Summary
Pharmacologically active compounds with known biological targets were evaluated for inhibition of SARS-CoV-2 infection in cell and tissue models to help identify potent classes of active small molecules and to better understand host-virus interactions. We evaluated 6,710 clinical and preclinical compounds targeting 2,183 host proteins by immunocytofluorescence-based screening to identify SARS-CoV-2 infection inhibitors. Computationally integrating relationships between small molecule structure, dose-response antiviral activity, host target, and cell interactome produced cellular networks important for infection. This analysis revealed 389 small molecules with micromolar to low nanomolar activities, representing >12 scaffold classes and 813 host targets. Representatives were evaluated for mechanism of action in stable and primary human cell models with SARS-CoV-2 variants and MERS-CoV. One promising candidate, obatoclax, significantly reduced SARS-CoV-2 viral lung load in mice. Ultimately, this work establishes a rigorous approach for future pharmacological and computational identification of host factor dependencies and treatments for viral diseases.
Subject areas: Bioinformatics, Pharmacoinformatics, Pharmacology, Virology
Graphical abstract
Highlights
-
•
Identified 389 SARS-CoV-2 inhibitors comprising >12 structurally similar groups
-
•
Drug-protein target network analyses reveal host dependencies
-
•
Mechanistic evaluation of virus variants in stable and primary cell cultures
-
•
Lead candidate decreases virus lung load in mouse model of disease
Bioinformatics; Pharmacoinformatics; Pharmacology; Virology
Introduction
In December 2019, an unidentified pneumonia was reported in Wuhan, China, and by early January 2020, the causative agent had been identified as a novel coronavirus now called SARS-CoV-2. SARS-CoV-2, of the family Coronaviridae, is a single-stranded, positive-sense RNA virus with a genome of approximately 29kb Wu et al., 2020. SARS-CoV-2 is the third coronavirus in recent history to produce an epidemic, after SARS-CoV and MERS-CoV (Fouchier et al., 2003; Memish et al., 2020). The virus has rapidly spread across the world causing over 570 million cases globally of COVID-19, as of July 2022, resulting in more than 6.4 million deaths (Dong et al., 2020) and causing economic contraction, mass unemployment, disruption of education, and increasing poverty levels (Stawicki et al., 2020). Fortunately, effective vaccines and first-generation therapies have become quickly available and are being distributed on an emergency basis. However, given the emergence of more contagious and potentially pathogenic variants, and ongoing issues of emerging resistance, the continued search for new therapeutics remains a priority (Firestone, 2021; Malden, 2022; Mwenda, 2021). Therapeutics discovery efforts for SARS-CoV-2 were launched by a series of repurposing screens of varying sizes. The largest were screens of a 12,000 compound library, termed ReFRAME (Riva et al., 2020), and a screen of a 3,000 compound library (Dittmar et al., 2021). Both studies identified PIKfyve inhibitors, numerous protease inhibitors, several classes of kinase inhibitors, and cyclosporin analogues. Whereas it was encouraging to see overlap between these studies, each also highlighted the interassay variability seen in the evaluation of small molecule inhibitors of SARS-CoV-2 replication to date and the need for additional, more detailed studies.
The current study focused on screening of the Drug Repurposing Hub (DRH) library, a collection of 6,710 compounds highly enriched with molecules that have been FDA approved, entered clinical trials (4.8% phase 1, 2.0% phase 2, 7.5% phase 3, 4.4% phase 4), or have been extensively pre-clinically characterized (Corsello et al., 2017). The library comprises clusters of structurally related molecules that are well annotated, targeting a spectrum of host proteins, as well as numerous compounds with overlapping targets, affording the opportunity to capture preliminary structure-similarity relationships of active molecules and identification of host targets of importance. The primary screen is the first to be conducted across multiple doses at this scale, providing a dataset that grades all compounds by activity and potency from strongest to least active. These data were used to expand our previous computational studies in establishing SARS-CoV-2-associated protein networks that may be enriched in host targets for future drug discovery efforts (Gysi et al., 2021). The most promising hits from the screening effort were assessed for efficacy in a series of orthogonal assays conducted in human cell lines (Huh7 and A549 cells) and human primary cell-based tissue models. Our studies confirmed the activity of several previously identified classes (inhibitors of PIKfyve, cathepsins, and protein synthesis) and additional unreported compound classes. The most promising small molecule treatment to emerge from the screen, obatoclax, demonstrated consistent activity across all cell-based assays and virus strains tested, including against MERS-CoV, and reduced SARS-CoV-2 titers by up to 10-fold in a mouse infection model using clinically achievable compound exposures.
Results
Immunofluorescence-based screening of small compound library identifies potent inhibitors of SARS-CoV-2 infecton of cells
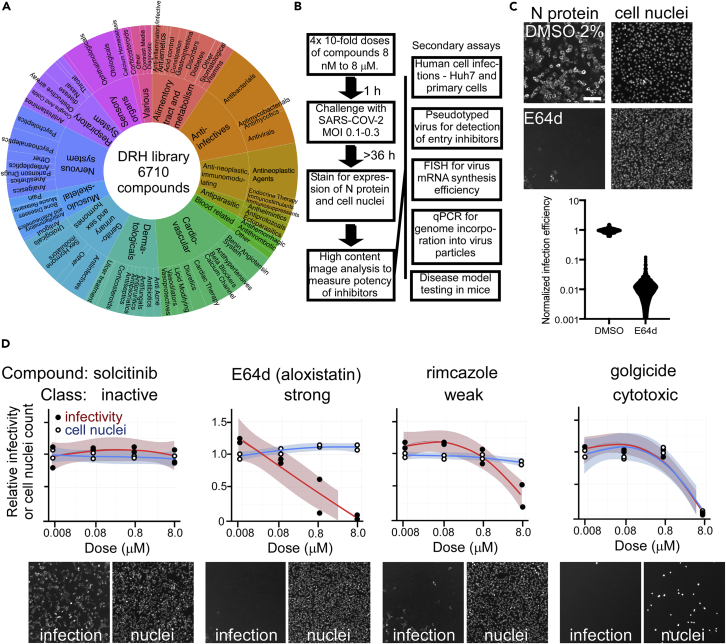
The DRH compound library (Figure 1A) was initially evaluated using an immunofluorescence-based assay to detect SARS-CoV-2 infection by N protein expression. The screen was followed by computational analyses to prioritize the best candidates for follow-up in mechanistic assays evaluating the impact on cell entry, genome replication, and egress of progeny viruses, as well as evaluation in primary human cell models (Figure 1B). In order to capture multiple rounds of viral replication (Ogando et al., 2020) and obtain strong N protein staining, the primary screen was conducted for 36–48 h. Whereas the suitability of other cell types was evaluated for screening, VeroE6 cells, derived from African green monkey kidney cells, supported the most robust and consistent infection compatible with high-throughput screening at maximum biocontainment. The assay gave an average of 80-fold difference in infection between untreated cells and cells treated with a known SARS-CoV-2 replication inhibitor, E64d Figure 1C, upper) (Hoffmann et al., 2020). Overall, this 384-well-based assay yielded a Z′ of 0.6, indicating suitability for use in screening in this format (Zhang et al., 1999) (Figure 1C, lower).
Figure 1.
HTS screen for inhibitors of SARS-CoV-2
(A) Composition of the Drug Repurposing Hub library based on ATC classifications for compounds (4,277 of 6710).
(B) Workflow of immunofluorescence-based infection focus assay developed for the screening of SARS-CoV-2 inhibitors and secondary mechanistic assays performed on active compounds.
(C) Dynamic range of the HTS assay evaluated using 5-μM E64d (Aloxistatin) versus 2% DMSO as vehicle. Upper panel: examples of microscope images of infected cells stained with SARS-CoV-2 N-specific antibody and cell nuclei stained with Hoechst 33342 for DMSO or E64d-treated cells. Scale bar is shown on left upper image and is 50 μm. Lower panel: outcomes for 190 replicate wells for each treatment are shown. The assay gave a Z-factor of 0.6 in 384 well plates and, thus, was suitable for HTS.
(D) Examples of concentration response curve classes seen in the screen and cell images with N protein staining on the left and cell nuclei on the right. The indicated compounds show no effect (solcitinib), strong activity with no cell loss (E64d), weak reduction in infectivity and no cell loss (rimcazole), and reduction in infection that parallels cell loss that is likely owing to cytotoxicity (golgicide). Scale is the same as for images in part C.
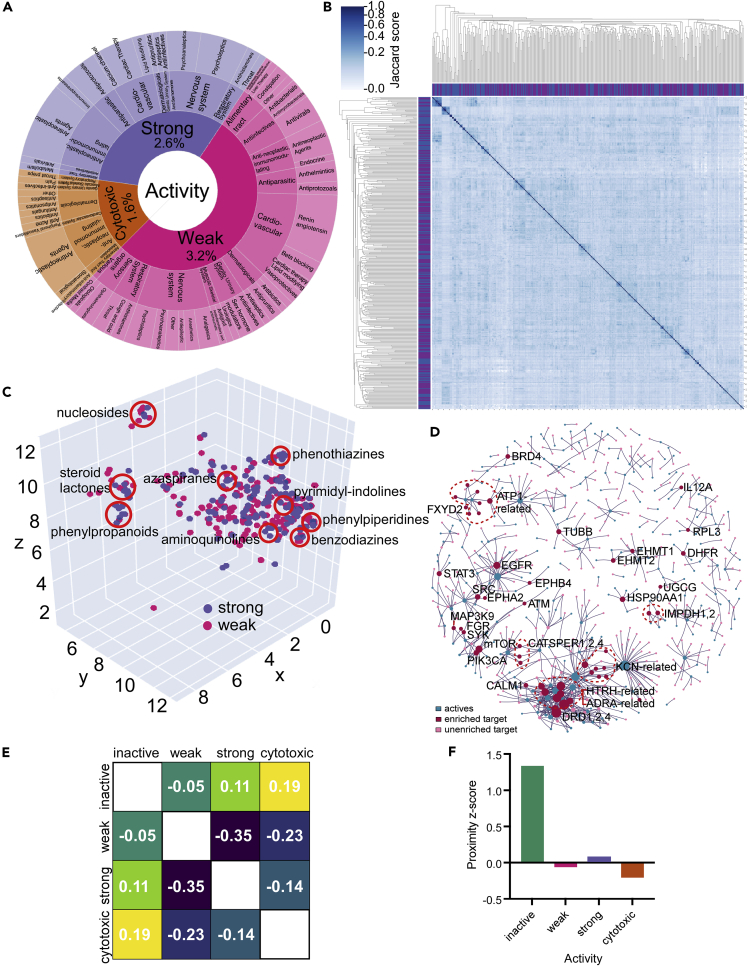
To identify compounds with dose responses, grade potency, and maximize the value of the full screening set for target discovery, four doses of each compound were tested, ranging from 8 nM to 8 μM in 10-fold increments. Compounds were initially grouped using a log-logistic regression for viral load (Wald test; padj <0.01; Bonferroni correction) to classify compounds (Table S1) on potency as strong (80% viral reduction, or Z-score >2.5), weak (50–80% viral reduction, Z-score 1.5–2.5), inactive (padj ≥ 0.01), or cytotoxic (>60% cell loss by nuclei count and Wald test on a log-logistic regression for cell count; padj <0.01; Bonferroni correction). We identified 172 compounds (2.56%) as strongly active, 217 (3.23%) as weakly active, and 1.6% as cytotoxic (examples shown in Figure 1D). Active compounds are summarized in Figure 2A, and the full dataset is deposited in the DRH database: https://www.broadinstitute.org/drug-repurposing-hub.
Figure 2.
Computational analysis of primary screen outcomes
(A) Chart showing the active compounds by ATC classifiers for strongly active, weakly active, and cytotoxic compounds. The fraction of compounds in each category is shown as a percentage of all compounds in the library.
(B) Pairwise comparison of active compounds based on computed structural similarity using Morgan fingerprints and Jaccard analysis.
(C) 3D representation of structural similarity of active molecules based on the reduced dimensionality of the molecular bit vectors. Localized clusters of major structural classes are indicated by red circles and are approximate locations in the plot. Refer to Figure S3 for an interactive version with higher detail.
(D) Relationship of enriched protein drug targets (red circles) or unenriched targets (pink circles) with active compounds (blue circles) for compounds with annotated targets in the DRH database.
(E) Separation heatmap between the targets of different activity classes within the human protein–protein interaction network. Negative network separation values reflect overlapping neighborhoods.
(F) Proximity Z-score between the drug targets of each category and the SARS-CoV-2 protein-binding host factors in the human protein-protein interaction network. Drugs with treatment activity have Z-scores close to zero, much lower than inactive drugs, indicating they hit targets in the network proximity of SARS-CoV-2-binding proteins.
Structure-similarity analysis reveals compound classes effective against SARS-CoV-2 replication
To reveal the structural characteristics that defined active compounds, structures were encoded using Morgan fingerprints (FP). Morgan FPs are a topological metric used routinely for structure-similarity analysis of different small molecules (Rogers and Hahn, 2010). To generate a Morgan FP, all substructures around the heavy atoms of a molecule within radius 3 were generated and assigned to unique identifiers. The substructure identifiers were then hashed to a binary vector of 8,192 bits (Table S2), which was used to perform similarity searches. Whereas the library showed diverse structural characteristics, active molecules showed a higher median of active bits (Figure S1). Hierarchical clustering of the library members according to the Jaccard similarity matrix reflected the diversity of the library with no clear connections for the entire library (Figure S2), as would be expected from a diverse library of this size, but actives showed small clusters of structurally related molecules appearing close to the diagonal (Figure 2B). Reducing the dimensionality of the bit vector representation to 3D space with UMAP (McInnes et al., 2018) revealed the presence of local clusters Figure 2C (and in interactive Figure S3) rather than global structural properties characterizing each experimental class. Of these, some have been previously reported, such as aminoquinolines (amodiaquine-related), nucleosides (Remdesivir-like), and phenothiazines (chlorpromazine-like antipsychotic drugs), but others have not been previously discussed in detail: pyrimidyl-indolines (similar to GSK2606414), azaspiranes, phenylpiperidines, benzodiazines, phenylpropanoids, and steroid lactones (cardiac glycosides).
We next evaluated if the drugs that presented structural similarity were also targeting the same or homologous proteins. Molecules with pairwise Jaccard similarity higher than 0.5 for which both had at least one protein target described in the Broad Library were used for the analysis. Of the 32 sets of pairwise similar drugs, 29 pairs shared at least one protein target (Figure S4). For the remaining three pairs, we compared the protein target sequences and found each was related. Only one pair (sertraline and indraline) shared structural similarity without sharing protein target similarity. This analysis suggested that the activity of structurally similar compounds corresponded to the targeting of homologous or similar protein targets.
Computational analysis reveals enriched drug targets and pathways that are involved in SARS-CoV-2 replication
Of the 6,710 compounds, 4,277 had attributable host targets annotated in the DRH database. GO enrichment showed a similar profile with active drugs enriched for processes related to rRNA modulation, microtubule regulation, collagen and proteoglycan binding, GPCR receptor activity, GTP metabolism, viral transcription processes, ion channel function, and protein folding (Table S3). It was interesting that angiotensin-related drugs were enriched, as SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as a receptor, though it should be noted patients taking ACE2 inhibitors derived no clinical benefit (Gurwitz, 2020). An effort was made to prevent oversampling of promiscuous compounds and over-represented targets, but GPCR-targeting compounds are one of the most abundant drug classes and, thus, must be interpreted with caution. Fisher exact test was used to identify if active compounds were enriched for known drug targets. Of the 389 active compounds, 813 targets were identified. Of these, 51 targets were enriched over the remainder, indicating a statistical association with inhibition of virus infection (Fisher exact test; p < 0.05). These included sodium ion export, membrane repolarization, and regularization of cardiac conduction (Table S4). The drug-target network analysis for these compounds revealed two large modules of drugs sharing targets, one comprising 61 drugs and 119 targets, and the other 43 drugs and 66 targets. We also identified 105 additional smaller modules suggesting compounds targeted a discrete group of host targets (Figure 2D).
As described in previous work (Gysi et al., 2021), we assembled a human protein–protein interaction (PPI) network by combining data assembled from (i) binary PPIs, (ii) literature mining, (iii) affinity purification followed by mass spectrometry, (iv) kinase–substrate interactions, (v) signaling interactions, and (vi) regulatory interactions.
Often, genes associated with a biological process are localized in the same region of the PPI graph or map (Menche et al., 2015), prompting us to investigate if the drug targets are co-localized in the PPI. We first calculated the largest connected component (LCC) targets in each outcome class and the significance of the module size using a degree-preserving approach (Guney et al., 2016), preventing the same high-degree nodes being selected repeatedly. We find that the LCC of the drug targets is statistically significantly larger than a random value for strong (149; 126.02 ± 10.91; Z-score 2.11) and weak (211; 173.84 ± 15.18; Z-score 2.45) classes. The combined class was also significant (311; 277 ± 18.69; Z-score 1.8) (Figure S5). Taken together, the analysis suggests clustering of drug targets within the interaction network.
To better understand the relationships of active compounds, the network separation between targets of each category was computed (Figure 2E). The separation measures the network overlap of two sets of genes and can be obtained by comparing the mean shortest distance within the respective sets of genes and the average shortest distance between them. Proximity and the extent of overlap of two network neighborhoods can be highly predictive of the pathological similarity of those diseases (Menche et al., 2015). Similar to our previous study (Gysi et al., 2021), targets of active compounds have a negative network separation ( = −0.35), indicating that each targets the same neighborhood in the human PPI network. In contrast, inactive compounds have close-to-zero or positive separation from the active compounds ( = −0.05, = 0.11), indicating inactive compounds target a different network neighborhood. Interestingly, when asked if targets were related to identified SARS-CoV-2 host protein interactions (Gordon et al., 2020) we found, similar to our previous study (Gysi et al., 2021), that the relative network proximity of each target module to the COVID-binding proteins is predictive of efficacy. Here, proximity is a network-based measure that calculates the shortest path length between drug targets and SARS-CoV-2-associated genes. Active compounds have z-scores close to zero (Figure 2F), whereas inactive compounds have a much stronger positive proximity Z-score, indicating their targets are farther from COVID-binding proteins than random expectation. Taken together, these data show that active and inactive compounds target distinct network neighborhoods in the human PPI network, and their network proximity to the COVID-binding proteins is predictive of drug efficacy.
In addition to analyzing gene targets, 6,150 of the compounds were annotated for the mechanism of action (MOA) in the DRH (Corsello et al., 2017). Using this information, we checked for over- and under-represented MOA that could be driving the drug response using a -test (p < 0.05, FDR-BH). Here, inhibitors of malaria, mammalian target of rapamycin (mTOR), ATPases, heat shock proteins (HSP), bromodomain and extraterminal (BET) proteins, and tubulin polymerization were significantly enriched. Furthermore, mapping of compounds to the Anatomical Therapeutic Chemical Classification (ATC) drug categories (Nahler, 2009) and chemical taxonomy identified compound classes that were over-represented in the hits (Table 1). These included previously identified classes of compounds active against SARS-CoV-2 in vitro, including phenothiazines, benzothiazines, and other antipsychotic agents, as well as new classes such as PERK inhibitors and cardiac glycosides. Each of these classes was also found clustered in the structural analysis (Figure 2C) supporting the relationship between drug class and host targets.
Table 1.
Relationship of active compounds based on the Anatomical Therapeutic Chemical (ATC) classification and drug categories
| Classification | Unadjusted p-value | |
|---|---|---|
| Drug category | Phenothiazines | 0.0004 |
| Antipsychotic Agents (First Generation [Typical]) | 0.0394 | |
| Level 2 ATC Code | Antimigraine Preparations | 0.0483 |
| Cardiac Glycosides | 0.0082 | |
| Antipsychotics | 0.0006 | |
| Antidepressants | 0.0321 | |
| Agents Against Amebiasis And Other Protozoal Diseases | 0.0303 | |
| Angiotensin II Receptor Blockers (Arbs), Combinations | 0.0483 | |
| Plant Alkaloids And Other Natural Products | 0.0235 | |
| Angiotensin II Receptor Blockers (Arbs), Plain | 0.0235 |
Drug category and therapeutic indication were evaluated for overrepresentation using a one-sided binomial exact test and Benjamini–Hochberg correction for multiple hypothesis testing to calculate the p-value.
Rescreening and orthogonal validation of hits with human cell lines
Based on the activity in the primary screen and compound classification using the structural similarity clustering and ATC classifications and, to a lesser extent, compound availability, 40 representative compounds were chosen for detailed potency determination and mechanistic analysis. The compounds were initially rescreened in VeroE6 cells to confirm activity before evaluation in human cell lines, and timing was set to capture limited virus replication. In general, most compounds yielded EC50 values similar to that expected from the primary screen potency determination (Table 2, third column, Figure S6 and examples in Figure 3A). For some compounds, such as methotrexate, a partial inhibition pattern was observed and so an EC50 was not calculated.
Table 2.
Effects of small molecule treatment on virus infection
| Compound | DRH mechanism of action target | EC50 (μM) in VeroE6 cells | gRNA release inhibitory concentration >50% (μM) | Virus mRNA synthesis inhibitory concentration >50% (μM) | mRNA:gRNA fold difference | Indicated site of actiona |
|---|---|---|---|---|---|---|
| Narasin | Antiprotozoal agent | 0.01 | 0.15 | 0.05 | 0.4 | |
| K-strophanthidin | ATPase | 0.75 | 0.15 | 0.45 | 3 | |
| Ouabain | ATPase | 0.045 | 0.15 | 0.15 | 1 | |
| VE-822 | ATR kinase | 1.33 | 0.45 | 1.30 | 2.9 | |
| Nanchangmycin | Autophagy | 0.033 | 0.15 | 0.15 | 1 | |
| Obatoclax | BCL | 0.048 | 0.02 | 0.05 | 3 | |
| BET-BAY-002 | BET | >4 | 0.15 | >10 | >10 | egress |
| BMS-986158 | BET | >4 | >10 | >10 | – | |
| CPI-0610 | BET | >4 | >10 | >10 | – | |
| Mivebresib | BET | >4 | 0.15 | >10 | >10 | egress |
| Calpeptin | Calpain protease | 2 | 1.30 | 4.00 | 3.1 | |
| Proscillaridin | Na+/K+ channel | 0.001 | 0.01 | 0.05 | 3.9 | |
| VBY-825 | Cathepsin protease | 1.8 | 1.30 | 1.30 | 1 | |
| Talniflumate | Cyclooxygenase | >4 | 1.30 | >10 | >10 | egress |
| Methotrexate | DHFR | a- | >10 | >10 | – | |
| Pralatrexate | DHFR | >4 | 0.15 | >10 | >10 | egress |
| BAY-2402234 | DHODH | 0.006 | 0.02 | 0.01 | 0.4 | |
| Sangivamycin | DNA synthesis | 0.045 | 0.02 | 0.05 | 3 | |
| Anisomycin | DNA synthesis | 0.049 | >10 | >10 | – | |
| Eliglustat | Glycosyl transferase | >4 | 4.00 | 12.00 | 3 | |
| A-485 | Histone acetyltransferase | 0.65 | 1.00 | 4.00 | 4 | |
| BIX-01294 | Histone lysine methyltransferase | 0.82 | 0.45 | >10 | >10 | egress |
| AT13387 | Heat shock protein | >4 | 0.15 | 0.15 | 1 | |
| Ganetespib | Heat shock protein | >4 | >10 | 0.45 | <0.1 | replication |
| NVP-HSP990 | Heat shock protein | >4 | >10 | >10 | – | |
| SNX-2112 | Heat shock protein | >4 | >10 | 0.45 | <0.1 | replication |
| Apilimod (STA-5326) | Interleukin synthesis | 0.1 | 0.45 | >10 | >10 | egress |
| Omipalisib (GSK2126458) | mTOR/PI3K | >4 | 0.05 | 0.15 | 3 | |
| Deslanoside | Na/K-ATPase | 0.165 | 0.45 | 0.45 | 1 | |
| Mepacrine | NFkB pathway | 1.33 | 0.05 | 0.45 | 9 | egress |
| APY0201 | Phosphoinositide dependent kinase | 0.0.01 | 1.30 | 1.30 | 1 | |
| MG-132 | Proteasome | 0.15 | 0.45 | 1.30 | 2.9 | |
| Bruceantin | Protein synthesis | 0.11 | 0.15 | 0.45 | 3 | |
| Emetine | Protein synthesis | 0.125 | 0.05 | 0.05 | 1 | |
| Harringtonine | Protein synthesis inhibitor | 0.12 | 0.01 | 0.05 | 5 | egress |
| Homoharringtonine | Protein synthesis | 0.43 | 0.02 | 0.15 | 8.9 | egress |
| Tioguanine | Purine antagonist | 1.2 | 0.45 | 4.00 | 8.9 | egress |
| Niclosamide | STAT/DNA replication | >4 | 1.30 | 4.00 | 3.1 | |
| Bafilomycin A1 | V-ATPase | 0.12 | 0.05 | 0.05 | 1 | |
| GSK-2606414 | PERK inhibitor | >0.36 | ND | ND | – |
ND – Not determined.
Shown is the annotated mechanism of action of each compound, the EC50 values obtained for infection inhibition in VeroE6 cells, and subsequent testing in Huh7 cells for virus mRNA production and virus-associated gRNA release into culture supernatant. For the VeroE6 cell tests, 10 concentrations from 2 nM up to 4 μM were used to construct dose-response curves (Figure S6) and EC50 values calculated. For the Huh7 cells, treatments started at this EC50 value 土4-fold and tested again at reduced concentrations if a higher-than-expected potency was seen. Measurements were performed in triplicate and concentrations giving the indicated reduction in signal are shown.
Gave a dose-response curve but did not cross the 50% infection threshold and so EC50 was not calculated. Site of action is based on the ratio of potencies for the inhibition of virus mRNA production in cells and gRNA release into culture supernatants. Potent inhibition of mRNA indicates the inhibition of RNA replication, whereas a lack of replication inhibition but more potent inhibition of gRNA release indicates selective inhibition of egress.
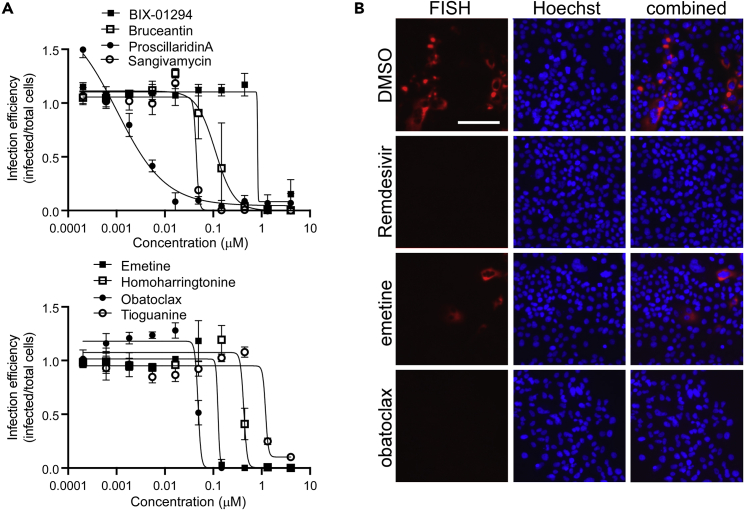
Figure 3.
Concentration response curves for active compounds and the measurement of small molecule effect on mRNA production from the virus subgenomic promoter by FISH assay in Huh7 cells
(A) Examples of response curves are shown for the indicated active compounds using treatment concentrations ranging from 4 μM down to 0.2 nM. Each concentration was repeated in triplicate and average and standard deviations shown and normalized to vehicle (DMSO)-treated controls.
(B) Virus mRNA production was measured by in situ oligonucleotide-based detection. Examples of images are shown for the indicated compounds. Cells were stained for production of virus mRNA encoding the N protein by the smiFISH method using virus gene-specific oligonucleotides and a Cy5-labeled oligonucleotide that bound to a shared complementary region on each (red). Cell nuclei were stained with Hoechst 33342 (blue). The scale bar in the top left image is 100 μm.
Compounds were then evaluated for efficacy using the human cell line Huh7, which is intrinsically susceptible to SARS-CoV-2 infection. Orthogonal assays measured cell-associated viral RNA levels by a FISH assay and extracellular viral RNA by qPCR. The FISH assay was performed on fixed cells and the qPCR was performed on virus-containing cell supernatants to measure the replication and release, respectively. As the qPCR and FISH assays have low throughput, initial concentrations of the compound were limited to the EC90 values from VeroE6 cell tests +/− 4-fold. For the PCR-based assay, strong and statistically significant effects of harringtonine, homoharringtonine, proscillaridin, BAY-2402234, obatoclax, and sangivamycin were seen at <20 nM, producing 74–93% (p < 0.01) decreases in viral genomic RNA (gRNA) in the culture supernatant (Table 2, fourth column). Whereas most of the compounds showed potencies similar to those in VeroE6 cells, the mTOR inhibitor, omipalisib, showed inhibition below the lowest concentration tested, indicating >10-fold stronger potency than seen in VeroE6 cells. Differences were also observed for two of the HSP90 and BET protein inhibitors that were classed as weak inhibitors in Vero cells. For the HSP90 inhibitor AT13387, virus infection was inhibited at the highest dose tested (150 nM). The BET inhibitors BET-BAY-002 and mivebresib were also inhibitory. Overall, such changes in activity suggest differences in dependency on BET and HSP90 proteins or drug action in Vero versus human Huh7 cells.
The impact of compounds on viral mRNA production was measured using a FISH assay, targeting RNA production from the viral subgenomic promoter that controls the expression of structural proteins (Figure 3B) and image analysis to score replication efficiency. In general, activity in this assay reflected that seen in the qPCR-based assay, with potency typically being within three-fold of each other (Table 2). The dihydroorate dehydrogenase (DHODH) inhibitor, BAY-2402234, was the most potent, inhibiting infection by 50% at 10 nM. The protein synthesis inhibitors emetine, harringtonine, and homoharringtonine were equally active, blocking >50% RNA production at 50 nM (Figure 3B). Other strongly active compounds were the nucleoside analog sangivamycin, as well as the mTOR and B-cell lymphoma 2 (BCL2) inhibitors, omipalisib and obatoclax, respectively. Cardiac glycosides, proscillaridin and ouabain, were also active at 50 and 150 nM, respectively. Of note was the large difference in activity of harringtonine and homoharringtonine, being over five- to eight-fold more potent in the genome release assay over the vRNA production assay, which would be consistent with the disruption of the virus assembly and may reflect the need for the balanced stoichiometry of structural proteins easily disrupted by protein synthesis inhibition. Similarly, differences were noted for compounds that inhibited genome release from cells in the qPCR assay but had a negligible impact on the viral mRNA signal. These compounds were the DHFR inhibitor pralatrexate, the cyclooxygenase (COX) inhibitor talniflumate, the BET inhibitors BET-BAY-002 and mivebresib, and the methyltransferase inhibitor BIX-01294, suggesting that each also worked to disrupt packaging of viral genomes into new virus particles and/or their release into the culture medium. A recent report indicated that methotrexate, a paralog of pralatrexate, can block the release of SARS-CoV-2 genomic RNA into the culture medium (Caruso et al., 2021). For the COX inhibitor, talniflumate, COX activity has also been shown to be important for the packaging of pseudorabies genomes into capsids (Ray et al., 2004). Furthermore, BET proteins were identified as interacting with SARS-CoV-2 E, a protein important for the maturation and release of virions from cells (Gordon et al., 2020) and were identified as a COVID-network-associated drug target class l analysis in our computational analysis (Figures 2E and 2F). To our knowledge, a role of methyltransferases in the production and packaging of virus genomes has not been reported and will require additional work to understand its role.
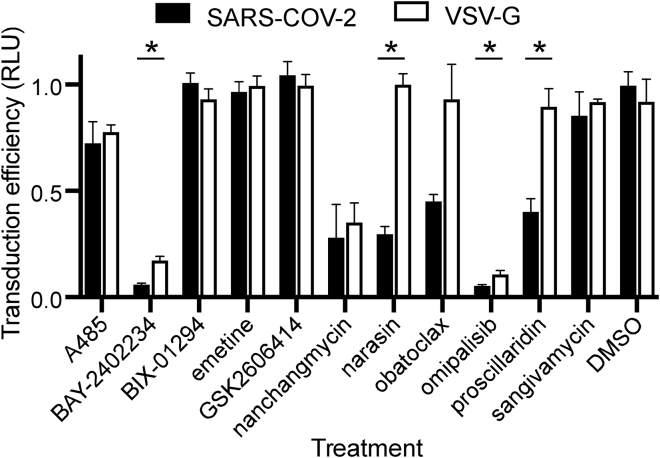
Pseudotype assay to test entry inhibition
We next sought to evaluate the impact of compounds that displayed the most potent effects on virus replication on SARS-CoV-2 spike glycoprotein (S)-mediated entry. SARS-CoV-2 S protein mediates virus entry upon binding to the angiotensin-converting enzyme 2 (ACE2) receptor and subsequent proteolytic activation by either cell surface expressed serine proteases, such as transmembrane protease, serine 2 (TMPRSS2) or endosomal cathepsins (CatL or CatS) to trigger fusion of viral and host membranes (Hoffmann et al., 2020). Eleven compounds were tested at the Vero E6 EC90 concentrations in A549 cells expressing recombinant ACE2 (Figure S7) and challenged with SARS-CoV-2 S pseudotyped lentiviral vectors (LV). Of the compounds evaluated, omipalisib and BAY-2402234 most potently inhibited SARS-CoV-2 S pseudotyped LV entry by >95%, p < 0.05 (Figure 4), though similar inhibition was also observed against vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped LV, suggesting these molecules likely targeted a post-entry step. Nanchangmycin also similarly affected both SARS-CoV-2 S and VSV-G pseudotype LV entry (75% inhibition, p < 0.05). In contrast, narasin displayed selective inhibition (∼80%) of SARS-CoV-2 S pseudotypes without significantly affecting VSV-G pseudotype LV entry. Salinomycin, a methylated analogue of narasin, has been shown to alter lysosome function (Baumert et al., 2019), which is suggestive of an effect of narasin on endosomal uptake of the virus. Similarly, proscillaridin also appeared to specifically block entry of SARS-CoV-2 S pseudotype LV. Proscillaridin was shown to reduce receptor availability for hepatitis B virus (Okuyama-Dobashi et al., 2015) and thus may also affect a membrane function important for SARS-CoV-2 infection. Obatoclax interestingly displayed partial inhibition of SARS-CoV-2 S pseudotype LV entry. Despite being characterized as a BCL inhibitor, obatoclax has been reported to interfere with endosomal acidification pathways needed for the entry of alpha and flaviviruses into cells (Varghese et al., 2017). The partial inhibition of SARS-CoV-2 S-mediated entry by proscillaridin and obatoclax compared with the almost complete inhibition of virus replication (robust decrease in N protein expression and viral RNA copy number in cell supernatants) suggests each drug may act both at entry and post-entry steps. The methyltransferase inhibitor BIX-01294, the PERK inhibitor GSK2606414, the CREBBP/p300 inhibitor A485, the nucleoside analog sangivamycin, and the protein synthesis inhibitor emetine, were each ineffective in blocking SARS-CoV-2 S-mediated entry, and based on activity in the replication assays, must act at steps after virus entry into cells.
Figure 4.
Effect of compounds on infection by virus glycoprotein pseudotyped viruses
A549 cells expressing recombinant ACE2 protein were pretreated with each indicated compound at the EC90 concentration from assays using a wild-type virus on VeroE6 cells. Lentivirus pseudotypes, encoding firefly luciferase, as a marker of infection, and bearing the SARS-CoV-2 or VSV glycoproteins (VSV-G) were used to transduce cells at 20 ng of p24 capsid equivalent per infection. Transduction efficiency was measured by the luciferase activity as relative light units (RLU) and normalized to DMSO-treated controls. Measurements used three replicates with the average and SD shown. Multiple t-tests compensating for false discovery (Q = 2) were used to identify significant differences (p < 0.05) between the VSV-G and SARS-CoV-2 pseudotype infection efficiencies indicated by ∗.
Prioritized compounds do not show phospholipidosis
Recently, it was reported that many compounds from repurposing libraries showing antiviral activity act by inducing phospholipidosis (PLD) and disrupting cell lipid metabolism (Gunesch et al., 2020; Tummino et al., 2021). As some of the compounds of interest had potentially protonatable amines, a hallmark of PLD inducers, we evaluated each by staining for accumulation of a fluorescently labeled lipid (NBD-PE), but found that none induced PLD (Figure S8), even when tested at >100 times higher than their antiviral active concentrations. We conclude that the compounds act through non-PLD mechanisms.
Confirmation of SARS-CoV-2 inhibition in a physiologically relevant 3D tissue model
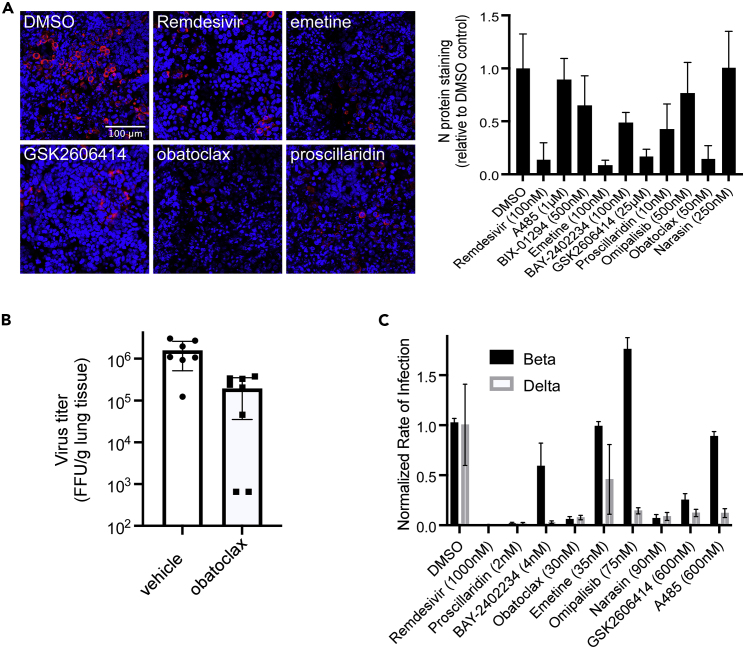
Nine of the most active treatments were evaluated using primary lung and gastrointestinal multicellular models. Whereas COVID-19 causes severe respiratory distress, several studies have reported over half of the patients also suffer from gastrointestinal symptoms, including diarrhea, abdominal pain, and vomiting. The gut is also a prominent site for virus replication (Lamers et al., 2020). Human donor-derived stratified epithelial cell models representing the airway and gut were obtained commercially (Markus et al., 2021). Similar to previous reports (Sims et al., 2008), infection in the lung model was variable between batches and not easily used for compound evaluation. However, the gastrointestinal model cells expressed ACE2 mRNA and protein (Figure S9) and gave robust, uniform infection (Figure 5A). By morphology, the infected cells appeared to be epithelial. The compounds tested in this model, including Remdesivir as an active control (Pruijssers et al., 2020), were used at the VeroE6 EC90 concentration. Emetine, GSK2606414, and obatoclax were as effective as Remdesivir, preventing >80% of cell infection. Proscillaridin was less effective, showing inhibition ranging from 50 to 80%. These observed differences helped further refine compound activities, with the most promising showing consistent activity across cell lines and human tissue models.
Figure 5.
Inhibition of infection in primary human cell model and efficacy testing in the K18 ACE2 mouse model of disease
(A) Primary intestinal epithelial cell cultures were challenged with SARS-CoV-2 and fixed after three days in formalin. Cells were stained for N protein using a specific antibody (red) and cell nuclei using Hoechst 33342 (blue) and examples of active compounds shown (left panel). Infection efficiency was measured by the total fluorescence relative to DMSO-treated controls and normalized to the cell nuclei signal (right panel). Each test was performed in triplicate with average and standard deviations shown. The scale bar in the top left image is 100 μm.
(B) Male and female mice were challenged by the intranasal route with SARS-CoV-2 and starting 6 h after a daily treatment with obatoclax at 3 mg/kg. Virus load in the lungs (FFU/g tissue) was measured four days post-challenge. For each mouse tissue sample, two measurements were made and averaged. Two treated female mice gave virus loads below and at the limit of detection for the assay, respectively. Each was plotted at 50% LOD.
(C) Beta and delta virus strains were tested for the susceptibility to small molecule treatments tested at the indicated doses (close to EC90) seen in the Washington strain using A549-ACE2 cells. Each was performed in triplicate and averages with standard deviations are shown.
In vivo evaluation in the mouse disease model
Obatoclax demonstrated potent and consistent antiviral activity across all in vitro assessments, and has been reported to have favorable pharmacokinetics and tolerability in mice (Nguyen et al., 2015). We used K18-hACE2 transgenic mice that express recombinant human ACE2 (hACE2) protein under the control of a K18 cytokeratin promoter, to evaluate the compound as a treatment for disease. Others have shown that challenge by the intranasal route results in virus replication in the lungs, heart, brain, kidneys, and gut (Hassan et al., 2020; Winkler et al., 2020), with titers increasing over 2–3 days and then subsiding after day 5 in most tissues, but with titers in the brain continuing to rise to >108 pfu/g tissue. The latter is thought to result in severe symptoms, which may not be relevant to human disease outcomes. To focus on treatments effective in the respiratory system, animals were treated daily for 4 days after virus challenge and virus load in the lungs was measured. Vehicle control animals reached titers consistent with previously reported levels of 1–3 × 106 FFU/g lung tissue. At a dose of 3 mg/kg, obatoclax gave a consistent and significant reduction in average virus load by >88% in both male and female mice (p = 0.0028). For two of the female-treated mice, lung virus load fell below the limits of detection of the assay, although one did have trace amounts of detectable virus (Figure 5B). This suggests that obatoclax may be useful as a COVID-19 treatment candidate.
Lastly, we tested the efficacy of each of the potent hits against Beta and Delta variants of SARS-CoV-2 Figure 5C) and the distantly related MERS-CoV virus (Figure S10). Surprisingly, the Beta variant showed several differences in susceptibility to drug treatment. Of note, omipalisib appeared to elevate infection, whereas A485 and BAY-2402234 showed little activity. In contrast, the Delta variant showed a similar profile to that seen for the Washington strain used for the initial screening. This suggests that the Beta variant may have a different dependency profile on cell processes disrupted by the compounds.
Discussion
Whereas multiple vaccine candidates and several targeted drugs are now available, alternative therapeutic avenues will be needed to control COVID-19. Here, we evaluated 6,710 small molecules for inhibition of SARS-CoV-2 infection using in vitro, ex vivo, and in vivo disease models. Of the approximately 200 strongly active compounds, multiple candidates had potencies that overlapped with their clinically achievable exposures at the EC90.
We performed a detailed mechanistic analysis for 40 inhibitors of SARS-CoV-2 replication with differing ascribed mechanisms of action, including proscillaridin, emetine, obatoclax, sangivamycin, omipalisib, GSK2606414, and BAY2402234. Each demonstrated in vitro potency that may be clinically useful given further evaluation in the context of disease. Structural analysis revealed distinct clusters of related molecules with conserved molecular motifs, suggesting potential pharmacophores that could be further developed into more highly active compounds. Further analysis of the reported mechanism of action and associated gene targets suggested discrete families of host factors involved in SARS-CoV-2 replication related to microtubule function, mTOR, ER kinases, protein synthesis and folding, and ion channel function. Mechanistic evaluation of the antiviral effects of the lead candidates revealed activities impacting each step of virus replication from cell entry through to egress and may provide new tools to better understand the infection cycle of this virus. These results highlight the host dependency factors necessary for SARS-CoV-2 replication and potential sites of virus vulnerability for further development of antiviral therapeutics.
Obatoclax consistently showed potent antiviral activity across all in vitro systems, as well efficacy in the mouse disease model. The 10-fold decrease in lung titers is promising given that a similar reduction of virus load has been associated with decreased mortality in COVID-19 patients (Fajnzylber et al., 2020). Obatoclax was originally developed as a broadly acting BCL-2 homology domain 3 inhibitor for cancer treatment (McCoy et al., 2010) with activities including inducing apoptosis and elevating autophagy (Campàs et al., 2006; McCoy et al., 2010), each potentially virucidal outcomes. However, the 7 other BCL-2 inhibitors that we tested were weak or inactive, indicating that BCL-2 inhibition is less likely to be a specific mechanistic target. Significantly, a previous report showed obatoclax, but not other BCL-2 antagonists, appeared to inhibit endosomal acidification. This was shown to be the likely cause of inhibition of alphavirus infection through preventing pH-dependent endosomal escape (Varghese et al., 2017). Similarly, SARS-CoV-2 is known to require endosomal acidification for triggering of fusion of the endosomal and virus membranes to release the virus capsid and genome into the cell cytosol to initiate replication. Consistent with this mechanism, the pseudotype infection assay (Figure 4) showed obatoclax partially blocking infection for the SARS-CoV-2 pseudotype. Indeed, a recent report demonstrated that this is the likely mechanism of action for this drug (Mao et al., 2022). However, another report showed that the influenza virus, which also requires endosomal acidification to infect cells, was unaffected by obatoclax (Park et al., 2020). Our data support a dominant role for obatoclax interfering with S function and therefore endosomal escape, but also suggests an additional entry-independent mechanism that will require more detailed work to be understood.
One of the most potent classes of inhibitors identified were the cardiac glycosides, such as proscillaridin, that have Na+/K+ channel blocking activity. These steroid-like molecules affect cardiac function (Hauptman and Kelly, 1999), have been reported to alter cell membrane fluidity (Manna et al., 2006), affect receptor function for a range of ligands, and can induce apoptosis (Fang et al., 2020) of cancer cells. Based on the potent cardiac effects and narrow therapeutic windows, they are unlikely to be viable for repurposing, but may provide information on virus infection mechanisms. The pseudotype entry assay suggested that proscillaridin, like obatoclax, only partially interfered with virus entry. Cardiac glycosides were reported to inhibit hepatitis B entry into cells by interfering with binding to its receptor, the sodium/bile acid cotransporter, SLC10A1/NTCP, as well as a post-entry replication step (Okuyama-Dobashi et al., 2015). Hepatitis B virus showed a similar spectrum of cardiac glycoside type and potency infection inhibition to that seen here for SARS-CoV-2, suggesting a similar mechanism may aid SARS-CoV-2 infection. In contrast, other cardiac glycosides, including ouabain, also a SARS-CoV-2 inhibitor, were found active against HIV-1 gene transcription by altering RNA processing (Wong et al., 2018) with disruption of mitogen-activated protein kinase kinase (MEK)/mitogen-activated protein kinase 3/1 (ERK1/2) signaling being responsible for infection inhibition. Another action of cardiac glycosides is to inhibit protein synthesis (Perne et al., 2009). Indeed, other known protein synthesis inhibitors, such as emetine, harringtonine, and homoharringtonine, rivalled the cardiac glycosides as potent inhibitors, in some cases showing efficacy at low nanomolar concentrations. These agents have previously been well characterized as broad-spectrum antivirals (Andersen et al., 2019) and likely reflect the importance of balanced protein production needed during virus replication and assembly.
Another class of compounds that have not been reported to affect SARS-CoV-2 infection are the BET inhibitors. These compounds, like mivebresib, affected SARS-CoV-2 infection in both cell types tested and were identified in our protein network analysis (Figure 2). Previous work showed two of the four BET proteins, BRD2 and BRD4, were bound by virus E protein (Gordon et al., 2020). The E protein is involved in the assembly and budding of newly formed coronaviruses from the cell. Our finding that the BET inhibitors block viral egress (Table 2) is consistent with this mechanism of action. Being able to identify such egress inhibitors also serves to demonstrate the utility of the secondary orthogonal assays to detect the effects of candidate compounds on the virus at different points in its life cycle.
In addition to identifying individual compounds that can inhibit SARS-CoV-2, we have demonstrated that the data from the high-throughput screen can be used to identify the potential importance of cellular pathways required for virus replication that in turn may lead to new drug targets not part of the screen. Recent publications have demonstrated a convergence of data obtained from large-scale screens, such as genome-wide CRISPR and small molecule screens. For example, both genetic and compound screens have repeatedly suggested endosomal trafficking as an important pathway for viral entry and replication, including for coronaviruses (Puhl et al., 2020; Wei et al., 2021; Wong et al., 2015). This view is bolstered by the reproducible activity of apilimod in our study and others (Kang et al., 2020), and the positive activity observed in our screen with APY0201, a second PIKfyve inhibitor. However, we also found distinct differences between genetically closely related virus strains, suggesting that even subtle changes in the virus genome can induce complicated changes in host factor dependency and replication behavior. This finding emphasizes the need to evaluate multiple virus strains for susceptibility to small molecule inhibitors.
The two largest compound screens reported to date are this study and the screen of the 12,000 compound ReFRAME library (Riva et al., 2020). Significant overlap was observed in the classes of molecules observed to have activity, but new active compounds were also identified. The main submicromolar potency leads identified in the ReFRAME study included apilimod, VBY-825, ONO 5334, Z LVG CHN2, and MDL 28170. Of these, apilimod and VBY-825 were present in the DRH library and gave similar activities in both studies. In the present study, we also identified additional active drug classes including the BET inhibitors. The main difference in the assay design of the two screens was the use of a single treatment dose (5 μM) and measuring infection by virus-induced cell death for the ReFRAME screen versus multiple concentrations and virus protein expression used here, which likely accounts for the differences in outcomes. A recent report indicated that many drug repurposing efforts for antivirals may enrich compounds that induce phospholipidosis (PLD) by disrupting cell lipid metabolism (Tummino et al., 2021). Most of these compounds have micromolar potencies for inhibition of virus infection that reflect the effects on PLD. By examining only the most potent compounds from our screening, we likely avoided such compounds, as indeed none showed PLD even when tested at over 100 times that needed to inhibit virus infection.
In summary, using a high-throughput drug screen, we have identified both potent antivirals and targeted host factors involved in the SARS-CoV-2 life cycle. We aimed here to provide both a more detailed understanding of how the virus hijacks host machinery to replicate, as well as methods to interrupt these dependencies. By making all the data publicly available through the DRH, we anticipate that both the active and inactive compounds will be useful for future studies on anti-SARS-CoV-2 drug development. We also provide a comprehensive roadmap for a more rigorous computational analysis of future antiviral drug screens. Owing to the continued need for antivirals to combat COVID-19, our findings will aid scientists and clinicians with identifying, prioritizing, and testing therapeutics, and help alleviate the burden this pandemic has placed on our society. Whereas these findings are encouraging and suggest drug-repurposing as a productive approach for rapidly identifying treatments, it is important to be mindful that unexpected drug-disease interactions may occur when used for a different purpose, necessitating prospective clinical trial assessments.
Limitations of the study
The work is dependent on the annotation of the drug database used. As more information is obtained on drug action and targets, the network models are expected to improve. Multiple cell types were tested in this study as it has been reported that drug activities can vary depending on the cell type. In this study, we focused on compounds that were active in multiple cell systems, thus increasing the confidence in outcomes. The mouse model of SARS-CoV-2 disease provides a good measurement of the effectiveness of a drug for preventing disease but the correlation to human disease is still being determined.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ACE2 Antibody | R&D Systems | Cat#AF933, RRID:AB_355722 |
| DPP4 Antibody | R&D Systems | Cat#AF1180, RRID:AB_354651 |
| SARS-CoV-2 N Protein Antibody | SinoBiological | Cat#40143-MM05, RRID Number: AB_2827977 |
| Goat anti-Mouse AF488 Secondary Antibody | ThermoFisher | Cat#A28175, RRID:AB_2536161 |
| Chicken anti-Goat AF488 | Invitrogen | Cat#A21467, RRID:AB_2535870 |
| p24gag Antibody, Clone 183-H12-5C | HIV Reagent Program (Contributor: Dr. Bruce Chesebro) | Cat#ARP3537, RRID:AB_2832923 |
| Goat anti-Mouse HRP-conjugated Antibody | Sigma | Cat#A2554, RRID:AB_258008 |
| Chicken anti-Goat AF488 Secondary Antibody | ThermoFisher | Cat#A-21467, RRID:AB_2535870 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WA (hCoV-19/USA-WA1/2020) | BEI | Cat#NR-52281 |
| SARS-CoV-2 Beta (B.1.351, hCoV-19/South Africa/KRISP-K005325/2020) | BEI | Cat#NR-55282 |
| SARS-CoV-2 Delta (B.1.617, hCoV-19/USA/MA-NEIDL-01399/2021) | Dr. John Connor | N/A |
| MERS-CoV (EMC/2012) | BEI | Cat#NR-44260 |
| 2019-nCoV_N_Positive Control | IDT | Cat#10006625 |
| Biological samples | ||
| EpiIntestinal | MatTek | Cat#SMI-100 |
| Chemicals, peptides, and recombinant proteins | ||
| Broad Institute Drug Repurposing Hub Compounds | SM Corsello, et al., 2017 | https://www.broadinstitute.org/drug-repurposing-hub |
| NBD-PE | Invitrogen | Cat#N360 |
| Obatoclax | MedChemExpress | Cat#HY-10969 |
| Formamide | ThermoFisher | Cat#17899 |
| NEBuffer 3 | NEB | Cat#7003S |
| RNAse-Free Bovine Serum Albumin | Millipore Sigma | Cat#B6917 |
| Ribonucleoside Vanadyl Complex | NEB | Cat#S1402S |
| tRNA from E. coli | Millipore Sigma | Cat#10109541001 |
| Critical commercial assays | ||
| Luna Universal Probe One-Step RT-qPCR mixture | NEB | Cat#E3006 |
| 2019-nCoV RUO Kit | IDT | Cat#10006713 |
| Monarch DNA Gel Extraction Kit | NEB | Cat#T1020 |
| HiScribe T7 High Yield RNA Synthesis Kit | NEB | Cat#E2040 |
| Monarch Cleanup Kit | NEB | Cat#T1030 |
| Bright-Glo Luciferase Assay | Promega | Cat#E2610 |
| LR Clonase II | ThermoFisher | Cat#11791020 |
| Deposited data | ||
| Screening results can be found at https://clue.io/data | Broad Institute | COVID#LINCS_COVID_PUBLIC |
| Additional data is deposited at Zenodo https://doi.org/10.5281/zenodo.6678298 | Zenodo | 10.5281/zenodo.6678298 |
| Experimental models: Cell lines | ||
| African Green Monkey Kidney: VeroE6 | ATCC | Cat#CRL-1586, RRID:CVCL_0574 |
| Human Lung Epithelial: A549 | ATCC | Cat#CCL-185 |
| Human Liver Epithelial: Huh7 | JCRB | Cat#JCRB0403, RRID:CVCL_0336 |
| Human Kidney Epithelial: HEK293FT | Invitrogen | Cat#R70007, RRID:CVCL_6911 |
| Experimental models: Organisms/strains | ||
| K18-hACE2 Mice: B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Labs | Strain #034860 |
| Oligonucleotides | ||
| 2019-nCoV_N2-F 5′-GACCCCAAAATCAGCGAAAT-3′ | IDT | Cat#10006713 |
| 2019-nCoV_N2-R 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | IDT | Cat#10006713 |
| probe 2019-nCoV_N2-P 5′-FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1-3′ | IDT | Cat#10006713 |
| T7 promoter-containing forward primer 5′-TAATACG ACTCACTATAGGGTAAAGGCCAACAACAACAAG-3′ |
Synthesized by GENEWIZ | N/A |
| T7 promoter-containing reverse primer 5′-GAGTCAG CACTGCTCATGGATTG-3′ |
Synthesized by GENEWIZ | N/A |
| FISH Oligonucleotides | See Table S5 | N/A |
| Recombinant DNA | ||
| pHAGE-EF1-ACE2 | Dr. Steve Elledge | N/A |
| pLEX307-DPP4-puro | Addgene | Cat#158451 |
| pLP1, pLP2, pLP/VSVG | Invitrogen | Cat#K497500 |
| SARS-CoV-2 S/gp41 | Dr. Nir Hacohen | N/A |
| RRL.sin.cPPT.SFFV/ACE2.IRES-puro | Addgene | Cat#145839 |
| psPAX2 | Addgene | Cat#12260 |
| pMD2.G | Addgene | Cat#12259 |
| Software and algorithms | ||
| CellProfiler | McQuin, C. et al. 2018 | https://cellprofiler.org/ |
| Oligostan | Tsanov et al. (2016). | https://bitbucket.org/muellerflorian/fish_quant/src/master/Oligostan/ |
| Other code is available through Zenodo https://doi.org/10.5281/zenodo.6678298 | Zenodo | https://doi.org/10.5281/zenodo.6678298 |
| Other | ||
| Cytation 1 | BioTek | https://www.biotek.com/products/imaging-microscopy-cell-imaging-multi-mode-readers/cytation-1-cell-imaging-multi-mode-reader/ |
| Ti2-E Eclipse Microscope | Nikon | https://www.microscope.healthcare.nikon.com/products/inverted-microscopes/eclipse-ti2-series |
| Prime BSI Camera | Photometrics | https://www.photometrics.com/products/prime-family/primebsi |
| LSM 700 Confocal Microscope | Zeiss | N/A |
| TissueLyser II | Qiagen | https://www.qiagen.com/us/products/human-id-and-forensics/automation/tissuelyser-ii/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Robert Davey (radavey@bu.edu).
Materials availability
All cell lines and plasmids not commercially available will be made available upon request.
Experimental model and subject details
Cell cultivation
Vero E6 (African green monkey kidney) and A549 (male lung epithelium) cells were obtained from ATCC (Manassas, VA, USA) and maintained in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified CO2 incubator. Huh7 (male liver epithelium-like) cells were obtained from JCRB (Ibaraki City, Osaka, Japan) and maintained as described above.
pHAGE-EF1-ACE2 was generated by Gateway recombination of pHAGE-EF1-DEST with an entry vector containing human ACE2 cDNA from the Ultimate ORF library using LR Clonase II per the manufacturer’s instructions. The construct was verified by Sanger sequencing. pHAGE -EF1-ACE2 was packaged into lentivirus using psPAX2 and pMD2.G. Cells used for testing SARS-CoV-2 Beta and Delta variants were made by transduction of A549 cells with pHAGE-EF1-ACE2. ACE2 expression measured by FACS after staining with ACE2 specific antibody at 0.25μg/106 cells for 1 h at room temperature, washing the cells with PBS, and chicken anti-goat Alexa Fluor 488 fluorescently labeled antibody at a 1:1000 dilution for 30 min. The cell pool was sorted by FACS without prior selection into four pools based on ACE2 surface expression levels. Cells were then passaged and ACE2 expression was confirmed via flow cytometry as described above. The lowest ACE2 expressing cells were used for testing efficacy of small molecules for inhibition of SARS-CoV-2 variants due to best infection and low formation of syncytia.
A549 cells stably expressing DPP4 were generated by transducing A549 cells (ATCC CCL-185) with DPP4 lentivirus. To generate DPP4 lentivirus, 293FT cells grown in DMEM supplemented with 10% FBS were transfected with pLEX307-DPP4-puro (Addgene plasmid #158451), pLP1, pLP2, and pLP/pVSV-G using calcium phosphate and incubated at 37°C. 16 h post transfection, supernatant was discarded and replaced with fresh media. 48 h after transfection, cell supernatant was collected, clarified by centrifugation, and used to inoculate A549 cells. Two days after transduction, cells were put under 1 μg/mL puromycin selection. For FACS, 5 × 106 transduced cells were stained with 1.25μg DPP4 antibody (R&D Systems AF1180) for 1 h at room temperature. After washing cells 2X with PBS, Chicken anti-goat-FITC secondary antibody (Invitrogen A21467) was applied for 45 min and then washed 2X with PBS before sorting. Cells were sorted into four bins based on DPP4 expression using a BD FACSAria-II SORP.
Virus cultivation
The SARS-CoV-2 strain USA-WA1/2020, was isolated from a traveler returning to Washington State, USA from Wuhan, China, and was obtained from BEI resources (Manassas, VA, USA). The virus stock was passaged twice on Vero E6 cells by challenging the cells at an MOI of less than 0.01 and incubating until cytopathology was seen (typically 3 days after inoculation). A sample of the culture supernatant was sequenced by NGS and was consistent with the original isolate without evidence of other viral or bacterial contaminants. The virus stock was stored at −80°C. SARS-CoV-2 B.1.351, designated Beta (hCoV-19/South Africa/KRISP-K005325/2020), and MERS-CoV (EMC/2012) were also obtained from BEI and grown as above. We thank Jacquelyn Turcinovic, Scott Seitz, and John H. Connor, NEIDL, Boston University, for isolation and sequence analysis of the Delta variant, SARS-CoV-2 B.1.617 designated as hCoV-19/USA/MA-NEIDL-01399/2021. The virus was cultivated by Devin Kenney and provided by Dr. Florian Douam, NEIDL, Boston University.
EpiIntestinal ex vivo Tissue model
Human small intestine epithelial cells were obtained and used to produce a reconstructed tissue model as described previously (Markus et al., 2021). Briefly, cryoprotected intestinal epithelial cells were seeded onto collagen coated cell culture inserts (MatTek Corporation, 0.6 cm2) in medium (SMI-100-MM, MatTek Corporation, Ashland, MA). All primary cells were derived from a single, healthy, female donor. Cells were cultivated submerged for 24 h, followed by cultivation for 13 days at the air-liquid interface (ALI) at 37°C, with 5% CO2 and 98% relative humidity. During the ALI culture period, tissues (designated SMI-100 or EpiIntestinal) were fed basolaterally through the membrane of the cell culture inserts of the 24-well plate. During this culture period, epithelial cells and fibroblasts self-assemble in the correct orientation. Under this culture condition, the organotypic tissues stratify, differentiate, and form a distinct apical-basolateral polarity. The polarized organotypic small intestinal full thickness tissues form “villi-like” tissue structure with an apical epithelial architecture on top of a fibroblast substrate. To complete cellular differentiation and stratification, cells were cultured for a total of 14 days prior to their shipment for the SARS-CoV-2 infection studies.
K18 mouse SARS-CoV-2 disease model
B6.Cg-Tg(K18-ACE2)2Prlmn/J (K18-hACE2) transgenic mice were purchased from Jackson laboratories (Farmington, CT). All experiments were pre-approved by the Boston University IACUC and is AAALAC accredited. Mice were housed according to AVMA standards in HEPA filtered microisolation cages. Mice were acclimated for at least 5 days before being challenged with virus and then monitored at least once per day for clinical signs of disease. All work was supervised by a veterinarian and performed by veterinary technicians.
Method details
Primary high throughput drug screening against SARS-CoV-2
A total of 6,710 compounds from the Broad Institute DRH (Cambridge, MA, USA) were Echo plated at 4 doses in 384 well plates by Broad Institute staff. The night prior to screening, 7 × 103 Vero E6 cells were seeded into each well of a 384 well plate. For evaluation of small molecule efficacy against infection with wild type SARS-CoV-2 virus, compounds were first dissolved in DMSO and then diluted into culture medium before addition to cells (final concentration of DMSO <0.5%). The cells were incubated for a minimum of 1 h, moved to the biocontainment laboratory, and challenged with virus at an MOI between 0.1 and 0.3. Dosing was at a final concentration of 8, 0.8, 0.08, and 0.008 μM. As a positive control, 5 μM E64d was used as it was previously reported to inhibit SARS-CoV-2 infection (Hoffmann et al., 2020). Negative controls were treated with DMSO at 0.5%. After 36 to 48 h, cells were submerged in 10% neutral buffered formalin for at least 6 h, removed from the containment lab, and washed in PBS. Cells were permeabilized in 0.1% (v/v) Triton X-100 for 15 min and blocked in 3.5% BSA for 1 h. Virus antigen was stained with SARS-CoV-2 specific antibody MM05 (Sino Biologicals, Beijing, China) overnight at 4 °C. Alexa Fluor 488-labeled goat anti-mouse antibody from ThermoFisher (Waltham, MA, USA) was added to cells for 2 h, cells were washed in PBS, and Hoechst 33342 dye was added to stain cell nuclei. Plates were imaged on a Biotek Cytation 1 automated imager and CellProfiler software (Broad Institute, MA, USA) was used for image analysis incorporating a customized processing pipeline (available on https://doi.org/10.5281/zenodo.6678298). Infection efficiency was calculated as the ratio of infected cells to total cell nuclei. Reduction of nuclei was used to flag treatments as indicative of potential cytotoxicity. The assay was performed in duplicate. Results from the assay will be available at https://repurposing.broadinstitute.org/AssayResultsViewer/Results.
Reconfirmation of top hits
To verify the results of the initial screen, small molecules were chosen for full dose-response evaluation based on initial potency and drug class. The night before the screen, 2 × 104 Vero E6 cells were seeded in each well of a 96 well plate. As before, compounds were diluted in culture medium and incubated on cells for a minimum of 1 h, with a range of final concentration from 4 μM to 0.2 nM in a 3-fold dilution series. Infection, fixation, staining, and analysis were performed as described above. The assay was performed in triplicate.
RT-qPCR detection of viral genomes from cell supernatants
To measure virus assembly and release, qPCR was used to detect viral genomic RNA in the cell culture supernatant from infected Huh7 cells using a method modified from Suzuki et al. (Suzuki et al., 2018). The cell supernatant was collected at 48 h post infection, and then mixed with 2x virus lysis buffer (0.25% Triton X-100, 50 mM KCl, 100 mM Tris-HCl pH 7.4, 40% glycerol) at an equal volume for 10 min at room temperature. Five microliters of the mixture was added to Luna Universal Probe One-Step RT-qPCR mixture (NEB, MA, USA) to a final volume of 20 μl. PCR amplification was detected and validated by CFX96 Touch Real-Time PCR Detection System (Bio-Rad, CA, USA). The cycling protocol used was 55°C for 10 min, 95°C for 1 min, followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s. The primer and probe sets were from 2019-nCoV RUO Kit (IDT, IA, USA) and nCoV_N2 forward and reverse primers. The primer sequences were forward primer 2019-nCoV_N2-F 5′-GACCCCAAAATCAGCGAAAT-3′, reverse primer 2019-nCoV_N2-R 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ and probe 2019-nCoV_N2-P 5′-FAM-ACC CCG CAT TAC GTT TGG TGG ACC-BHQ1-3’.
As a positive control for RT-qPCR of SARS-CoV-2 RNA, an RNA fragment was synthesized. In brief, 2019-nCoV_N_Positive Control (IDT) DNA was amplified using T7 promoter-containing forward primer 5′-TAATACGACTCACTATAGGGTAAAGGCCAACAACAACAAG-3′ and reverse primer 5′-GAGTCAGCACTGCTCATGGATTG-3′ from GENEWIZ (MA, USA). After electrophoresis and gel extraction by Monarch DNA Gel Extraction Kit (NEB), the PCR product was transcribed using HiScribe T7 High Yield RNA Synthesis Kit (NEB) according to the manufacturer’s instructions, followed by DNA template removal by DNase I (NEB) treatment and purification using Monarch Cleanup Kit (NEB). RNA copy number was calculated from its molecular weight and absorbance measured by a NanoDrop 1000 (Thermo Fisher Scientific, MA, USA). A standard curve was generated using dilutions of RNA to relate genome copy number to qPCR cycle threshold (Ct). For each qPCR reaction set, 4 of the standards were included to ensure assay performance.
smiFISH detection of viral mRNA
RNA fluorescence in situ hybridization (FISH) was used to measure the amount of virus subgenomic mRNA being produced and its localization in cells following a method adapted from Tsanov et al. (2016) and optimized for SARS-CoV-2. Thirty oligonucleotide probes were designed to hybridize to S and ORF3a genes using Oligostan software (see Table S5). S and the adjacent ORF3a were targeted as each is transcribed off the subgenomic promoter at higher levels than other virus genes. Each had a common tail that bound to a complementary sequence on an oligonucleotide conjugated to a Cy5 fluor. Huh7 cell nuclei were stained with Hoechst 33342 dye and samples were imaged on a Cytation 1 microscope (Biotek, VT, USA). Images were quantified using CellProfiler (McQuin et al., 2018), (using pipelines available on https://doi.org/10.5281/zenodo.6678298) and infection efficiencies calculated from the percentage of RNAFISH positive cells in each sample.
Pseudotype evaluation of treatment effects on cell entry
Single round SARS-CoV-2 S or VSV-G pseudotyped luciferase-expressing lentivectors were generated via calcium phosphate mediated transient co-transfection of HEK293T (female embryonic kidney) cells with HIVΔenv/luc and SARS-CoV-2 S/gp41 or VSV-G expression plasmids (Akiyama et al., 2018). The SARS-CoV-2 S/gp41 expression plasmid was a gracious gift of Dr. Nir Hacohen (Broad Institute), that expresses a codon-optimized version of CoV-2 S and modified to include the eight most membrane-proximal residues of the HIV-1 envelope glycoprotein cytoplasmic domain after residue 1246 of the S protein. Virus-containing cell supernatants were harvested 2 days post transfection and filtered using 0.45μm syringe filters, aliquoted, and stored at −80°C until further use. The p24gag content of the virus stocks was quantified using a p24gag ELISA, as described before (Miller et al., 2017). Briefly, virus particle containing cell supernatants were added to HIV-Ig coated wells and detected with an anti-p24gag monoclonal antibody and HRP-conjugated goat anti-mouse secondary antibody. ACE2-expressing lentivectors were generated via transient co-transfection of HEK293T cells with RRL.sin.cPPT.SFFV/Ace2.IRES-puro, HIV-1 packaging plasmid psPAX2, and VSV-G. A549 cells were transduced with ACE2 lentivectors, selected for ACE2 expression by culturing in puromycin-containing media, and cell surface expression of ACE2 was confirmed by FACS. For entry inhibition assays, A549/ACE2 cells (1 × 104 cells per well of 96 well flat bottom plate) were pretreated with the compounds at the indicated concentrations for 1 h prior to spinoculation with 20 ng of p24gag equivalent SARS-CoV-2 S or VSV-G pseudotyped lentivirus particles. Cells were lysed with Bright Glo-lysis buffer at 48 h post infection, and cell lysates used for quantification of luciferase activity.
Phospholipidosis assay
A549-ACE2 cells were plated in 96-well plates in RPMI supplemented with 10% FBS and allowed to attach overnight. NBD-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzooxadiazol-4-yl) dissolved in DMSO was added to cells for a final concentration of 17μM, and cells were incubated with reagent for 2 h at 37⁰C. After 2 hours, compound dose curves, beginning at 10μM and following a 5-fold 9-point serial dilution series, were added to cells and cells were incubated overnight at 37⁰C. Amodiaquine served as a positive control. After 22 to 23 h post compound addition, cells were fixed in 10% neutral buffered formalin for 30 min at room temperature, then stained with Hoechst at 1:10,000 in PBS. Plates were imaged at 4x on a Cytation Cell Imaging Multimode Reader using Gen 3.08 acquisition software. Phospholipidosis (PLD) was quantified using a CellProfiler pipeline. Briefly, Hoechst-stained nuclei were expanded to encompass approximate cell area and PLD, defined as GFP signal above background, was classified as an object. PLD falling within the cell area was attached to each cell to define a new object, and the total intensity of PLD-positive cells in each image was measured. PLD was normalized to fold change over DMSO treatment and compared to amodiaquine (a known PLD inducer) to determine PLD activity.
Reconstructed in vitro 3D tissue model of small intestine
Tissues were supplied growing on permeable membranes in transwell cultures. Tissues were maintained at the conditions described above, being fed every other day with 5mL of fresh medium. Tissues were assayed well within their recognized lifetime. Tissues were treated 1 h prior to infection both apically and basolaterally. The tissues were challenged with SARS-CoV-2 WA on the apical and basolateral sides at an MOI of 1 (estimated from virus titer on Vero E6 cells and 106 cells in the tissue model). After 1 h, the inoculum was washed off with PBS and the culture incubated for an additional 3 days. Tissues were then fixed in 10% formalin, and stained for SARS-CoV-2 N protein and Hoechst 33342 as above. The tissue was mounted on a glass slide and imaged using a Zeiss LSM 700 laser scanning confocal microscope.
Drug testing against variants of concern and MERS-CoV
The evening prior to infection, 1.5 × 104 A549 cells expressing ACE2 or DPP4 were seeded into 96 well plates. Compounds were dosed out at three concentrations-their respective IC90s, plus 5-fold above and below. After a 1 h incubation, virus was added to the pretreated cells (approximately 100 FFUs/well of SARS-CoV-2 Beta, 200 FFUs/well of SARS-CoV-2 Delta, and 200 PFUs/well of MERS-CoV). The SARS-CoV-2 infected cells were fixed after 36-48 h, immunostained for viral infection, and quantified as described above. The MERS-CoV infected cells were allowed to incubate for five days prior to fixation. Cell nuclei were stained with Hoechst and counted. Treated/untreated and infected wells were normalized to untreated and uninfected wells to determine if the addition of drug rescued cell counts.
Mouse model treatment and infection focus assay detection of virus load in lungs
Mice were challenged at 10-15 weeks of age in groups of 4 or 5 by the intranasal route with 105 FFU SARS-CoV-2 per nare. Starting at 6 h after virus challenge and once each day, animals were dosed by the intraperitoneal route with the indicated treatment. Four days after challenge, mice were euthanized and lungs removed at necropsy. A biopsy punch (4 mm) was used to collect 2 samples of lung tissue and was stored frozen in PBS. For evaluation of virus load, the lung tissue was homogenized using a TissueLyser II (Qiagen, MD, USA) for two cycles at 30 Hz for 2 min. Debris was pelleted for 10 min at 16,800 xg in a centrifuge (Eppendorf, CT, USA). The supernatant was titrated onto Vero E6 cells and after 1 h overlaid with a 5% solution of methylcellulose in DMEM and incubated overnight. Cells were fixed in 10% formalin and stained with the N specific antibody used for screening. Foci of infected cells were counted and titers calculated.
Drug-response classifier
In order to classify the drug response by potency, the infection efficiency and total cell nuclei count were normalized per plate to the average of in-plate untreated control wells. Infection efficiency was calculated as the ratio of total infected cells divided by the total cell nuclei count per well. Wells with an infection efficiency greater than one were not analyzed further as these indicated a fault in the imaging. Treatments that resulted in >60% loss of cell nuclei compared to untreated controls were flagged as being potentially cytotoxic and were de-prioritized. Treatments resulting in >80% cell loss were not evaluated as the virus MOI would be significantly altered together with a loss in calculation accuracy.
The classification of the drug-response outcomes was performed using a drug response curve (DRC) model. We used the R package drc (Ritz et al., 2015) to calculate the DRCs using a log-logistic model that estimates four parameters (Hill slope, IC50, min, and max). Each drug-response was classified by inspecting cell nuclei count, and then evaluating the drug effect on the inhibition of viral infection efficiency. Each drug-response was classified in two steps: first inspecting toxicity by nuclei count and then evaluating the drug effect on the inhibition of viral proliferation using the model given by (Equation 1).
| (Equation 1) |
(Equation 1) DRC log-logistic Model with four parameters, where b is hill, c is the min value, d is the max value and e is the.
To inspect the cell nuclei count for each drug, we first estimated the model parameters using as response variable the normalized nuclei count in the treated cells. We tested the dose-response effect for all drugs using a test for goodness of fit, and treatments with p < 0.01 (FDR-Bonferroni correction) were defined as potentially cytotoxic causing substantial nuclei loss. If a drug had a nuclei count reduction less than 40% of untreated, or if toxicity was observed only at the highest (8 μM) concentration it was not considered cytotoxic. To evaluate inhibition of viral replication, we used as response for the DRC model the normalized number of infected cells in the treated well. A drug was considered to have a dose-response effect by using a test for goodness of fit (p < 0.01, FDR-Bonferroni correction), and the significant drugs were defined as strong (Z score >2.5 corresponding to >80% inhibition), weak (Z-score 1.5-2.5 with 50-80% inhibition), or not effective (Z score <1.5 and <50% inhibition) over the range of concentrations. Z-scores were calculated based on the standard deviation and mean signals of vehicle treated cells from all plates.
Treatments that showed cell nuclei loss greater than infection inhibition for at least half of doses tested were classified as being potentially cytotoxic and were de-prioritized for follow-up studies.
Accuracy
All outcomes were manually inspected in order to validate the two step-model and annotation compared to the computational model derived outcome.
GO enrichment
Using all targets within each drug category, we performed a GO enrichment analysis in the biological process and molecular function mode using the enrichR tool.
Gene-target network analysis
Using genes that were statistically significantly enriched in each category, we built a bipartite network, from drugs and targets for each category.
Network effect
In order to understand where the drug-targets were placed in a human protein-protein interaction network, we first calculated the largest connected component (LCC) of each drug category and calculated the significance of the module size using a degree-preserving approach (Guney et al., 2016), preventing the same high degree nodes from being selected repeatedly by choosing 100 bins in 1,000 simulations. For that purpose, the R package NetSci was used.
Target enrichment
To understand if there was an overrepresentation of any gene as a drug target in each category, we used a Fisher’s exact test, and call enriched a gene has its p-value is lower than 0.05 (complete results can be found in Table S3). For each set of genes, we performed a biological function enrichment using Enrichr (Chen et al., 2013).
Pathway analysis
We aggregated the targets of the compounds classified in the different outcome categories (e.g., strong, weak) and performed pathway enrichment analysis (Reactome) using the R package ReactomePA (Yu and He, 2016).
Mechanism of action
We retrieved mechanism of action annotations for 6,150 drugs from the Drug Repurposing Hub. (Corsello et al., 2017). For each mechanism of action, we checked over- or under-representation of drugs in the different drug outcomes (e.g., strong, weak) by using a Chi-Square test (p < 0.05, FDR-BH).
Chemical structural relationship analysis
To evaluate structure similarity between molecules, similarity searches and clustering were performed using Morgan molecular fingerprints (FPs). The python package RDKit (Schneider et al., 2015) was used to standardize SMILES and InChIKeys associated with each drug tested in the experiments, and generate Morgan FPs (also known as extended-connectivity fingerprints). All molecular substructures up to radius 3 were assigned to unique identifiers and hashed to vectors of 8,192 bits in order to capture fragments of bigger size and reduce the potential bit collision. For all molecules with experimental outcome, structural similarity was quantified by Jaccard (or Tanimoto) similarity. We further leveraged the Jaccard metric to reduce the dimensionality of the bit vector representation to 3D space with UMAP (McInnes et al., 2018). In the bit representation of Morgan FPs each bit is mapped to multiple structural fragments appearing in molecules. We evaluated the significance of each bit for ligand binding by calculating the average number of non-hydrogen atoms in the associated fragments, finding that in the overall database the median size of the expected chemical fragment associated with each bit is 9. The enrichment of each experimental class in active bits (hypergeometric test, BH multiple testing correction with ) was then calculated. As shown in Figure S1, the significant bits for strong drugs appear to be moderately bigger and potentially more meaningful for binding, with 22 bits in the Strong Class with average size greater than the database median, compared to only 1 bit for ‘Cyto’, cytotoxic class.
Quantification and statistical analysis
Aside from the tests described above and within the text, statistical significance between treatments and across experiments was calculated using Student’s T test or one-way ANOVA with Tukey’s post-test for comparison of multiple groups. For the animal experiments, nested ANOVA was used to compare multiple replicates for different samples within a single experiment. Dose-response curves were generated and IC50 values calculated by fitting a four parameter [Inhibitor] versus response with variable slope curve to the data. Each test and curve fitting were performed using GraphPad Prism version 8.0.0 (GraphPad Software, GraphPad Software, San Diego, California USA). Where an ∗ is indicated in figures, this indicates p < 0.05.
Acknowledgments
J.J.P., C.J.D., H.M., P.K., K.G.B., R.B., S.H.S., and R.A.D. were supported by NIH grants P01AI120943, 5R01AI128364-02, 5R01AI125453-05, UC7AI095321 and by a grant from the Massachusetts Consortium on Pathogen Readiness. J.L.B., S.J., and S.G. were supported by NIH grants 3RO1AI064099-15. A.L.B. is supported by NIH grant 1P01HL132825, American Heart Association grant 151708, and ERC grant 810115-DYNASET. M.Z. is supported, in part, by NSF grants IIS-2030459 and IIS-2033384, and by Harvard Data Science Initiative. J.L. was supported, in part, by NIH grants HG007690, HL108630, and HL119145, and by American Heart Association grants D700382 and CV-19; M.N. and S.J.E. were supported by a grant from the Massachusetts Consortium on Pathogen Readiness. S.J.E. is an investigator with the Howard Hughes Medical Institute. We acknowledge the hard work and dedication of the NEIDL veterinary services team who performed disease treatment testing in the high biological containment laboratory under institutionally approved IACUC protocols to RAD. We thank the Center for the Development of Therapeutics and Repurposing Hub at the Broad Institute for providing the compound library. We thank Tim Mitchison for critical reading of the manuscript. We thank John H. Connor, Jacquelyn Turcinovic, Scott Seitz, Florian Douam, and Devin Kenney for the isolation and production of SARS-CoV-2 Delta (B.1.617), and Anthony Griffiths and Anna Honko for the production of MERS-CoV. The graphical abstract was created with BioRender.com.
Author contributions
Conceptualization, R.A.D., M.N.N., J.J.P., A.L.B., F.W., S.A., S.J.E., D.A., J.L., M.Z., S.G.; Investigation, J.J.P., P.T.K., D.M-G., G.M., H.M., C.J.D., X.G., I.d.V., K.G-B., M.A., R.B., J.L.B, S.H.S., T.A., C.O., S.J.; Writing–Original Draft, J.J.P., M.N.N., M.Z., D.M-G., G.M. and R.A.D.; Writing–Review &Editing, J.J.P., D.M-G., G.M., R.A.D., M.N.N., J.L.; Funding Acquisition, M.N.N.; Supervision, R.A.D., A.L.B., F.W., S.A., D.A., S.J.E., S.G.
Declaration of interests
There are no interests to declare.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104925.
Supplemental information
Data and code availability
-
•
All data generated in this paper is available at https://doi.org/10.5281/zenodo.6678298.
-
•
All original code has been deposited at https://doi.org/10.5281/zenodo.6678298 and is publicly available. DOIs are listed in the Key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Supporting data for this study are shown or available in the Supplemental information. The primary screening data is also available through the Drug Repurposing Hub portal: https://www.broadinstitute.org/drug-repurposing-hub or https://clue.io/data/COVID#LINCS_COVID_PUBLIC. The data as well as computer coding scripts for image and data analysis are available on https://doi.org/10.5281/zenodo.6678298.
References
- Akiyama H., Miller C.M., Ettinger C.R., Belkina A.C., Snyder-Cappione J.E., Gummuluru S. HIV-1 intron-containing RNA expression induces innate immune activation and T cell dysfunction. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-05899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P.I., Krpina K., Ianevski A., Shtaida N., Jo E., Yang J., Koit S., Tenson T., Hukkanen V., Anthonsen M.W., et al. Novel antiviral activities of obatoclax, emetine, niclosamide, brequinar, and homoharringtonine. Viruses. 2019;11 doi: 10.3390/v11100964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert T.F., Berg T., Lim J.K., Nelson D.R. Status of direct-acting antiviral therapy for HCV infection and remaining challenges. Gastroenterology. 2019;156:431–445. doi: 10.1053/j.gastro.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campàs C., Cosialls A.M., Barragán M., Iglesias-Serret D., Santidrián A.F., Coll-Mulet L., de Frias M., Domingo A., Pons G., Gil J. Bcl-2 inhibitors induce apoptosis in chronic lymphocytic leukemia cells. Exp. Hematol. 2006;34:1663–1669. doi: 10.1016/j.exphem.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Caruso A., Caccuri F., Bugatti A., Zani A., Vanoni M., Bonfanti P., Cazzaniga M.E., Perno C.F., Messa C., Alberghina L. Methotrexate inhibits SARS-CoV-2 virus replication “in vitro. J. Med. Virol. 2021 doi: 10.1002/jmv.26512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M., et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Truitt R., Yang W., et al. Drug repurposing screens reveal cell-type specific entry pathways and FDA approved drugs active against SARS-Cov-2. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S., Tao H., Xia K., Guo W. Proscillaridin A induces apoptosis and inhibits the metastasis of osteosarcoma in vitro and in vivo. Biochem. Biophys. Res. Commun. 2020;521:880–886. doi: 10.1016/j.bbrc.2019.11.012. [DOI] [PubMed] [Google Scholar]
- Firestone M.J. First identified cases of SARS-CoV-2 variant B.1.1.7 in Minnesota — december 2020–january 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70 doi: 10.15585/mmwr.mm7008e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A.M., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J.J., van den Hoogen B.G., Peiris M., Lim W., Stöhr K., Osterhaus A.D.M.E. Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R., et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 2020 doi: 10.1101/2020.03.22.002386. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesch A.P., Zapatero-Belinchón F.J., Pinkert L., Steinmann E., Manns M.P., Schneider G., Pietschmann T., Brönstrup M., von Hahn T. Filovirus antiviral activity of cationic amphiphilic drugs is associated with lipophilicity and ability to induce phospholipidosis. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00143-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney E., Menche J., Vidal M., Barábasi A.-L. Network-based in silico drug efficacy screening. Nat. Commun. 2016;7:10331. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev. Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gysi D.M., Valle Í. do, Zitnik M., Ameli A., Gan X., Varol O., Ghiassian S.D., Patten J.J., Davey R.A., Loscalzo J., Barabási A.-L. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauptman P.J., Kelly R.A. Digitalis. Circulation. 1999;99:1265–1270. doi: 10.1161/01.cir.99.9.1265. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-L., Chou Y.-Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malden D.E. Hospitalization and emergency department encounters for COVID-19 after paxlovid treatment — California, December 2021–May 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71 doi: 10.15585/mmwr.mm7125e2. [DOI] [PubMed] [Google Scholar]
- Manna S.K., Sreenivasan Y., Sarkar A. Cardiac glycoside inhibits IL-8-induced biological responses by downregulating IL-8 receptors through altering membrane fluidity. J. Cell. Physiol. 2006;207:195–207. doi: 10.1002/jcp.20555. [DOI] [PubMed] [Google Scholar]
- Mao B., Le-Trilling V.T.K., Wang K., Mennerich D., Hu J., Zhao Z., Zheng J., Deng Y., Katschinski B., Xu S., et al. Obatoclax inhibits SARS-CoV-2 entry by altered endosomal acidification and impaired cathepsin and furin activity in vitro. Emerg. Microbes Infect. 2022;11:483–497. doi: 10.1080/22221751.2022.2026739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus J., Landry T., Stevens Z., Scott H., Llanos P., Debatis M., Armento A., Klausner M., Ayehunie S. Human small intestinal organotypic culture model for drug permeation, inflammation, and toxicity assays. In Vitro Cell. Dev. Biol. Anim. 2021;57:160–173. doi: 10.1007/s11626-020-00526-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy F., Hurwitz J., McTavish N., Paul I., Barnes C., O’Hagan B., Odrzywol K., Murray J., Longley D., McKerr G., Fennell D.A. Obatoclax induces Atg7-dependent autophagy independent of beclin-1 and BAX/BAK. Cell Death Dis. 2010;1:e108. doi: 10.1038/cddis.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. [Google Scholar]
- McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet Lond. Engl. 2020;395:1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.-L. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.M., Akiyama H., Agosto L.M., Emery A., Ettinger C.R., Swanstrom R.I., Henderson A.J., Gummuluru S. Virion-associated vpr alleviates a postintegration block to HIV-1 infection of dendritic cells. J. Virol. 2017;91 doi: 10.1128/JVI.00051-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwenda M. Detection of B.1.351 SARS-CoV-2 variant strain — Zambia, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70 doi: 10.15585/mmwr.mm7008e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahler G. In: Dictionary of Pharmaceutical Medicine. Nahler G., editor. Springer; 2009. Anatomical therapeutic chemical classification system (ATC) p. 8. [DOI] [Google Scholar]
- Nguyen M., Cencic R., Ertel F., Bernier C., Pelletier J., Roulston A., Silvius J.R., Shore G.C. Obatoclax is a direct and potent antagonist of membrane-restricted Mcl-1 and is synthetic lethal with treatment that induces Bim. BMC Cancer. 2015;15 doi: 10.1186/s12885-015-1582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama-Dobashi K., Kasai H., Tanaka T., Yamashita A., Yasumoto J., Chen W., Okamoto T., Maekawa S., Watashi K., Wakita T., et al. Hepatitis B virus efficiently infects non-adherent hepatoma cells via human sodium taurocholate cotransporting polypeptide. Sci. Rep. 2015;5:17047. doi: 10.1038/srep17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-G., Ávila-Pérez G., Nogales A., Blanco-Lobo P., Torre J.C. de la, Martínez-Sobrido L. Identification and characterization of novel compounds with broad-spectrum antiviral activity against influenza A and B viruses. J. Virol. 2020;94 doi: 10.1128/JVI.02149-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perne A., Muellner M.K., Steinrueck M., Craig-Mueller N., Mayerhofer J., Schwarzinger I., Sloane M., Uras I.Z., Hoermann G., Nijman S.M.B., Mayerhofer M. Cardiac glycosides induce cell death in human cells by inhibiting general protein synthesis. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijssers A.J., George A.S., Schäfer A., Leist S.R., Gralinksi L.E., Dinnon K.H., Yount B.L., Agostini M.L., Stevens L.J., Chappell J.D., et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl A.C., Fritch E.J., Lane T.R., Tse L.V., Yount B.L., Sacramento C.Q., Tavella T.A., Costa F.T.M., Weston S., Logue J., et al. Repurposing the ebola and marburg virus inhibitors tilorone, quinacrine and pyronaridine: in vitro activity against SARS-CoV-2 and potential mechanisms. BioRxiv. 2020 doi: 10.1101/2020.12.01.407361. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N., Bisher M.E., Enquist L.W. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J. Virol. 2004;78:12964–12974. doi: 10.1128/JVI.78.23.12964-12974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz C., Baty F., Streibig J.C., Gerhard D. Dose-response analysis using R. PLoS One. 2015;10:e0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Schneider N., Sayle R.A., Landrum G.A. Get your atoms in order--an open-source implementation of a novel and robust molecular canonicalization algorithm. J. Chem. Inf. Model. 2015;55:2111–2120. doi: 10.1021/acs.jcim.5b00543. [DOI] [PubMed] [Google Scholar]
- Sims A.C., Burkett S.E., Yount B., Pickles R.J. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawicki S.P., Jeanmonod R., Miller A.C., Paladino L., Gaieski D.F., Yaffee A.Q., De Wulf A., Grover J., Papadimos T.J., Bloem C., et al. The 2019–2020 novel coronavirus (severe acute respiratory syndrome coronavirus 2) pandemic: a joint American College of academic international medicine-world academic council of emergency medicine multidisciplinary COVID-19 working group consensus paper. J. Glob. Infect. Dis. 2020;12:47–93. doi: 10.4103/jgid.jgid_86_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki ., Kotoura M., Yashima S., Wu H., Nakano T., Sano K. Measuring dengue virus RNA in the culture supernatant of infected cells by real-time quantitative polymerase chain reaction. J. Vis. Exp. 2018;58407 doi: 10.3791/58407. [DOI] [PubMed] [Google Scholar]
- Tsanov N., Samacoits A., Chouaib R., Traboulsi A.-M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M., et al. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O’Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021 doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese F.S., Rausalu K., Hakanen M., Saul S., Kümmerer B.M., Susi P., Merits A., Ahola T. Obatoclax inhibits alphavirus membrane fusion by neutralizing the acidic environment of endocytic compartments. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.-M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.H., Kumar P., Tay F.P.L., Moreau D., Liu D.X., Bard F. Genome-wide screen reveals valosin-containing protein requirement for coronavirus exit from endosomes. J. Virol. 2015;89:11116–11128. doi: 10.1128/JVI.01360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R.W., Lingwood C.A., Ostrowski M.A., Cabral T., Cochrane A. Cardiac glycoside/aglycones inhibit HIV-1 gene expression by a mechanism requiring MEK1/2-ERK1/2 signaling. Sci. Rep. 2018;8:850. doi: 10.1038/s41598-018-19298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., He Q.-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- Zhang null, Chung null, Oldenburg null. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data generated in this paper is available at https://doi.org/10.5281/zenodo.6678298.
-
•
All original code has been deposited at https://doi.org/10.5281/zenodo.6678298 and is publicly available. DOIs are listed in the Key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
Supporting data for this study are shown or available in the Supplemental information. The primary screening data is also available through the Drug Repurposing Hub portal: https://www.broadinstitute.org/drug-repurposing-hub or https://clue.io/data/COVID#LINCS_COVID_PUBLIC. The data as well as computer coding scripts for image and data analysis are available on https://doi.org/10.5281/zenodo.6678298.