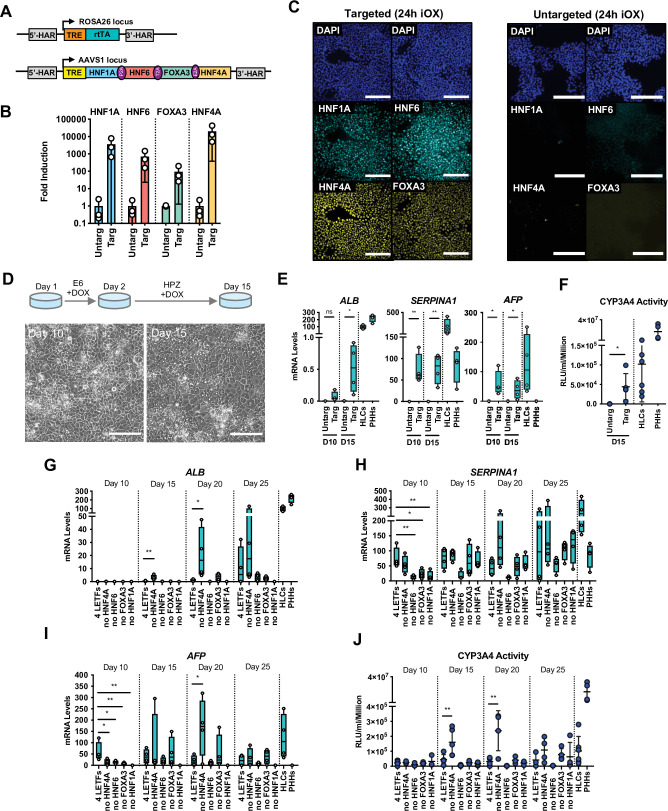
Figure 1. Forward programming of human pluripotent stem cells (hPSCs) into hepatocytes with four and three liver-enriched transcription factors (LETFs).
(A) Schematic representation of the two sequentially targeted loci. The human ROSA26 was targeted with a constitutively expressed reverse tetracycline transactivator (rtTA). The AAVS1 locus was targeted with the four LETFs HNF1A, HNF6, FOXA3, and HNF4A downstream of a TET-responsive element (TET). (B) mRNA induction levels of the four factors in targeted human embryonic stem cells (hESCs) (Targ) relative to untargeted (Untarg) hESCs stimulated with doxycycline (dox) for 24 hr (n=3). Data is shown relative to the untargeted control. (C) Immunofluorescence staining of the four LETFs in targeted and untargeted hESCs after 24 hr of inducible overexpression (iOX) with dox confirming transgene induction. Nuclei were counterstained with DAPI (blue). Scale bar, 200µm. (D) Schematic representation of the iOX culture conditions for forward programming. Phase contrast images of hESCs targeted with the four LETFs after 10 and 15 days of forward programming. Scale bar, 200µm. (E) mRNA levels of hepatocyte markers (ALB, SERPINA1, and AFP) in hESCs targeted with the four LETFs after 10 and 15 days of forward programming. Untargeted hESCs treated with the same protocol as in (D) were used as control (n=4). Statistical difference was calculated with unpaired t-test against untargeted. (F) CYP3A4 activity levels normalised per cell number (millions) in untargeted and targeted hESCs with the four LETFs after 15 days of forward programming (n=5) . Statistical difference between targeted and untargeted cells was calculated with unpaired t-test. (G,H,I) mRNA levels of hepatocyte markers (ALB, SERPINA1, and AFP) in hESCs targeted with the four LETFs and with combinations of three LETFs (n=4). The factor removed from each construct is indicated. Expression levels were determined after 10, 15, 20, and 25 days of forward programming. Statistical differences were calculated with one-way ANOVA, corrected for multiple comparisons compared to four LETFs. All mRNA levels were normalised to the average of two housekeeping genes (PBGD and RPLP0). (J) CYP3A4 activity levels normalised per cell number (millions) in hESCs targeted with the four LETFs and combinations of three LETFs after 10, 15, 20, and 25 days of forward programming (n=3–5). Statistical differences were calculated with one-way ANOVA, corrected for multiple comparisons compared to four LETFs. In all plots, bars represent mean with SD, and individual datapoints are shown for all biological replicates. Hepatocyte-like cells (HLCs) generated by direct differentiation and primary human hepatocytes (PHHs) where plotted as controls for all CYP3A4 activity and expression data. Significant p-values are shown at each comparison and indicated as *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.