.
We also assessed three robust training methods using DeepDerm’s training data: GroupDRO, CORAL, and CDANN, which have been shown to reduce the differential performance of AI models across subpopulations of data (13–15). These methods require us to partition our dataset into the groups we would like to be robust across. We use the source of each image to define the groups, since the data source can capture confounders, artifacts, and imbalances across skin color that we would like the AI model to be robust to. As we have 5 source datasets and 2 classes (benign and malignant), we define 10 groups in total (5 sources × 2 classes).
Each of these robust training methods is then trained using the same data augmentation strategies and optimization algorithm as described above, with the exception that we perform balanced sampling across these 10 groups. GroupDRO minimizes the maximum expected loss across the 10 groups to be robust to the worst-case training loss across groups; it additionally adds a regularization penalty to be more effective in the deep learning setting. CORAL minimizes the standard cross entropy loss but adds an additional penalty to force the intermediate image embedding to be similar across groups. In particular, CORAL penalizes differences in mean and covariance of the image embedding between all group pairs. CDANN also attempts to enforce domain-invariant image embeddings but uses a discriminator network instead that is trained to distinguish between domains. The loss from this network forces embeddings to be similar across domains. We train these robust training algorithms across five seeds and calculate the mean and 95% CI ROC-AUC values.
Data analysis
We performed each analysis with all 656 images, which included a mix of common and uncommon skin diseases. To assess the effects of uncommon disease on performance when assessing benign versus malignant lesions, we removed any diagnosis that was reported in the literature to have an incidence less than 1 in 10,000 in the entire population or, in the absence of relevant literature, were determined to be uncommon by three board-certified dermatologists and dermatopathologists (29–37). Consistent with a previously reported taxonomy, we considered diseases as a whole and not based on subtypes (for example, acral lentiginous melanoma is considered as part of melanoma in considering incidence) (4). Three board-certified dermatologists independent from the labelers (R.D., R.N., and A.C.) assessed the diseases to ensure that diseases labeled as “uncommon” were considered uncommon among dermatologists.
Benign diagnoses that were considered common included abrasions, ulcerations, and physical injuries; abscess; acne (cystic); acral melanotic macule; acrochordon; actinic keratosis; angioma; atypical lymphocytic infiltrate; benign keratosis; blue nevus; cherry angioma; clear cell acanthoma; condyloma acuminatum; dermatofibroma; dysplastic nevus; eczema/spongiotic dermatitis; epidermal cyst; epidermal nevus; fibrous papule; folliculitis; foreign body granuloma; hematoma; hyperpigmentation; keloid; lichenoid keratosis; lipoma; melanocytic nevi; molluscum contagiosum; neurofibroma; neuroma; onychomycosis; prurigo nodularis; pyogenic granuloma; reactive lymphoid hyperplasia; scar; seborrheic keratosis; solar lentigo; tinea pedis; trichilemmoma; and verruca vulgaris/wart.
Benign diagnoses that were considered uncommon included acquired digital fibrokeratoma (38); angioleiomyoma (39); arteriovenous hemangioma; cellular neurothekeoma; chondroid syringoma (32); cutaneous coccidioidomycosis (31); dermatomyositis (33); eccrine poroma (30); focal acral hyperkeratosis; glomangioma; graft-vs-host disease; inverted follicular keratosis (36); morphea (40); nevus lipomatosus superficialis (41); pigmented spindle cell nevus of Reed; syringocystadenoma papilliferum; trichofolliculoma; verruciform xanthoma (42); and xanthogranuloma (43).
Malignant diagnoses that were considered common included basal cell carcinoma; melanoma; melanoma in situ; squamous cell carcinoma; squamous cell carcinoma; keratoacanthoma type; and squamous cell carcinoma in situ.
Malignant diagnoses that were considered uncommon included atypical compound melanocytic proliferation (features of atypical spindle cell nevus of Reed with diffuse P16 loss requiring re-excision); blastic plasmacytoid dendritic cell neoplasm (44); Kaposi’s sarcoma (45); leukemia cutis; metastatic carcinoma (cutaneous metastases); mycosis fungoides (46); sebaceous carcinoma (47); and subcutaneous T cell lymphoma (48).
For calculating ROC-AUC, we used the probabilities generated by each algorithm. For the ensemble dermatologist ROC-AUC reported in table S2, we used probabilities generated by summing the votes and dividing by the total number of dermatologists. In Fig. 1, we show the consensus ensemble dermatologist performance using the majority vote. We calculated a 95% CI using bootstrapping with 50,000 iterations. We assess for drop-offs in sensitivity between FST I–II and FST V–VI using Fisher’s exact test to compare the proportion of true positives and false negatives.
Datasets of dermatology images can have extraneous artifacts such as rulers and markings. The presence of these artifacts did not correlate with a diagnosis of malignancy (Pearson’s correlation, r = −0.005, P = 0.89). In addition, predicting the correct label (benign or malignant) did not correlate with the presence of markings for dermatologists (Pearson’s correlation, r = −0.03, P = 0.50), ModelDerm (Pearson’s correlation, r = −0.01, P = 0.91), or HAM10000 (Pearson’s correlation, r = −0.01, P = 0.80). DeepDerm performance was negatively correlated with the presence of markings (Pearson’s correlation, r = −0.20, P = 3.2 × 10−5). Because FST V–VI had fewer images with markers and rulers, these artifacts do not explain why DeepDerm performed worse on the FST V-VI dataset.
Fine-tuning DeepDerm and HAM10000 using DDI
We fine-tuned the DeepDerm and HAM10000 algorithms on the DDI FST I–II and V–VI data. We were not able to fine-tune ModelDerm since we can only access it through an API. We perform a random 60-20-20 split into train, validation, and test sets. The split is randomized across skin tone and disease classification groups (e.g., so the ratio of malignant FST I–II images is the same across all three groups). During training, we augment our dataset using random rotations, vertical flips, resize and cropping, color jitter (perturbations to brightness, contrast, and saturation), and Gaussian blurring using torchvision.transforms. Details are provided in table S6. We also use Mixup data augmentation during training with parameter alpha = 1 (49). Mixup generates synthetic data by taking interpolations of pairs of training data and has been shown to improve model generalizability (49, 50). We fine-tune all layers of our model using the Adam optimizer with a learning rate of 5 × 10−2 and weight regularization of 1 × 10−4. We train for up to 500 epochs and use the validation loss to select the model weights we keep. During testing, we select a center crop of the image. This training procedure is repeated for 20 random seeds, where we randomly sample the 60-20-20 data split differently for each seed. We report the average test AUC across these 20 seeds along with 95% CIs in Fig. 2. We calculated the 95% CI using bootstrapping across the 20 seeds we train on; this procedure is repeated for baseline data to ensure a fair comparison. The same experiment was repeated for DeepDerm using only FST I–II images for fine-tuning (fig. S2).
Acknowledgments
Funding: J.Z. is supported by NSF CAREER 1942926. A.S.C. is supported by a Dermatology Foundation Medical Dermatology Career Development Award. A.S.C., R.A.N., J.K., S.M.S., and O.G. are supported by the Melanoma Research Alliance’s L’Oreal Dermatological Beauty Brands-MRA Team Science Award. R.D. is supported by 5T32AR007422-38. K.V. is supported by an NSF graduate research fellowship and a Stanford Graduate Fellowship award. V.R. reports serving as an expert advisor for Inhabit Brands Inc.
Author contributions: Concept and design: R.D., K.V., R.A.N., M.J., W.L., V.R., J.K., J.Z., and A.S.C. Acquisition, analysis, or interpretation of data: All authors. Drafting of manuscript: R.D., K.V., R.A.N., J.Z., and A.S.C. Critical revision of the manuscript for important intellectual content: R.D., K.V., R.A.N., W.L., V.R., J.K., S.M.S., E.E.B., J.Z., and A.S.C. Statistical analysis: R.D., K.V., W.L., and J.Z. Obtained funding: R.A.N., J.K., S.M.S., J.Z., and A.S.C. Administrative, technical, or material support: J.Z. and A.S.C. Supervision: J.Z. and A.S.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The DDI dataset is available at the Stanford AIMI repository: https://stanfordaimi.azurewebsites.net/datasets/35866158-8196-48d8-87bf-50dca81df965. Our trained AI models are available at https://zenodo.org/record/6784279#.Yr3mT-zMLFo. Our website is https://ddi-dataset.github.io.
Supplementary Materials
This PDF file includes:
Tables S1 to S6
Figs. S1 to S3
REFERENCES AND NOTES
- 1.Coustasse A., Sarkar R., Abodunde B., Metzger B. J., Slater C. M., Use of teledermatology to improve dermatological access in rural areas. Telemed. J. E Health 25, 1022–1032 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Tsang M. W., Resneck J. S., Even patients with changing moles face long dermatology appointment wait-times: A study of simulated patient calls to dermatologists. J. Am. Acad. Dermatol. 55, 54–58 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Tschandl P., Rinner C., Apalla Z., Argenziano G., Codella N., Halpern A., Janda M., Lallas A., Longo C., Malvehy J., Paoli J., Puig S., Rosendahl C., Soyer H. P., Zalaudek I., Kittler H., Human-computer collaboration for skin cancer recognition. Nat. Med. 26, 1229–1234 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Esteva A., Kuprel B., Novoa R. A., Ko J., Swetter S. M., Blau H. M., Thrun S., Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daneshjou R., Smith M. P., Sun M. D., Rotemberg V., Zou J., Lack of transparency and potential bias in artificial intelligence data sets and algorithms: A scoping review. JAMA Dermatol. 157, 1362–1369 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freeman K., Dinnes J., Chuchu N., Takwoingi Y., Bayliss S. E., Matin R. N., Jain A., Walter F. M., Williams H. C., Deeks J. J., Algorithm based smartphone apps to assess risk of skin cancer in adults: Systematic review of diagnostic accuracy studies. BMJ 368, m127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tschandl P., Rosendahl C., Kittler H., The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 5, 180161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.N. Codella, V. Rotemberg, P. Tschandl, M. Emre Celebi, S. Dusza, D. Gutman, B. Helba, A. Kalloo, K. Liopyris, M. Marchetti, H. Kittler, A. Halpern, Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC); https://arxiv.org/abs/1902.03368 (2018).
- 9.N. M. Kinyanjui, T. Odonga, C. Cintas, N. C. F. Codella, R. Panda, P. Sattigeri, K. R. Varshney, Fairness of classifiers across skin tones in dermatology. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. October 4–8, 2020, Lima, Peru (Springer, 2020), pp. 320–329. [Google Scholar]
- 10.M. Groh, C. Harris, L. Soenksen, F. Lau, R. Han, A. Kim, A. Koochek, O. Badri, Evaluating deep neural networks trained on clinical images in dermatology with the Fitzpatrick 17k dataset; https://arxiv.org/abs/2104.09957 (2021).
- 11.Daneshjou R., Barata C., Betz-Stablein B., Celebi M. E., Codella N., Combalia M., Guitera P., Gutman D., Halpern A., Helba B., Kittler H., Kose K., Liopyris K., Malvehy J., Seog H. S., Soyer H. P., Tkaczyk E. R., Tschandl P., Rotemberg V., Checklist for evaluation of image-based artificial intelligence reports in dermatology: CLEAR derm consensus guidelines from the international skin imaging collaboration artificial intelligence working group. JAMA Dermatol. 158, 90–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han S. S., Park I., Eun Chang S., Lim W., Kim M. S., Park G. H., Chae J. B., Huh C. H., Na J.-I., Augmented intelligence dermatology: Deep neural networks empower medical professionals in diagnosing skin cancer and predicting treatment options for 134 skin disorders. J. Invest. Dermatol. 140, 1753–1761 (2020). [DOI] [PubMed] [Google Scholar]
- 13.S. Sagawa, P. W. Koh, T. B. Hashimoto, P. Liang, Distributionally robust neural networks for group shifts: On the importance of regularization for worst-case generalization; https://arxiv.org/abs/1911.08731 (2019).
- 14.B. Sun, K. Saenko, Deep coral: Correlation alignment for deep domain adaptation, paper presented at the European Conference on Computer Vision, October 8-16, 2016, Amsterdam, Netherlands (Springer, 2016), pp. 443–450. [Google Scholar]
- 15.Y. Li, X. Tian, M. Gong, Y. Liu, T. Liu, K. Zhang, D. Tao, Deep domain generalization via conditional invariant adversarial networks, in European Conference on Computer Vision, September 8–14, Munich, Germany (Springer, 2018), pp. 647–663. [Google Scholar]
- 16.Han S. S., Kim Y. J., Moon I. J., Jung J. M., Lee M. Y., Lee W. J., Won C. H., Lee M. W., Kim S. H., Navarrete-Dechent C., Chang S. E., Evaluation of artificial intelligence-assisted diagnosis of skin neoplasms: A single-center, paralleled, unmasked, randomized controlled trial. J Invest Dermatol. S0022-202X, 00122–00121 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Sierro T. J., Blumenthal L. Y., Hekmatjah J., Chat V. S., Kassardjian A. A., Read C., Armstrong A. W., Differences in health care resource utilization and costs for keratinocyte carcinoma among racioethnic groups: A population-based study. J. Am. Acad. Dermatol. 86, 373–378 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Agbai O. N., Buster K., Sanchez M., Hernandez C., Kundu R. V., Chiu M., Roberts W. E., Draelos Z. D., Bhushan R., Taylor S. C., Lim H. W., Skin cancer and photoprotection in people of color: A review and recommendations for physicians and the public. J. Am. Acad. Dermatol. 70, 748–762 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Jain A., Eng C., Way D. H., Lee K., Bui P., Kanada K., de Oliveira Marinho G., Gallegos J., Gabriele S., Gupta V., Singh N., Natarajan V., Hofmann-Wellenhof R., Corrado G. S., Peng L. H., Webster D. R., Ai D., Huang S. J., Liu Y., Dunn R. C., Coz D., A deep learning system for differential diagnosis of skin diseases. Nat. Med. 26, 900–908 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Hekler A., Kather J. N., Krieghoff-Henning E., Utikal J. S., Meier F., Gellrich F. F., Upmeier zu Belzen J., French L., Schlager J. G., Ghoreschi K., Wilhelm T., Kutzner H., Berking C., Heppt M. V., Haferkamp S., Sondermann W., Schadendorf D., Schilling B., Izar B., Maron R., Schmitt M., Fröhling S., Lipka D. B., Brinker T. J., Effects of label noise on deep learning-based skin cancer classification. Front Med. (Lausanne) 7, 177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lester J. C., Clark L. Jr., Linos E., Daneshjou R., Clinical photography in skin of colour: Tips and best practices. Br. J. Dermatol. 184, 1177–1179 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Adelekun A., Onyekaba G., Lipoff J. B., Skin color in dermatology textbooks: An updated evaluation and analysis. J. Am. Acad. Dermatol. 84, 194–196 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Diao J. A., Adamson A. S., Representation and misdiagnosis of dark skin in a large-scale visual diagnostic challenge. J. Am. Acad. Dermatol. 86, 950–951 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Gupta R., Ibraheim M. K., Dao H. Jr., Patel A. B., Koshelev M., Assessing dermatology resident confidence in caring for patients with skin of color. Clin. Dermatol. 39, 873–878 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Haenssle H. A., Fink C., Schneiderbauer R., Toberer F., Buhl T., Blum A., Kalloo A., Hassen A. B. H., Thomas L., Enk A., Uhlmann L.; Reader study level-I and level-II Groups, Alt C., Arenbergerova M., Bakos R., Baltzer A., Bertlich I., Blum A., Bokor-Billmann T., Bowling J., Braghiroli N., Braun R., Buder-Bakhaya K., Buhl T., Cabo H., Cabrijan L., Cevic N., Classen A., Deltgen D., Fink C., Georgieva I., Hakim-Meibodi L. E., Hanner S., Hartmann F., Hartmann J., Haus G., Hoxha E., Karls R., Koga H., Kreusch J., Lallas A., Majenka P., Marghoob A., Massone C., Mekokishvili L., Mestel D., Meyer V., Neuberger A., Nielsen K., Oliviero M., Pampena R., Paoli J., Pawlik E., Rao B., Rendon A., Russo T., Sadek A., Samhaber K., Schneiderbauer R., Schweizer A., Toberer F., Trennheuser L., Vlahova L., Wald A., Winkler J., Wölbing P., Zalaudek I., Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 29, 1836–1842 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Okoji U. K., Taylor S. C., Lipoff J. B., Equity in skin typing: Why it is time to replace the Fitzpatrick scale. Br. J. Dermatol. 185, 198–199 (2021). [DOI] [PubMed] [Google Scholar]
- 27.K. Vodrahalli, R. Daneshjou, R. A. Novoa, A. Chiou, J. M. Ko, J. Zou, TrueImage: A machine learning algorithm to improve the quality of telehealth photos, Pacific Symposium on Biocomputing 2020, Big Island, Hawaii, January 3–7, 2021. Pacific Symposium on Biocomputing 26, 220-231 (2021); http://psb.stanford.edu/psb-online/proceedings/psb21/vodrahalli.pdf. [PubMed]
- 28.D. P. Kingma, J. Ba, Adam: A method for stochastic optimization; https://arxiv.org/abs/1412.6980 (2014).
- 29.Mehregan A. H., Infundibular tumors of the skin. J. Cutan. Pathol. 11, 387–395 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Rao E. M., Knackstedt T. J., A systematic review of periungual eccrine neoplasms. Int. J. Dermatol. 61, 812–820 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Shiu J., Thai M., Elsensohn A. N., Nguyen N. Q., Lin K. Y., Cassarino D. S., A case series of primary cutaneous coccidioidomycosis after a record-breaking rainy season. JAAD Case Rep. 4, 412–414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yavuzer R., Başterzi Y., Sari A., Bir F., Sezer C., Chondroid syringoma: A diagnosis more frequent than expected. Dermatol. Surg. 29, 179–181 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Bendewald M. J., Wetter D. A., Li X., Davis M. D. P., Incidence of dermatomyositis and clinically amyopathic dermatomyositis: A population-based study in Olmsted County, Minnesota. Arch Dermatol. 146, 26–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boukovalas S., Rogers H., Boroumand N., Cole E. L., Cellular neurothekeoma: A rare tumor with a common clinical presentation. Plast. Reconstr. Surg. Glob. Open 4, e1006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sulochana S., Manoharan M., Anitha, Chondroid syringoma-An unusual presentation. J. Clin. Diagn. Res. 8, FD13–FD14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Llambrich A., Zaballos P., Taberner R., Terrasa F., Bañuls J., Pizarro A., Malvehy J., Puig S., Dermoscopy of inverted follicular keratosis: Study of 12 cases. Clin. Exp. Dermatol. 41, 468–473 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Hernández-San Martín M. J., Vargas-Mora P., Aranibar L., Juvenile xanthogranuloma: An entity with a wide clinical spectrum. Actas Dermosifiliogr (Engl Ed). 111, 725–733 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Shih S., Khachemoune A., Acquired digital fibrokeratoma: Review of its clinical and dermoscopic features and differential diagnosis. Int J Dermatol. 58, 151–158 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Malik K., Patel P., Chen J., Khachemoune A., Leiomyoma cutis: A focused review on presentation, management, and association with malignancy. Am. J. Clin. Dermatol. 16, 35–46 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Sapra A., Dix R., Bhandari P., Mohammed A., Ranjit E., A case of extensive debilitating generalized morphea. Cureus 12, e8117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima C. D. S., Issa M. C. A., Souza M. B., Góes H. F. O., Santos T. B. P., Vilar E. A. G., Nevus lipomatosus cutaneous superficialis. An. Bras. Dermatol. 92, 711–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipsen H. P., Reichart P. A., Takata T., Ogawa I., Verruciform xanthoma—Biological profile of 282 oral lesions based on a literature survey with nine new cases from Japan. Oral Oncol. 39, 325–336 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Liu D. T., Choi P. C. L., Chan A. Y. K., Juvenile xanthogranuloma in childhood and adolescence: A clinicopathologic study of 129 patients from the Kiel pediatric tumor registry. Am. J. Surg. Pathol. 29, 1117 (2005). [PubMed] [Google Scholar]
- 44.Deconinck E., Blastic plasmacytoid dendritic cell neoplasm. Hematol. Oncol. Clin. North Am. 34, 613–620 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Grabar S., Costagliola D., Epidemiology of Kaposi’s Sarcoma. Cancers (Basel) 13, 5692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amorim G. M., Niemeyer-Corbellini J. P., Quintella D. C., Cuzzi T., Ramos-e-Silva M., Clinical and epidemiological profile of patients with early stage mycosis fungoides. An. Bras. Dermatol. 93, 546–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi R., Chen Z., Li L., Bordeaux J. S., Incidence and survival of sebaceous carcinoma in the United States. J. Am. Acad. Dermatol. 75, 1210–1215 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Criscione V. D., Weinstock M. A., Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch. Dermatol. 143, 854–859 (2007). [DOI] [PubMed] [Google Scholar]
- 49.H. Zhang, M. Cisse, Y. N. Dauphin, D. Lopez-Paz, mixup: Beyond empirical risk minimization; https://arxiv.org/abs/1710.09412 (2017).
- 50.L. Zhang, Z. Deng, K. Kawaguchi, A. Ghorbani, J. Zou, How does mixup help with robustness and generalization? https://arxiv.org/abs/2010.04819 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S6
Figs. S1 to S3